This paper provides an analysis of the origin of multiple RNA recognition motifs (RRMs) within single RNA-binding proteins. One main finding is that such RRMs (sibling RRMs) have significantly higher similarity than expected by chance. These results suggest that sibling RRMs arose through domain duplication.
Keywords: RNA binding proteins, RNA recognition motif, domain duplication, RNA processing
Abstract
The sequence-specific recognition of RNA by proteins is mediated through various RNA binding domains, with the RNA recognition motif (RRM) being the most frequent and present in >50% of RNA-binding proteins (RBPs). Many RBPs contain multiple RRMs, and it is unclear how each RRM contributes to the binding specificity of the entire protein. We found that RRMs within the same RBP (i.e., sibling RRMs) tend to have significantly higher similarity than expected by chance. Sibling RRM pairs from RBPs shared by multiple species tend to have lower similarity than those found only in a single species, suggesting that multiple RRMs within the same protein might arise from domain duplication followed by divergence through random mutations. This finding is exemplified by a recent RRM domain duplication in DAZ proteins and an ancient duplication in PABP proteins. Additionally, we found that different similarities between sibling RRMs are associated with distinct functions of an RBP and that the RBPs tend to contain repetitive sequences with low complexity. Taken together, this study suggests that the number of RBPs with multiple RRMs has expanded in mammals and that the multiple sibling RRMs may recognize similar target motifs in a cooperative manner.
INTRODUCTION
Specific interactions between RNAs and proteins play an essential role in regulating mRNA processing, including RNA splicing, polyadenylation, translocation, and degradation (Janga and Mittal 2011). Altering the level or activity of RNA-binding proteins (RBPs) has a dramatic impact on various RNA-related cellular functions, with aberrant RBP function leading to human diseases (Lukong et al. 2008). For example, many RBPs specifically recognize regulatory cis-elements in pre-mRNA and thereby inhibit or promote use of nearby splicing sites (Black 2003; Wang and Burge 2008). The binding between these splicing factors and their RNA target is crucial to many cellular processes, as most human genes undergo alternative splicing to produce multiple isoforms with distinct functions. Therefore, examining the interactions between different RBPs and their RNA targets is an important component in understanding various gene regulation pathways.
The sequence-specific interaction between RBPs and single-stranded RNAs is usually mediated through various RNA binding domains (RBDs) including the RNA recognition motif (RRM), the pentatricopeptide repeat (PPR), the K homology (KH), the zinc-finger, the Pumilio/FBF (PUF), and the cold-shock (CSD) domains (Ray et al. 2013). Although protein sequence elements outside of the RBD may contact RNA and affect RNA binding (Hamy et al. 1993; Shen et al. 2004), the RBD is the key determinant of RNA binding specificity (Auweter et al. 2006). Among them, the RRM is the most abundant and present in over 50% of RBPs in humans (Maris et al. 2005). A typical RRM contains 80–90 aa that fold into a β1α1β2β3α2β4 topology, where the four anti-parallel β-sheets and the two additional α helices create ample surface that interacts with RNA (Birney et al. 1993; Maris et al. 2005). The most conserved region of the RRM consists of two short sites (6–8 aa) in β1 and β3 (named RNP-2 and RNP-1, respectively) that are crucial for RNA interaction (Bentley and Keene 1991; Birney et al. 1993; Caceres and Krainer 1993). However, recent structures of various RRMs bound by their cognate RNA show that RRMs may interact with RNA through diverse mechanisms (Oberstrass et al. 2005; Hargous et al. 2006; Sickmier et al. 2006). For example, hnRNP I (poly-pyrimidine tract binding protein or PTB) has four RRMs with similar specificities. The β3 of each RRM contributes only weakly to RNA binding, whereas the hydrophobic side chains in β2 are responsible for binding to RNA bases through hydrophobic interactions (Oberstrass et al. 2005). In other cases, like hnRNP F, interactions between the RNA target and the RRM were found mainly in the loop region rather than in the β-sheet of the RRM (Dominguez and Allain 2006; Dominguez et al. 2010).
RRMs usually recognize a short RNA element of 2–5 nt, and some RBPs contain multiple RRMs. The tandem RRMs in the same RBP can either bind to similar RNA sequences and function cooperatively (Oberstrass et al. 2005; Sickmier et al. 2006; Dominguez et al. 2010) or have very different RNA binding activities/specificities (Burd et al. 1991), or only one/some of the RRMs are functional while the others do not exhibit RNA binding (Safaee et al. 2012). Therefore, for RBPs with multiple RRMs, the general rules for how each RRM contributes to binding specificity are largely unclear.
We conducted a detailed sequence analysis of the RRM-containing RBPs in humans and other organisms. Surprisingly, we found a strong trend indicating that RRMs within the same protein (hereafter referred to as “sibling RRMs”) have higher sequence similarity to each other than the RRM pairs from different proteins. In addition, sibling RRMs within the RBPs specific to a single species have higher similarity than those shared by multiple species. Together, these findings suggest that prevalent domain duplications of RRMs have occurred within many RBPs during evolution. This result is further illustrated by cases of both a recent and an ancient RRM duplication. In addition, we found that the RBPs with similar sibling RRMs are more likely to bind to the 3′ UTR than those proteins having more divergent sibling RRMs and that the RBP sequence regions outside RRMs have a strong bias for low complexity and/or repetitive sequences. Altogether, these analyses reveal important implications regarding RBP evolution.
RESULTS
Increased numbers of RBPs in mammals
The number of proteins with canonical RBDs has expanded significantly in mammals. In Table 1, we list the common RBDs and the number of proteins containing common RNA-binding domains from five different organisms whose proteomes are thoroughly annotated. Humans have the most RBPs among all species examined, and there is a large expansion in the number of RBPs in mammals with the exception of PAZ domain-containing proteins. In addition, we found that the number of RRMs within a single RBP has increased in mammals compared to other low-complexity organisms when examining the RRM-containing RBPs across different species (Supplemental Fig. S1). These observations lead to intriguing fundamental questions such as why do humans need so many RBPs and why is it that many RBPs contain multiple RBDs? One possible explanation could be that multiple RBDs allow RBPs to bind RNA with higher sequence specificity and/or affinity than those with a single binding domain. Another possible reason is that multiple domains may help RBPs to bind to longer RNA sequences. On average, a single RBD binds to 4–6 nt; thus, multiple RBDs may have provided some selective advantage for increased binding specificity and affinity and/or also facilitate binding to longer RNA targets.
TABLE 1.
Number of proteins containing different RBDs in five species as reported from Ensembl biomart on 07/09/13
Sibling RRMs are more similar to each other
To study these questions, we analyzed RRM-containing RBPs at a proteome-wide scale across multiple species. We applied a series of filters to obtain unique human RBPs that have well-defined RRM domains and extracted the sequence of each RRM using the consensus annotation from three domain annotation databases (Fig. 1A). This process was repeated for three other species (Mus musculus, Drosophila melanogaster, Caenorhabditis elegans), and both the species-specific and conserved RBPs were extracted by the same set of filters for further analyses (see Materials and Methods). After filtering for gene duplication and database redundancy, we extracted 453 unique RRMs from human RBPs.
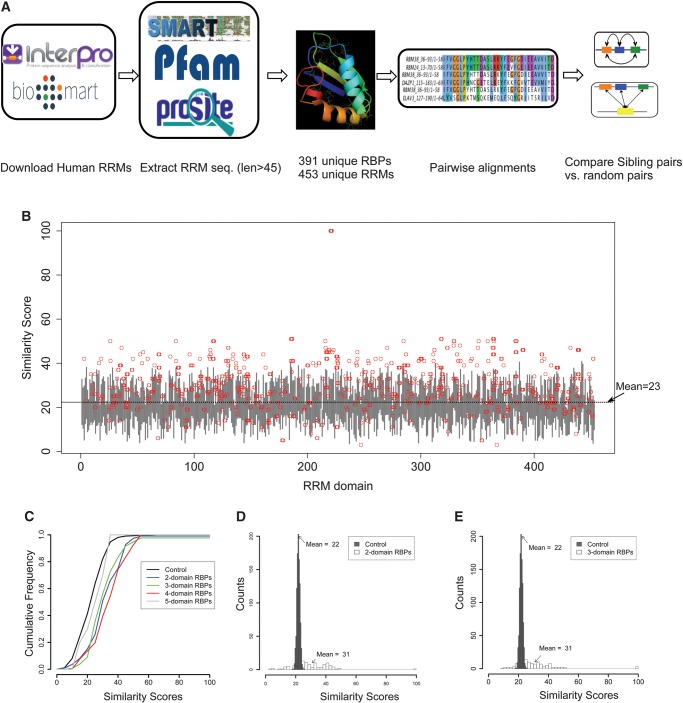
FIGURE 1.
Elevated sequence similarity between sibling RRMs in human RBPs. (A) Workflow of the analyses. The human proteins containing RRMs were obtained from the InterPro database, and the RRM sequences were extracted according to the consensus annotations from three different databases. After filtering out the duplicated sequence, 453 RRMs from 391 unique RBPs were analyzed through sequence comparison. (B) Similarity scores between all RRM pairs in human RBPs. Each RRM was aligned with all other 452 RRMs, where the distribution of similarity score is represented by a gray vertical line spanning the mean ± 1× standard derivation. The similarity score between sibling RRMs was represented as a red circle. The order of RRMs along the x-axis is arbitrary. (C) The cumulative frequency of similarity scores between sibling RRM pairs in proteins with 2, 3, 4, or 5 RRMs. As a control, we randomly selected 1000 RRM pairs and computed the cumulative frequency of their similarity scores. (D) Sibling RRMs are more conserved than the shuffled pairs. The histograms of similarity scores between sibling RRM pairs from 112 RBPs that contain two RRMs were plotted (open boxes). As controls, we shuffled the order of these RRMs to generate a simulated set of 112 RBPs with matched sequence composition. The shuffle was repeated 1000 times with replacement, and the mean similarity scores of RRM pairs were plotted as filled boxes. (E) Same as panel D, except 44 RBPs with three RRMs were analyzed.
We aligned each of the 453 unique human RRMs to all others to calculate sequence similarity scores and plotted the mean score ± standard deviation (1 × SD) as vertical gray bars (Fig. 1B), obtaining an average similarity score of ∼23. However, similarity scores between sibling RRMs appear to be skewed toward higher similarity (denoted by red circles in Fig. 1B), indicating that the sibling RRMs within the same RBPs have significantly higher sequence similarity to each other than what is expected by chance (P = 2.4 × 10−20 by Kolmogorov-Smirnov test, or P = 2.4 × 10−17 by t-test if assuming normal distribution) (Fig. 1B). In particular, among the 1186 sibling RRM pairs, 467 pairs (39.4%) had similarity scores higher than the mean plus 1 × SD, whereas 38 pairs (3.2%) scored below mean −1 × SD. This skewed distribution was not dependent on the score system that we used in measuring similarity, as we observed similar results using additional score methods and matrices (Supplemental Fig. S2). Further analysis suggested that the increased similarity between sibling RRMs was unrelated to the length of the peptide between these domains, as we did not find any correlation between the RRM similarities and their distances (Supplemental Fig. S3). This increased similarity is not limited to a single species, as the same results were obtained when we analyzed the sibling RRMs in the D. melanogaster genome (Supplemental Fig. S4A). In addition to RRM, we also analyzed KH and C2H2 zinc finger domains, both of which are commonly found in human RBPs. While comparing the similarity scores of sibling domain pairs to those of all other pairs (i.e., nonsibling pairs), we again found a higher sequence similarity in sibling pairs in both sibling KH and zinc finger domains (Supplemental Fig. S4B), suggesting the increased similarity between sibling RNA binding domains is a common feature for different types of RBPs.
There is a possibility that some proteins are under a global selection to preserve certain sequence bias, resulting in the increased sequence similarity between sibling RRM pairs within a single protein compared to random pairs. To measure the potential sequence bias, we calculated the average frequency of each amino acid in RRMs for different RBP groups with 2, 3, 4, and 5 RRM domains (Supplemental Fig. S5A). These groups include 112 proteins with two RRMs (112 sibling pairs), 44 proteins with three RRMs (132 sibling pairs), 11 proteins with four RRMs (66 sibling pairs), and one protein with five RRMs (10 sibling pairs). Overall, we found that the RRMs from different groups or within the same group have similar sequence composition. Five out of 20 aa have significant differences in mean of frequency between groups as judged by the ANOVA F-statistic (P-value < 0.01). Nevertheless, to better control the subtle sequence bias, we generated control groups of RRM pairs with matched composition distance to the real sibling RRM pairs for the rest of our analyses (see Materials and Methods section for details and Supplemental Fig. S5B).
We further analyzed the RBPs containing multiple RRMs and compared the cumulative distributions of RRM similarity scores in proteins with different numbers of RRMs. When compared to a control set of 1000 randomly selected RRM pairings, we found that the different sets of RBPs all have higher similarity between their sibling RRMs than the randomly chosen RRM controls (except the 5-domain RBP set that contains a single member), as judged by the right shifts of plots (P = 6.3 × 10−13, 4 × 10−12, 2.2 × 10−14, and 0.8 for control vs. 2-, 3-, 4-, and 5-domain RBPs, respectively, by the Kolmogorov–Smirnov test) (Fig. 1C). This result suggests that the higher similarity between sibling RRMs (Fig. 1B) is a common property for all RBPs with different numbers of RRMs.
A potential explanation of these observations is that the sibling RRMs resulted from domain duplication during evolution (Bjorklund et al. 2006). However, there is an alternative explanation that all the RRMs in proteins with multiple RRMs might be more conserved (i.e., similar to each other) regardless of whether they coexist in the same protein. To address this possibility, we selected the set of RBPs with two or three RRMs and shuffled the sibling relationship of these RRMs within each set. This shuffling of sibling relationships was conducted by randomly selecting two or three RRMs to form a simulated RBP (112 proteins with two RRMs and 44 proteins with three RRMs were generated in each shuffle), and this simulation was repeated 1000 times. We found that the mean similarity scores for shuffled RRM pairs were significantly less than the real sibling pairs (P = 0.001 by a rank test) (Fig. 1D,E), suggesting that the higher similarity observed is, indeed, due to a sibling (duplication) relationship rather than the natural sequence bias between the sets of the “singleton RRMs” and the RRMs with siblings. Consistently, the similarity scores of random pairs of RRMs with siblings (mean = 22 for RBPs with two or three RRMs) (Fig. 1D,E) are similar to those of random pairs of all RRMs (mean = 23) (Fig. 1B).
Sibling RRM pairs in species-specific RBPs are more similar to each other
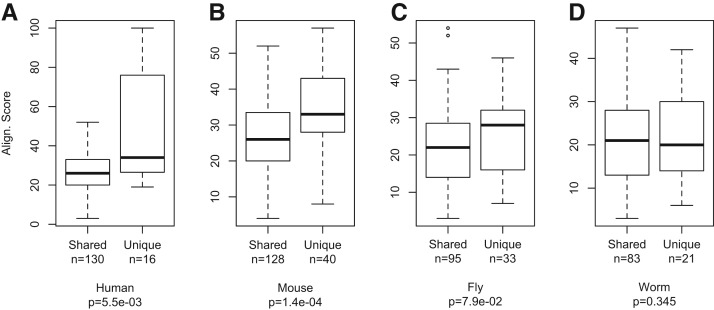
We further examined the sequence conservation of sibling RRM pairs from different species whose proteomes are well annotated. For each of four species (human, mouse, fruit fly, and worm), we selected the RBPs shared among all species and the RBPs found only in one species (see Materials and Methods) and compared the similarity between sibling RRMs within the same protein. We found that, in all species except worms, the similarity between sibling RRMs is significantly higher in the species-specific RBPs as compared to that of sibling RRMs in RBPs shared across all four species. Generally, genes conserved across multiple species are more ancient, as they appeared before speciation, whereas genes unique to certain species are more recently evolved. According to this simple assumption, our finding suggests that the RRM sibling pairs in “younger” (i.e., species-specific) proteins have higher sequence similarities than those in “older” proteins (i.e., conserved across distant species). A simple explanation is again that most sibling RRMs arose from domain duplication during evolution, which was then followed by sequence drift in each species through random mutations. The sibling RRMs in older proteins resulted from more ancient duplication and, therefore, would be expected to have higher sequence divergence. In particular, such increased similarity was more obvious between the sibling RRMs specific to human and mouse (Fig. 2A,B), suggesting an extensive RRM duplication in mammals. We are aware that our explanation is based on a usual assumption in gene evolution; however, there is an alternative but less likely scenario that the unique genes could have existed in the common ancestor but were subsequently lost in all species except one.
FIGURE 2.
Sibling RRM pairs in species-specific proteins are more conserved. (A) Human RBPs with multiple RRMs were divided into two classes: the proteins shared among four different species (H. sapiens, M. musculus, D. melanogaster, and C. elegans) and the proteins found only in human. The similarity scores between sibling RRMs were calculated for each class and represented as a box plot. The score distributions were compared by t-test with P value indicated. The same analyses were also carried out using RBPs from M. musculus (B), D. melanogaster (C), and C. elegans (D).
Recent RRM duplication in DAZ proteins
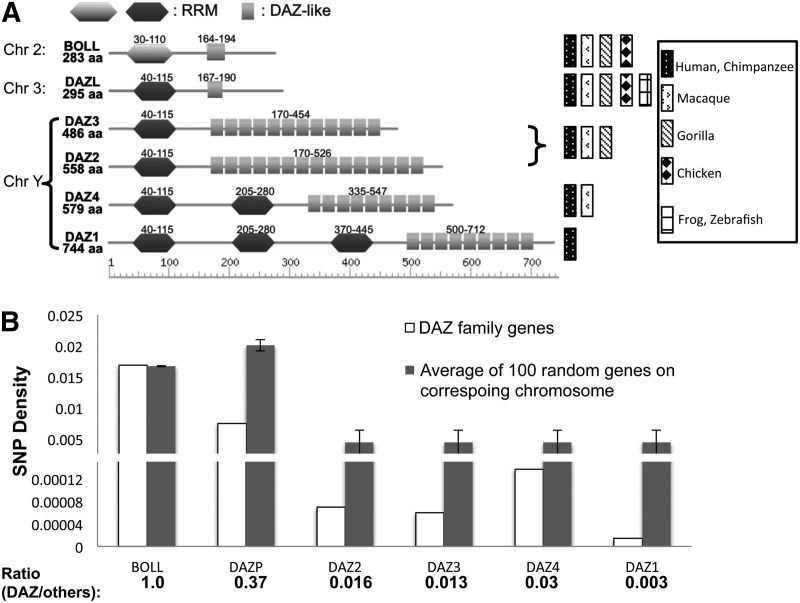
We observed an outlier with similarity score of 100 between RRMs of human DAZ proteins (i.e., completely identical). The DAZ proteins have four paralogs on the Y chromosome: DAZ1 (3 RRMs), DAZ2 (1 RRM), DAZ3 (1 RRM), and DAZ4 (2 RRMs); one paralog on chromosome 3: DAZL (DAZ like) (1 RRM), and one on chromosome 2: BOLL (1 RRM) (Fig. 3A). Among these six proteins, the RRMs in four DAZ proteins and DAZL are completely identical, whereas the RRM in BOLL has 53% identity with the others. Previous sequence analyses suggests that at least two gene duplication events were required to generate this protein family: The first duplication gave rise to DAZL and BOLL, which was followed by a second duplication of DAZL to generate Y chromosome-specific DAZ proteins (Xu et al. 2009; Eirin-Lopez and Ausio 2011; Li et al. 2011). The second duplication could either be a single duplication that generated four DAZ proteins, or alternatively, several sequential duplications that happened within a short time window so as to produce four proteins.
FIGURE 3.
An RBP family with recent RRM duplications. (A) The members in the human DAZ protein family contain one or more RRMs and DAZ-like domains. All RRMs in DAZ1, DAZ2, DAZ3, DAZ4, and DAZL are identical, whereas the RRM in BOLL has 53% sequence identity with the other RRM. The DAZ1 to DAZ4 are in the Y chromosome, while BOLL and DAZL are in chromosomes 2 and 3. The ortholog genes in other species were identified by a combination of inparanoid annotation and blast search, and species that contain various DAZ proteins were represented with different boxes. The DAZ proteins with multiple RRMs were only found in certain primates. (B) The SNP density of each human DAZ protein was compared with the average density of other genes in the same chromosome. The SNP density ratios between DAZ genes relative to other genes in the same chromosome are indicated. The genes in the Y chromosome encoding DAZ proteins have lower SNP density, suggesting that they are more recently diverged genes.
Among the six proteins within the DAZ family, only human DAZ1 and DAZ4 have multiple RRMs. To improve the annotation of this family, the sequences of the six human DAZ family proteins were compared against the genomes of chimpanzee, macaque, gorilla, chicken, frog, and zebra fish (Fig. 3A). Such reannotation is necessary, since the nomenclature does not necessarily reflect the real evolutionary route of these proteins in some species (e.g., Dazl in worm is the ortholog of human Boll). The single RRM proteins DAZL and BOLL can be found in all species tested, whereas DAZ proteins with multiple RRMs can only be identified in certain primates (human, chimpanzee, and macaque, but not in gorilla) (Fig. 3A). This result suggests that there was an RRM domain duplication following the second gene duplication on the Y chromosome, generating new DAZ family members with multiple RRMs. This domain duplication appears to be a recent event that happened only in a subgroup of primates including humans. It is also possible that such domain duplication happens in multiple steps, as the DAZ proteins with multiple RRMs were detected in macaque but not gorilla. Alternatively, assembly errors in this repetitive region of the Y chromosome could also prevent the detection of DAZ proteins with multiple RRMs in gorilla.
To examine their evolution over a more recent time frame, we further determined the SNP density within the DAZ protein family (Fig. 3B). We calculated the SNP density (number of SNPs/gene length) for each DAZ gene, as well as the average SNP density of 100 genes randomly selected from the same chromosome (gray bars). The SNP density of BOLL is similar to that of other genes randomly selected from chromosome 2, while the SNP density of DAZL is slightly lower than that of the randomly selected genes on chromosome 3. However, the SNP densities of the four DAZ genes are two orders of magnitude less than the densities of other randomly selected genes on the Y chromosome. Since the majority of gene variation observed in a population is due to random drift of neutral (or nearly neutral) mutations, as proposed by the neutral theory of molecular evolution (Kimura 1989), the SNP density is correlated with the functional importance and evolution time of the gene (Zhao et al. 2003). Our observation of SNP densities is consistent with the hypothesis that there has been at least one very recent RRM domain duplication event that generated DAZ1 with multiple RRMs.
Ancient RRM duplications in PABPs
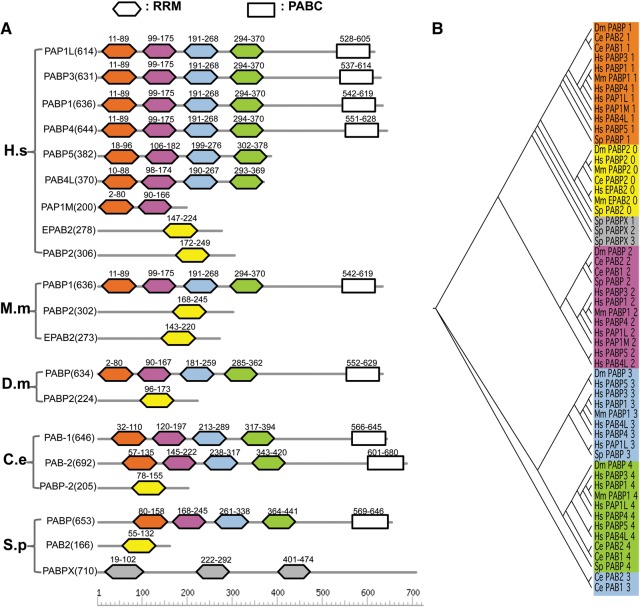
In addition to recent domain duplication, we also found a case of ancient duplication of RRMs in the human genome. Human polyadenylate-binding proteins (PABPs) belong to a conserved protein family that binds to the poly(A) tail of mRNA through RRMs (Goss and Kleiman 2013). Six PABP paralogs in humans (PABP1, PABP3, PABP4, PABP5, PAP1L, and PAP4L) contain four RRM domains, with some members containing an additional C-terminal domain called PABC. In addition, the human PABP2 and EPAB2 (embryonic PABP2) contain a single RRM, and PAP1M contains two RRMs (Fig. 4A). The family of PABP proteins in other species (M. musculus, D. melanogaster, C. elegans, and Schizosaccharomyces pombe) contains members with one RRM or four RRMs, with the exception of a yeast protein (PABX) that contains three RRMs. Through multiple sequence alignments of all 21 RRMs in different species, we clustered these RRMs according to similarity and found that these RRMs clustered predominantly by the relative locations in a protein rather than by the species (Fig. 4B). For example, the first of the four RRMs in all PABPs across five species has higher similarity to each other than to its sibling RRMs, thus forming a monophyletic clade. The same observation is also valid for the second, third, and fourth RRM in different proteins across all species. This relationship was clearly demonstrated in Figure 4B, where we color-coded the RRMs by different positions and observed that the RRMs of the same color were mostly clustered together (forming a monophyletic clade) in the phylogenetic tree (Fig. 4B). The proteins with single RRMs are also clustered with each other across different species, and this clade is more similar to the first of the four RRMs in other proteins. This conservation pattern suggests that the domain duplication generating four sibling RRMs had most likely happened in the common ancestor of all these species (additional duplications might also occur in human and nematode), producing a larger family of PABPs. We speculate that there may be additional ancient domain duplications similar to PABPs, but such events are difficult to identify due to the lack of reliable measurement to distinguish ancient duplication vs. nonduplicated RRMs. For the future work, we may be able to compare the ages of all genes vs. all potentially duplicated RRM domains (with a correct background model for age of the individual domain and entire protein) and thus to determine if there is a correlation between the similarity score of sibling RRMs and the approximate age of the duplication.
FIGURE 4.
An RBP family with ancient RRM duplications. (A) The diagram of PABP proteins from five species (H. sapiens, M. musculus, D. melanogaster, C. elegans, and S. pombe). The members in this protein family contain one to four RRMs, and some also contain a C-terminal PABC domain. Each RRM is colored according to their relative positions within the protein. (B) The phylogenetic tree of RRMs in the PABP family was visualized via TreeView. The RRMs are colored in the same scheme as in panel A, and the RRMs in the same position are more similar to each other across all species.
These two specific examples in DAZ and PABP families represent both a recent and an ancient RRM duplication, strongly supporting our finding in analyzing all sibling RRMs (Fig. 1B). Taken together, our results suggest a model wherein RRM duplication has happened frequently during evolution, followed by random evolutionary drift that introduces additional sequence variation. This simple model is consistent with the finding that the number of proteins with multiple RRMs has expanded in humans and other mammals (Supplemental Fig. S1).
Similarity between sibling RRMs is associated with RBP functions
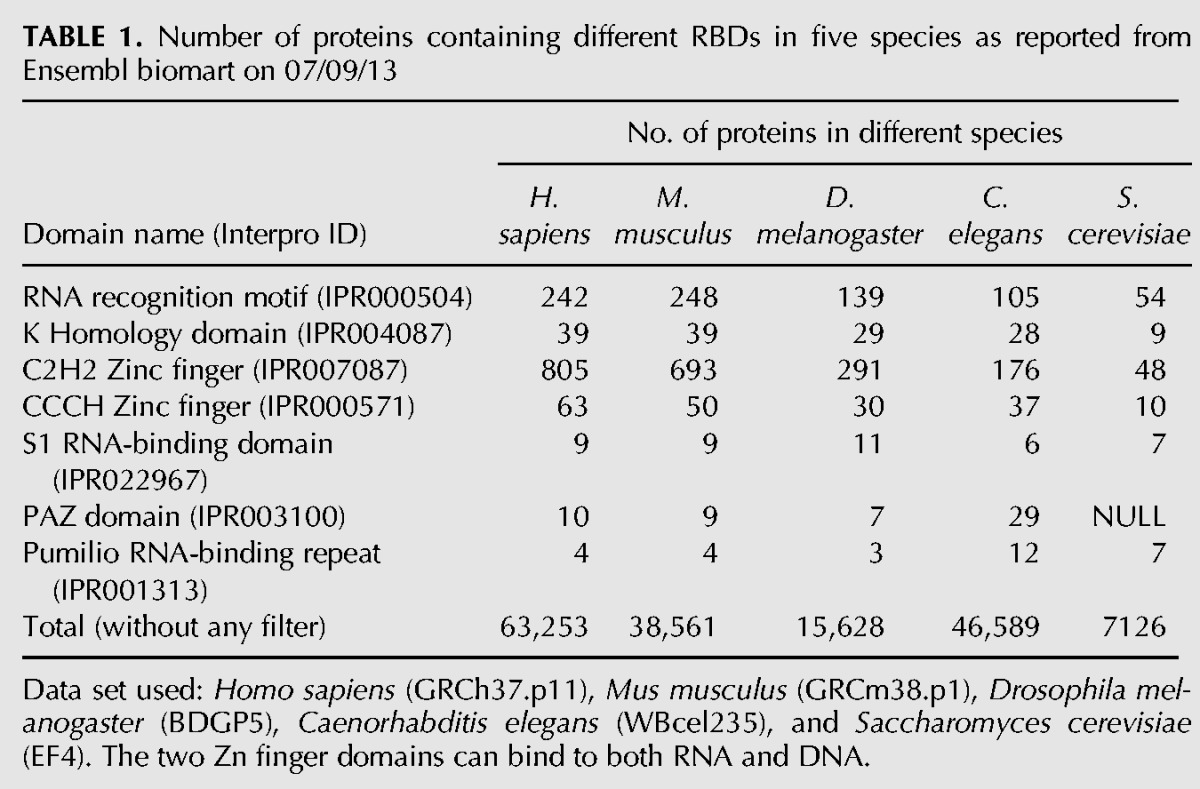
In addition to the time since duplication, other features might also contribute to the similarities between sibling RRMs. For example, evolutionary constraints can also affect how fast the sequence drifts through random mutations after domain duplication. To study if the similarities between sibling RRMs are associated with certain functional preferences of RBPs, we conducted a survey of functional differences in the RBPs with multiple RRMs. We observed a general trend that the proteins that bind to polyadenylated RNA in the 3′ UTR tend to have more similar sibling RRM pairs, whereas the proteins that bind to the 5′ UTR tend to have dissimilar sibling RRMs (Fig. 5A), suggesting there may be some association between the similarity of sibling RRM and the RBP function.
FIGURE 5.
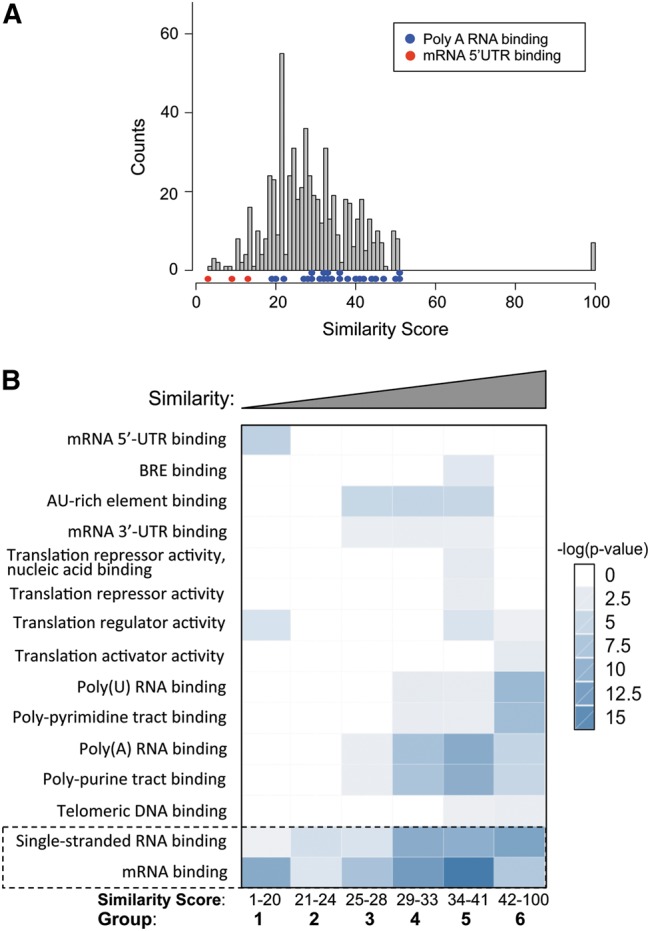
Gene Ontology analysis of human RBPs with multiple RRMs. (A) Sibling RRMs with different similarities tend to bind distinct regions of mRNA. The similarities between sibling RRM pairs are represented with a histogram (gray), with the colored dots indicating the gene ontology (GO) terms enriched in the genes from different bins of the histogram. (B) According to the domain similarity score between sibling RRMs, all RBPs were divided into six groups as equally as possible: 1–20 (108 pairs), 21–24 (92 pairs), 25–28 (106 pairs), 29–33 (104 pairs), 34–41 (97 pairs), and 42–100 (93 pairs). The GO analyses were carried out, and the enriched functional terms in each bin are represented with a heat map to indicate the significance of enrichment. The functions common to all groups are marked.
To further study this potential relationship, we conducted a gene ontology analysis on all human RBPs having multiple RRMs. According to the similarity scores between each RRM pair, we divided all pairs into six groups, each containing ∼100 RRM pairs. The corresponding proteins in each group were subjected to functional enrichment analysis by the DAVID annotation tool (http://david.abcc.ncifcrf.gov/) (Huang da et al. 2009), and the results were compared across all groups (Fig. 5B). As expected, the function of “single-stranded RNA binding” and “mRNA binding” are significantly enriched across all groups (Fig. 5B, bottom), serving as a positive control. Consistent with the earlier observation, we also found a significant enrichment of “mRNA 5′-UTR binding” (P = 1.8 × 10−5, fold enrichment = 406) in proteins with dissimilar sibling RRMs (group 1: similarity score = 1–20). In contrast, enrichment of “polyadenylated RNA binding” (P = 5.1 × 10−5, fold enrichment = 256) occurred in proteins having sibling RRM pairs with the highest similarity (group 6: similarity score = 42–100). In addition, the RBPs with similar sibling RRMs were also found to be enriched in poly(U) RNA binding, poly-pyrimidine track binding, and poly-purine track binding, suggesting that these RRMs are more likely to bind repetitive RNA elements (groups 4–6) (Fig. 5B). This finding is consistent with the notion that the requirement of binding to repetitive targets may impose additional selective pressure on these RBPs after RRM duplication. Individual RRMs are known to specifically recognize short sequences (usually 2–5 nt), and thus, RBPs with similar sibling RRMs could be expected to facilitate the binding to longer RNA targets containing repetitive elements.
Compared to other regions of mRNA, the 5′-UTR region usually contains binding sites for factors that affect the translation efficiency of mRNA (Zimmer et al. 2008). On the other hand, the 3′ UTR usually contains more repetitive sequences used to control RNA stability (e.g., AU-rich elements) (Barreau et al. 2005; Matoulkova et al. 2012). As expected, the RBPs with dissimilar sibling RRMs (group 1) are enriched only in 5′-UTR binding and translation regulation (Fig. 5B). Conversely, proteins with similar sibling RRMs have a small bias toward binding to the 3′ UTR. Recently, a comprehensive identification of the binding motifs for RBPs suggested that the RBDs with higher protein similarity are more likely to bind to similar RNA motifs (Ray et al. 2013). Our data raise an interesting prediction that mRNA metabolism is controlled by more diverse elements in the 5′ UTR but by more repetitive elements in the 3′ UTR. This hypothesis seems to be true for translation control and RNA degradation through AU-rich elements, but its generality remains to be examined.
RRM-containing RBPs are enriched with repetitive motifs
In addition to the RRMs, other sequence motifs may also contribute to RRM-containing RBP function or even RNA binding affinity/specificity (Shen and Green 2006). Thus, we analyzed the non-RRM fragments of RBPs to determine their characteristics that may contribute to function. We removed RRM sequences from the RBPs and calculated the frequency of each amino acid in the remaining fragments. To estimate the enrichment of each amino acid, we computed logarithm value for the ratio of amino acid frequency in these fragments vs. that in all human proteins and found that amino acids A, G, P, Q, R, S, and Y were highly enriched (Fig. 6A). We further searched for the frequent words flanking these enriched amino acids (five residues up- and downstream) (Fig. 6B). As expected, we found that RS di-peptides were highly enriched in this data set because the Ser/Arg-rich proteins (SR proteins) are a major class of splicing factors that recognize RNA targets through RRMs. In addition, we found a high frequency of GY di-peptides as well as many other low-complexity poly-G and poly-P sequences. These repetitive motifs were represented by a word cloud plot (Fig. 6C), where the occurrences of all possible di-, tri- (with arbitrary second amino acid), and tetra-peptides (the second and third amino acids could be any amino acid) were computed after removing the RRM from the RBP sequences. We found that the Gly-rich, Pro-rich, Ser-rich, and Ala-rich sequences often co-occurred with the RRMs (Fig. 6C); some of these repetitive motifs were also reported in an unbiased identification of all mammalian RBPs (Castello et al. 2012). To determine whether these repetitive sequences are specific to RRM-containing proteins, we analyzed sequences of RBPs containing the KH or zinc finger C2H2 domain (Supplemental Figs. S6, S7). All RBPs with RRM, KH, and zinc finger C2H2 domains have low complexity poly-G and poly-P motifs. Furthermore, we found the RS di-peptide repeats were only found in RRM-containing proteins, whereas the poly-S was found to be enriched in RBPs with the KH and zinc finger C2H2 domain (cf. Fig. 6B and Supplemental Figs. S6B, S7B).
FIGURE 6.
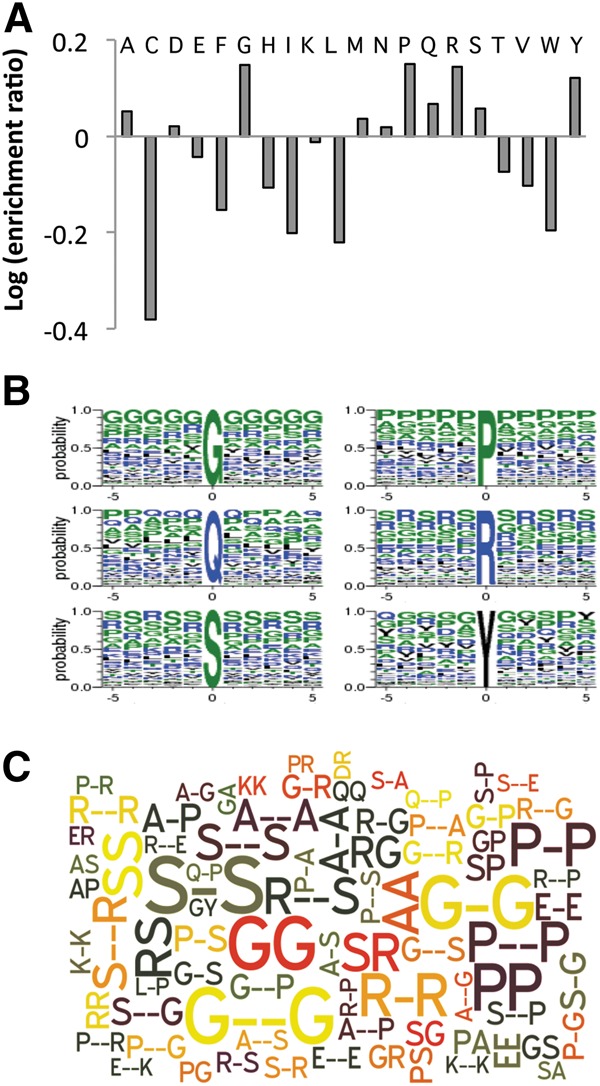
Sequence motifs enriched in the RRM-containing RBPs. (A) We removed the RRM sequence from the RBPs and analyzed the remaining sequence for amino acid propensities. For all 20 amino acids, their frequencies within non-RRM regions were compared to other proteins in the human proteome and the relative ratio is plotted. (B) Sequence logos around the most enriched amino acid residues in RBPs. The height of each single-letter amino acid code corresponds to the probability of occurrence at each position. (C) Repetitive sequence patterns that significantly co-occur with RRM in all human proteins. The size of each pattern corresponds to the number of occurrence. The word cloud was generated with the Wordle online tool. The top 80 motifs are shown.
DISCUSSION
Proteins that specifically bind to single-stranded RNA play critical roles in regulating various RNA processing pathways; thus a detailed sequence analysis of these RBPs will provide key insights into gene regulation at the RNA level. This study suggests extensive domain duplications of RRM. Such duplications are probably followed by random evolutionary drift that introduces additional sequence variation, leading to the observed higher degree of sequence divergence in old proteins with ancient RRM duplications (Figs. 2, 4). This domain duplication may play a significant role in the function of RBPs. One possible consequence could be that multiple RRMs allow a protein to bind RNA with higher sequence specificity and/or affinity than those RBPs with a single binding domain. Another consequence could be that multiple RRM domains may help RBPs to bind to longer RNA sequences. Typically a single RRM recognizes 2–5 nt; thus tandem RRMs may provide some selective advantage to increase binding specificity and bind to longer RNA targets. Consistent with this notion, the sibling RRMs in several RBPs, for example, PTB (Oberstrass et al. 2005), were found to recognize similar RNA motifs. The domain duplication of RRMs might provide a possible explanation of why the RRM-containing proteins are so abundant in the human genome.
The extensive RRM duplication during evolution raises some fundamental questions in RNA biology. The human genome (or mammals, in general) has the highest number of RBPs with RRM duplications, and this RRM expansion probably contributed to the increased complexity of RNA processing pathways in mammals. For example, the majority of human genes undergo alternative splicing, and a predominant fraction of splicing factors are RBPs with multiple RRM domains. In fact, we observed that the RBPs with different similarities in their sibling RRMs are functionally separated from each other (Fig. 5). The proteins with very similar sibling RRMs tend to bind the 3′ end of mRNA and might function in RNA polyadenylation, whereas the RBPs with more divergent sibling RRMs tend to bind the 5′ UTR of mRNA and might affect the RNA translation. We speculate that RRM duplication, together with their diverging RNA binding targets in the transcriptome, allows the mutual selection in RNA–protein interaction and eventually leads to the functional divergence of RBPs.
We also found that, compared to all other human proteins, the RRM-containing RBPs are more likely to have repetitive sequences in the regions outside the RRMs. These repetitive sequences frequently mediate protein–protein interactions, as RBPs with low-complexity domains tend to aggregate to form protein fibers (Kato et al. 2012). The association of RRM-containing proteins with repetitive sequences (encoded by low complexity DNAs) raises an interesting possibility that these sequences may provide a mechanism for domain duplication, as the repetitive DNA sequences are less stable during replication and tend to cause local DNA duplication/expansions (Thomas 2005; Jurka et al. 2007). Alternatively, such repetitive sequences could be a result of RRM duplication that is caused by local DNA duplication; however, the RBPs with a single RRM also contain low-complexity sequences. Nevertheless, the mechanism of domain duplication is an interesting question emerging from our study.
We described a systematic analysis of RBPs, focusing on the proteins with the RRM as their RNA-recognition domain. Surprisingly, we found an increase in the number of RBPs containing multiple RRMs in mammals (Supplemental Fig. S1) and that the sibling RRMs within these proteins are more similar to each other than what would be predicted by controls (Fig. 1). In addition, the sibling RRM pairs are more similar to each other in the species-specific RBPs when compared to the ancient RBPs shared by multiple species, suggesting a general RRM duplication in many genes of the mammalian genome. Such domain duplication is further supported by two extreme examples: In the case of the DAZ protein family, a very recent RRM duplication appears to have happened in humans and several primate species, generating multiple RBPs containing identical sibling RRMs (Fig. 3). In another case, the RRM duplications within the PABP proteins probably happened in the common ancestor of all eukaryotes, as similar duplication was found from yeast to human (Fig. 4). Taken together, these results suggested a new and simple model wherein RRM duplication happened frequently during evolution, resulting in increased numbers of RBPs with multiple RRMs.
MATERIALS AND METHODS
RNA binding proteins and RNA recognition motifs
We extracted the RRM sequences according to the scheme in Figure 1A. First, the RRM sequences were downloaded from InterPro Biomart (http://www.ebi.ac.uk/interpro/biomart/martview/) with the following configuration: DATABASE: “InterPro BioMart,” DATASET: “InterPro Entry Annotation,” Filters: “InterPro,” Entry ID: “IPR000504,” and Source Signature Database: “Pfam, SMART, and Prosite.” We selected Pfam annotation if there was inconsistency between Source Signature Databases. Unless specified, both Swiss-Prot and TrEMBL proteins were included, but only unique sequences were used. As a result, 453 unique RRMs with peptide sequence length ≥45 amino acids were included (Supplemental Table S1). Data of three other species, M. musculus, D. melanogaster, and C. elegans were also downloaded for ortholog analysis (Supplemental Tables S2, S3). Other protein attributes, such as Gene Ontology, Gene Orthologs, and Gene IDs, used in other databases, were downloaded from Ensembl Biomart (www.ensembl.org/biomart/martview). Because protein IDs are not standardized between or even within some databases, we performed a protein ID conversion as well as manual curation to combine our data sources.
Sequence similarity score calculation
ClustalW2 was used to compute all the pairwise alignment scores for every RRM pair. The similarity score was calculated by calibrating the number of identities between the two sequences with the length of alignment, and it is represented as a percentage, i.e., 0–100. The default protein weight matrix (Gonnet 250) was used for all the pairwise alignments in the main text. However, we also compared the similarity scores generated by using Gonnet 250 with BLOSUM30 (Supplemental Fig. S2A) and PAM350 (Supplemental Fig. S2B). We also repeated Figure 1B by using BLOSUM 30 as the weight matrix and obtained similar results (Supplemental Fig. S2C).
Sequence composition and composition distance
We calculated the sequence composition as the frequency of the 20 amino acids in each RRM sequence. Therefore, a sequence composition for an RRM is a vector with 20 dimensions. To measure the similarity of sequence compositions between two RRMs, we used the city block distance between two vectors (i.e., sum of the frequency difference of each amino acid). We named such measurement the “composition distance,” which ranges from 0 to 2.
To control for the sequence bias when choosing random RRM pairs, we use those with the composition distance matched to the real sibling pairs. For example, in Figure 1C, the 1000 control RRM pairs were randomly picked from all RRM pairs with composition distances within the 0.37–0.60 range (i.e., mean ± 1 SD of the composition distance from real sibling pairs) (see Supplemental Fig. S5B). In Figure 1, D and E, all control RRM-pairs have a composition distance within 0.41–0.62 and 0.34–0.57, respectively.
RBP orthologs
Homo sapiens, M. musculus, D. melanogaster, and C. elegans ortholog data were downloaded from the inparanoid database (http://inparanoid.sbc.su.se/download/current/sqltables/) (O'Brien et al. 2005). We downloaded six files, each containing orthologs between two species. We combined all the files and gathered more than 3000 proteins with orthologs found in all four species and thousands of species-specific proteins. These protein sequences were submitted to Pfam for domain analysis with an E-value cutoff of 0.1 (http://pfam.sanger.ac.uk/search#tabview=tab1). Only proteins with more than one predicted RRM were used to calculate the sequence similarity scores. Among the >3000 orthologs between the four species, 80 are RNA-binding proteins, among which 41 human RBPs, 41 mouse RBPs, 34 fly RBPs and 33 worm RBPs contain more than one RRM. We then extracted RBPs that are unique to the individual species and obtained 9, 12, 19, and 12 species-specific RBPs for human, mouse, fly, and worm, respectively. For the sequence similarity score calculation, the sequence pair of RRM from the same RBP were aligned to each other using ClustaW2. All proteins used are listed in Supplementary Tables S2 and S3. The location of the RRM sequence and the sequence similarity score were also included in the table.
Analyses of DAZ and PABP protein family
To obtain orthologs of the human DAZ protein family, we used inparanoid version 8.0 (http://inparanoid.sbc.su.se/download/8.0_current/Orthologs/). Six species were examined: chimpanzee, macaque, gorilla, chicken, frog, and zebrafish. When no ortholog was annotated in the inparanoid database for selected species, we manually searched the protein sequence database by blast to identify potential orthologs. If there are one DAZ domain and multiple DAZ-like repeats, we classified it as an ortholog of either DAZ3 or DAZ2, since orthologs of these two proteins are hard to distinguish. When the occurrence of the RRM is two, we consider it as a DAZ4 ortholog. If the occurrence of RRM is three, we count it as a DAZ1 ortholog. All the orthologs identified by both inparanoid and manual searches are listed in Supplemental Table S4 with their scores and bootstrap probabilities.
The protein sequences of polyadenylate-binding proteins of five species (H. sapiens, M. musculus, D. melanogaster, C. elegans, and S. pombe) were downloaded from uniprot, and their RRM sequences were extracted. We used ClustaW2 to build the phylogenetic tree according to multiple sequence alignments (default parameters were used, i.e., Protein Weight Matrix: gonnet, Clustering type: Neighbor-joining). The ClustaW2-generated guide tree file was then visualized via theTreeView program.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Dr. Alain Laederach, Dr. Todd Vision, and Daniel Dominguez for critical reading of the manuscript. This work was supported by the National Institutes of Health (R01-CA158283 to Z.W.) and the Jefferson Pilot Award to Z.W.
REFERENCES
- Auweter SD, Oberstrass FC, Allain FH 2006. Sequence-specific binding of single-stranded RNA: Is there a code for recognition? Nucleic Acids Res 34: 4943–4959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C, Paillard L, Osborne HB 2005. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res 33: 7138–7150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley RC, Keene JD 1991. Recognition of U1 and U2 small nuclear RNAs can be altered by a 5-amino-acid segment in the U2 small nuclear ribonucleoprotein particle (snRNP) B″ protein and through interactions with U2 snRNP-A′ protein. Mol Cell Biol 11: 1829–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birney E, Kumar S, Krainer AR 1993. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res 21: 5803–5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorklund AK, Ekman D, Elofsson A 2006. Expansion of protein domain repeats. PLoS Comput Biol 2: e114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Burd CG, Matunis EL, Dreyfuss G 1991. The multiple RNA-binding domains of the mRNA poly(A)-binding protein have different RNA-binding activities. Mol Cell Biol 11: 3419–3424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caceres JF, Krainer AR 1993. Functional analysis of pre-mRNA splicing factor SF2/ASF structural domains. EMBO J 12: 4715–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. 2012. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149: 1393–1406 [DOI] [PubMed] [Google Scholar]
- Dominguez C, Allain FH 2006. NMR structure of the three quasi RNA recognition motifs (qRRMs) of human hnRNP F and interaction studies with Bcl-x G-tract RNA: a novel mode of RNA recognition. Nucleic Acids Res 34: 3634–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez C, Fisette JF, Chabot B, Allain FH 2010. Structural basis of G-tract recognition and encaging by hnRNP F quasi-RRMs. Nat Struct Mol Biol 17: 853–861 [DOI] [PubMed] [Google Scholar]
- Eirin-Lopez JM, Ausio J 2011. Boule and the evolutionary origin of metazoan gametogenesis: a grandpa's tale. Int J Evol Biol 2011: 972457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss DJ, Kleiman FE 2013. Poly(A) binding proteins: Are they all created equal? Wiley Interdiscip Rev RNA 4: 167–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamy F, Asseline U, Grasby J, Iwai S, Pritchard C, Slim G, Butler PJ, Karn J, Gait MJ 1993. Hydrogen-bonding contacts in the major groove are required for human immunodeficiency virus type-1 tat protein recognition of TAR RNA. J Mol Biol 230: 111–123 [DOI] [PubMed] [Google Scholar]
- Hargous Y, Hautbergue GM, Tintaru AM, Skrisovska L, Golovanov AP, Stevenin J, Lian LY, Wilson SA, Allain FH 2006. Molecular basis of RNA recognition and TAP binding by the SR proteins SRp20 and 9G8. EMBO J 25: 5126–5137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janga SC, Mittal N 2011. Construction, structure and dynamics of post-transcriptional regulatory network directed by RNA-binding proteins. Adv Exp Med Biol 722: 103–117 [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Kohany O, Jurka MV 2007. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet 8: 241–259 [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. 2012. Cell-free formation of RNA granules: Low complexity sequence domains form dynamic fibers within hydrogels. Cell 149: 753–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M 1989. The neutral theory of molecular evolution and the world view of the neutralists. Genome 31: 24–31 [DOI] [PubMed] [Google Scholar]
- Li M, Shen Q, Xu H, Wong FM, Cui J, Li Z, Hong N, Wang L, Zhao H, Ma B, et al. 2011. Differential conservation and divergence of fertility genes boule and dazl in the rainbow trout. PLoS One 6: e15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Chang KW, Khandjian EW, Richard S 2008. RNA-binding proteins in human genetic disease. Trends Genet 24: 416–425 [DOI] [PubMed] [Google Scholar]
- Maris C, Dominguez C, Allain FH 2005. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Matoulkova E, Michalova E, Vojtesek B, Hrstka R 2012. The role of the 3′ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol 9: 563–576 [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, Reymond L, Amir-Ahmady B, Pitsch S, Black DL, et al. 2005. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science 309: 2054–2057 [DOI] [PubMed] [Google Scholar]
- O'Brien KP, Remm M, Sonnhammer EL 2005. Inparanoid: a comprehensive database of eukaryotic orthologs. Nucleic Acids Res 33: D476–D480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaee N, Kozlov G, Noronha AM, Xie J, Wilds CJ, Gehring K 2012. Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell 48: 375–386 [DOI] [PubMed] [Google Scholar]
- Shen H, Green MR 2006. RS domains contact splicing signals and promote splicing by a common mechanism in yeast through humans. Genes Dev 20: 1755–1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H, Kan JL, Green MR 2004. Arginine-serine-rich domains bound at splicing enhancers contact the branchpoint to promote prespliceosome assembly. Mol Cell 13: 367–376 [DOI] [PubMed] [Google Scholar]
- Sickmier EA, Frato KE, Shen H, Paranawithana SR, Green MR, Kielkopf CL 2006. Structural basis for poly-pyrimidine tract recognition by the essential pre-mRNA splicing factor U2AF65. Mol Cell 23: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas EE 2005. Short, local duplications in eukaryotic genomes. Curr Opin Genet Dev 15: 640–644 [DOI] [PubMed] [Google Scholar]
- Wang Z, Burge CB 2008. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA 14: 802–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Li Z, Li M, Wang L, Hong Y 2009. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One 4: e6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Fu YX, Hewett-Emmett D, Boerwinkle E 2003. Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution. Gene 312: 207–213 [DOI] [PubMed] [Google Scholar]
- Zimmer M, Ebert BL, Neil C, Brenner K, Papaioannou I, Melas A, Tolliday N, Lamb J, Pantopoulos K, Golub T, et al. 2008. Small-molecule inhibitors of HIF-2a translation link its 5′UTR iron-responsive element to oxygen sensing. Mol Cell 32: 838–848 [DOI] [PMC free article] [PubMed] [Google Scholar]