Abstract
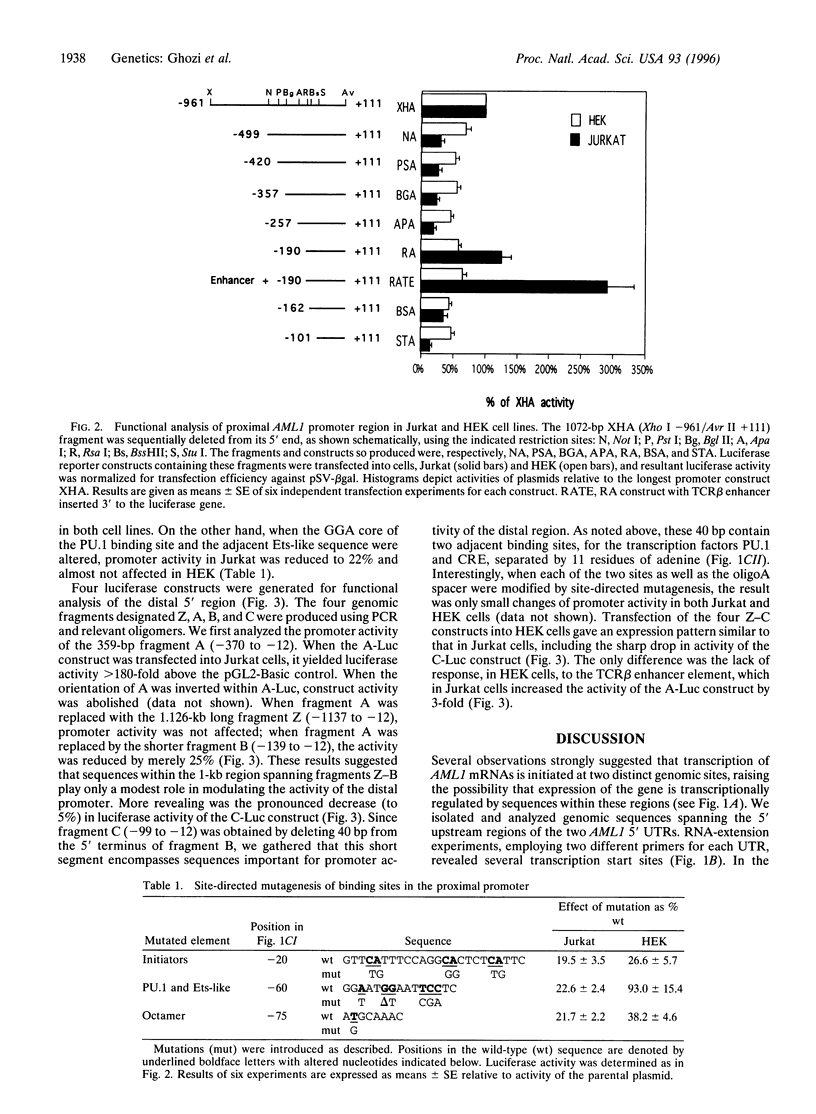
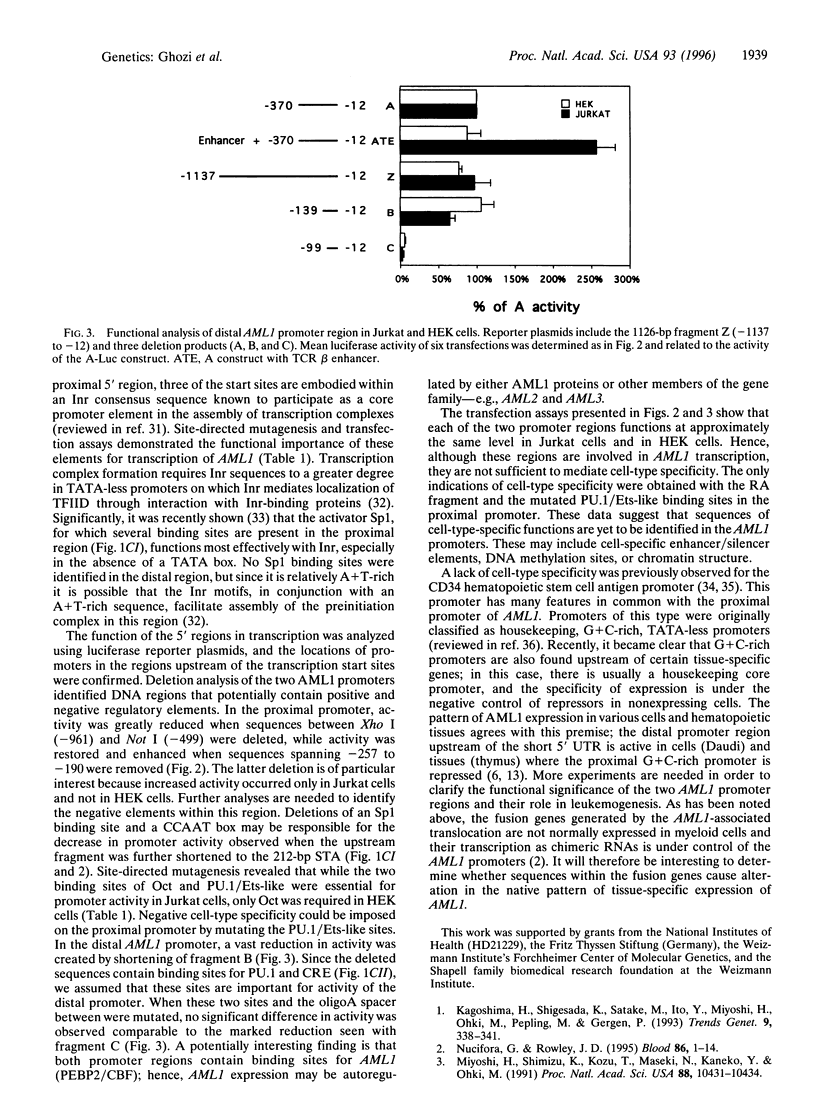
The human chromosome 21 AML1 gene is expressed predominantly in the hematopoietic system. In several leukemia-associated translocations AML1 is fused to other genes and transcription of the fused regions is mediated by upstream sequences that normally regulate the expression of AML1. The 5' genomic region of AML1 was cloned and sequenced. The two 5' untranslated regions (UTRs) previously identified in AML1 cDNAs were located in this region and the distance between them was established. The distal 5' UTR maps over 7 kb upstream of the proximal one. Using primer extension with mRNA, transcription start sites were identified at two distinct sites above these 5' uTRs. Sequence analysis revealed the absence of a TATA motif and the presence of Sp1, PU.1, Oct, CRE, Myb, Ets, and Ets-like binding sites in both upstream regions. Several initiator elements (Inr) that overlap the transcription start sites were also identified. These proximal and distal upstream regions and their deletion mutants were cloned in front of a luciferase reporter gene and used in transfection assays. We demonstrate that both upstream regions function as promoters in hematopoietic (Jurkat) and nonhematopoietic (HEK) cell lines. The activity of both promoters was orientation dependent and was enhanced, in a cell-type specific manner, by a heterologous enhancer sequence. These results indicate that additional control elements, either negative or positive, regulate the tissue-specific expression of AML1.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avraham K. B., Levanon D., Negreanu V., Bernstein Y., Groner Y., Copeland N. G., Jenkins N. A. Mapping of the mouse homolog of the human runt domain gene, AML2, to the distal region of mouse chromosome 4. Genomics. 1995 Jan 20;25(2):603–605. doi: 10.1016/0888-7543(95)80073-u. [DOI] [PubMed] [Google Scholar]
- Azizkhan J. C., Jensen D. E., Pierce A. J., Wade M. Transcription from TATA-less promoters: dihydrofolate reductase as a model. Crit Rev Eukaryot Gene Expr. 1993;3(4):229–254. [PubMed] [Google Scholar]
- Bae S. C., Takahashi E., Zhang Y. W., Ogawa E., Shigesada K., Namba Y., Satake M., Ito Y. Cloning, mapping and expression of PEBP2 alpha C, a third gene encoding the mammalian Runt domain. Gene. 1995 Jul 4;159(2):245–248. doi: 10.1016/0378-1119(95)00060-j. [DOI] [PubMed] [Google Scholar]
- Bae S. C., Yamaguchi-Iwai Y., Ogawa E., Maruyama M., Inuzuka M., Kagoshima H., Shigesada K., Satake M., Ito Y. Isolation of PEBP2 alpha B cDNA representing the mouse homolog of human acute myeloid leukemia gene, AML1. Oncogene. 1993 Mar;8(3):809–814. [PubMed] [Google Scholar]
- Burn T. C., Satterthwaite A. B., Tenen D. G. The human CD34 hematopoietic stem cell antigen promoter and a 3' enhancer direct hematopoietic expression in tissue culture. Blood. 1992 Dec 15;80(12):3051–3059. [PubMed] [Google Scholar]
- Colgan J., Manley J. L. Cooperation between core promoter elements influences transcriptional activity in vivo. Proc Natl Acad Sci U S A. 1995 Mar 14;92(6):1955–1959. doi: 10.1073/pnas.92.6.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin H. A., Diaz M., Bradley C. M., Le Beau M. M., Rowley J. D., Patterson D. Isolation and analysis of the 21q+ chromosome in the acute myelogenous leukemia 8;21 translocation: evidence that c-mos is not translocated. Proc Natl Acad Sci U S A. 1985 Jan;82(2):464–468. doi: 10.1073/pnas.82.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eustice D. C., Feldman P. A., Colberg-Poley A. M., Buckery R. M., Neubauer R. H. A sensitive method for the detection of beta-galactosidase in transfected mammalian cells. Biotechniques. 1991 Dec;11(6):739-40, 742-3. [PubMed] [Google Scholar]
- Faisst S., Meyer S. Compilation of vertebrate-encoded transcription factors. Nucleic Acids Res. 1992 Jan 11;20(1):3–26. doi: 10.1093/nar/20.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris N. L., Senapathy P. Distribution and consensus of branch point signals in eukaryotic genes: a computerized statistical analysis. Nucleic Acids Res. 1990 May 25;18(10):3015–3019. doi: 10.1093/nar/18.10.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X. Y., Cockerill P. N., Cen D., Davis B. R. Transcriptional regulation and chromatin structure of the human CD34 gene promoter region. Blood. 1994 Apr 1;83(7):1822–1830. [PubMed] [Google Scholar]
- Javahery R., Khachi A., Lo K., Zenzie-Gregory B., Smale S. T. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994 Jan;14(1):116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagoshima H., Shigesada K., Satake M., Ito Y., Miyoshi H., Ohki M., Pepling M., Gergen P. The Runt domain identifies a new family of heteromeric transcriptional regulators. Trends Genet. 1993 Oct;9(10):338–341. doi: 10.1016/0168-9525(93)90026-e. [DOI] [PubMed] [Google Scholar]
- Kaufmann J., Smale S. T. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes Dev. 1994 Apr 1;8(7):821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A. The macrophage and B cell-specific transcription factor PU.1 is related to the ets oncogene. Cell. 1990 Apr 6;61(1):113–124. doi: 10.1016/0092-8674(90)90219-5. [DOI] [PubMed] [Google Scholar]
- Kollmar R., Farnham P. J. Site-specific initiation of transcription by RNA polymerase II. Proc Soc Exp Biol Med. 1993 Jun;203(2):127–139. doi: 10.3181/00379727-203-43583. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P., de Jong R., Uematsu Y., Dembic Z., Ryser S., von Boehmer H., Steinmetz M., Berns A. Transcription of T cell receptor beta-chain genes is controlled by a downstream regulatory element. EMBO J. 1988 Mar;7(3):745–750. doi: 10.1002/j.1460-2075.1988.tb02871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiden J. M., Thompson C. B. Transcriptional regulation of T-cell genes during T-cell development. Curr Opin Immunol. 1994 Apr;6(2):231–237. doi: 10.1016/0952-7915(94)90096-5. [DOI] [PubMed] [Google Scholar]
- Levanon D., Negreanu V., Bernstein Y., Bar-Am I., Avivi L., Groner Y. AML1, AML2, and AML3, the human members of the runt domain gene-family: cDNA structure, expression, and chromosomal localization. Genomics. 1994 Sep 15;23(2):425–432. doi: 10.1006/geno.1994.1519. [DOI] [PubMed] [Google Scholar]
- Meyers S., Lenny N., Hiebert S. W. The t(8;21) fusion protein interferes with AML-1B-dependent transcriptional activation. Mol Cell Biol. 1995 Apr;15(4):1974–1982. doi: 10.1128/mcb.15.4.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Ohira M., Shimizu K., Mitani K., Hirai H., Imai T., Yokoyama K., Soeda E., Ohki M. Alternative splicing and genomic structure of the AML1 gene involved in acute myeloid leukemia. Nucleic Acids Res. 1995 Jul 25;23(14):2762–2769. doi: 10.1093/nar/23.14.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H., Shimizu K., Kozu T., Maseki N., Kaneko Y., Ohki M. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10431–10434. doi: 10.1073/pnas.88.23.10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora G., Begy C. R., Erickson P., Drabkin H. A., Rowley J. D. The 3;21 translocation in myelodysplasia results in a fusion transcript between the AML1 gene and the gene for EAP, a highly conserved protein associated with the Epstein-Barr virus small RNA EBER 1. Proc Natl Acad Sci U S A. 1993 Aug 15;90(16):7784–7788. doi: 10.1073/pnas.90.16.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nucifora G., Birn D. J., Espinosa R., 3rd, Erickson P., LeBeau M. M., Roulston D., McKeithan T. W., Drabkin H., Rowley J. D. Involvement of the AML1 gene in the t(3;21) in therapy-related leukemia and in chronic myeloid leukemia in blast crisis. Blood. 1993 May 15;81(10):2728–2734. [PubMed] [Google Scholar]
- Nucifora G., Rowley J. D. AML1 and the 8;21 and 3;21 translocations in acute and chronic myeloid leukemia. Blood. 1995 Jul 1;86(1):1–14. [PubMed] [Google Scholar]
- Ogawa E., Maruyama M., Kagoshima H., Inuzuka M., Lu J., Satake M., Shigesada K., Ito Y. PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci U S A. 1993 Jul 15;90(14):6859–6863. doi: 10.1073/pnas.90.14.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohki M. Molecular basis of the t(8;21) translocation in acute myeloid leukaemia. Semin Cancer Biol. 1993 Dec;4(6):369–375. [PubMed] [Google Scholar]
- Perez C., Coeffier E., Moreau-Gachelin F., Wietzerbin J., Benech P. D. Involvement of the transcription factor PU.1/Spi-1 in myeloid cell-restricted expression of an interferon-inducible gene encoding the human high-affinity Fc gamma receptor. Mol Cell Biol. 1994 Aug;14(8):5023–5031. doi: 10.1128/mcb.14.8.5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge D. S. SIGNAL SCAN: a computer program that scans DNA sequences for eukaryotic transcriptional elements. Comput Appl Biosci. 1991 Apr;7(2):203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Rowley J. D. Down Syndrome and acute leukaemia: increased risk may be due to trisomy 21. Lancet. 1981 Nov 7;2(8254):1020–1022. doi: 10.1016/s0140-6736(81)91218-6. [DOI] [PubMed] [Google Scholar]
- Sacchi N., Nisson P. E., Watkins P. C., Faustinella F., Wijsman J., Hagemeijer A. AML1 fusion transcripts in t(3;21) positive leukemia: evidence of molecular heterogeneity and usage of splicing sites frequently involved in the generation of normal AML1 transcripts. Genes Chromosomes Cancer. 1994 Dec;11(4):226–236. doi: 10.1002/gcc.2870110405. [DOI] [PubMed] [Google Scholar]
- Tanaka T., Tanaka K., Ogawa S., Kurokawa M., Mitani K., Nishida J., Shibata Y., Yazaki Y., Hirai H. An acute myeloid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms. EMBO J. 1995 Jan 16;14(2):341–350. doi: 10.1002/j.1460-2075.1995.tb07008.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B., Hahn S. L., Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993 Jan 15;211(1-2):7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Wijmenga C., Speck N. A., Dracopoli N. C., Hofker M. H., Liu P., Collins F. S. Identification of a new murine runt domain-containing gene, Cbfa3, and localization of the human homolog, CBFA3, to chromosome 1p35-pter. Genomics. 1995 Apr 10;26(3):611–614. doi: 10.1016/0888-7543(95)80185-o. [DOI] [PubMed] [Google Scholar]
- Zawel L., Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]




