Abstract
Although Neanderthals are extinct, fragments of their genomes persist in contemporary humans. Here we show that while the genome-wide frequency of Neanderthal-like sites is approximately constant across all contemporary out-of-Africa populations, genes involved in lipid catabolism contain more than threefold excess of such sites in contemporary humans of European descent. Evolutionally, these genes show significant association with signatures of recent positive selection in the contemporary European, but not Asian or African populations. Functionally, the excess of Neanderthal-like sites in lipid catabolism genes can be linked with a greater divergence of lipid concentrations and enzyme expression levels within this pathway, seen in contemporary Europeans, but not in the other populations. We conclude that sequence variants that evolved in Neanderthals may have given a selective advantage to anatomically modern humans that settled in the same geographical areas.
 Modern human genomes contain Neanderthal sequences, but it is unclear whether these were selected. Here, Khrameeva et al. show that Neanderthal sequences associated with lipid catabolism are three times more frequent in Europe, suggesting that these sequences might have been beneficial to Europeans.
Modern human genomes contain Neanderthal sequences, but it is unclear whether these were selected. Here, Khrameeva et al. show that Neanderthal sequences associated with lipid catabolism are three times more frequent in Europe, suggesting that these sequences might have been beneficial to Europeans.
The ancestors of Neanderthals and modern humans diverged from a common ancestral population ~800–400 thousand years ago (KYA)1,2,3,4,5. Subsequently, Neanderthals evolved in Europe and Central Asia and possibly spread further east into the Asian continent6. The first successful migration of modern humans out of Africa can be traced back in the archaeological and genetic records to ~70–60 KYA7. After that, modern humans spread quickly across all continents and could have coexisted with Neanderthals in Europe and Central Asia for thousands of years before the Neanderthal extinction 45–30 KYA7,8. Studies comparing the complete nuclear genome sequences of Neanderthals and contemporary modern humans indicate that out-of-Africa human populations, but not sub-Saharan African populations, contain genomic regions with unusually high similarity to the Neanderthal genome9,10. In each individual, these regions were reported to occupy 1–4% of the total genome sequence10. This phenomenon implies the occurrence of gene flow from Neanderthals into those human populations that migrated from the African continent11,12.
In this study, we asked whether genomic regions with high sequence similarity to the Neanderthal genome are distributed randomly across the genomes of contemporary modern humans. If certain regions in the modern human genome show a decreased Neanderthal gene flow, these regions may contain genetic changes essential to the modern human phenotype that are undergoing purifying selection against the Neanderthal alleles. By contrast, the presence of genomic regions experiencing excessive gene flow from Neanderthal may indicate that genetic changes that evolved in Neanderthals gave modern humans carrying the Neanderthal genotype a selective advantage.
We show that genetic variants shared between modern humans and Neanderthals, but distinct from chimpanzees, are specifically enriched in genes involved in lipid catabolism in contemporary humans of European, but not East Asian descent. Excess of Neanderthal variants in lipid catabolism genes was further associated with signatures of recent positive selection. To assess whether Neanderthal variants resulted in adaptive lipid catabolism changes specific to Europeans, we measured lipidome and transcriptome compositions of the brain tissue in contemporary humans of European, East Asian and African descent, as well as in chimpanzees representing the ancestral state. In agreement with observations made at the genome level, we show significant excess of lipid concentration and gene expression divergence in lipid catabolism pathways in Europeans, but not the other population groups. Furthermore, lipid catabolism genes showing increased expression divergence in Europeans contain an even higher proportion of Neanderthal sites than other lipid catabolism genes. Taken together, these observations indicate that lipid catabolism of contemporary Europeans was partially shaped by the genetic variants shared with Neanderthals. We further speculate that these variants provided European ancestors with adaptations to the geographic environment where Neanderthals had evolved and where Neanderthals and archaic Europeans later coexisted.
Results
Neanderthal ancestry in contemporary human populations
We searched for regional similarities to the Neanderthal genome in the genomes of 11 contemporary human populations, which have the best genome coverage in the 1,000 genomes project: three populations of African ancestry—HapMap African ancestry individuals from South West of the United States (ASW), Luhya individuals (LWK) and Yoruba individuals (YRI); three populations of East Asian ancestry—Han Chinese in Beijing (CHB), Han Chinese from South China (CHS) and Japanese (JPT); and five populations of European ancestry—CEPH individuals (CEU), HapMap Finnish individuals from Finland (FIN), British individuals from England and Scotland (GBR), Iberian populations in Spain (IBS) and Toscan individuals (TSI)13. For each pair of the human populations, the human genome sequences were compared with the Neanderthal and the chimpanzee genome sequences. We used the high-coverage Neanderthal genome sequence, obtained from the bone of one individual excavated in 2008 in the east gallery of Denisova Cave in the Altai mountains (Altai)14,15, combined with the low-coverage Neanderthal genome sequence, obtained from three individuals (Vindjia)10, to reconstruct a consensus Neanderthal genome sequence by excluding all sites with sequence variation among individuals. We further used the reference chimpanzee genome in combination with genome sequence data from 10 chimpanzees16 to exclude sites variable among chimpanzees. Only single nucleotide sites displaying sequence differences between the chimpanzee16,17 and the two reference Neanderthal genomes10,15 were used in the analysis (n=1,158,559).
Similar to Green et al.10, we used the D statistic to detect potential Neanderthal gene flow. For each pair of human populations, we defined two configurations for genomic sites: sites where the sequence of an individual from one human population (population A) matched the Neanderthal rather than chimpanzee genotype were assigned to the ABBA configuration; and sites where the sequence of the other human population (population B) matched the Neanderthal rather than chimpanzee genotype were assigned to the BABA configuration (Fig. 1a). For each population, the D statistic, which we will refer to as the fraction of Neanderthal-like sites (NLS), was calculated as the ratio of (#ABBA−#BABA) to (#ABBA+#BABA), where # stands for the number of genomic sites in the specific genotype configuration (ABBA or BABA; see Methods). While D-statistic values reflect relative similarity between the Neanderthal and the modern human genomes tested, they do not provide a quantitative estimate of Neanderthal ancestry, that is, a 5% D-statistic value reflects a higher similarity between population A and Neanderthal compared with that for population B, but does not signify a 5% level of Neanderthal ancestry in the population A genome.
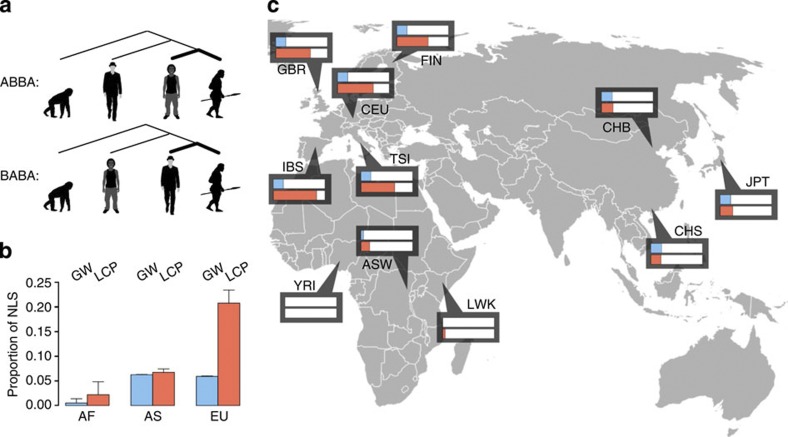
Figure 1. Proportions of NLS in contemporary human populations.
(a) Schematic representation of genomic distance calculations between contemporary human populations and Neanderthals. The genomes of out-of-Africa individuals were compared with the genomes of individuals of purely African ancestry (YRI). Single nucleotide differences from the Neanderthal genotype in an African genome were referred to as ‘ABBA’, while sites with the Neanderthal genotype in an out-of-Africa genome were referred to as ‘BABA’. (b) Average proportions of NLS in contemporary African (AF), European (EU) and Asian (AS) populations calculated based on sequence data from the 1,000 genomes project13; blue: genome wide (n=1,158,559 sites), red: LCP genes (n=498 sites). The error bars show the s.d. of the NLS proportion estimates. (c) Genomic distances between 11 contemporary human populations and Neanderthals; blue, genome wide; red, LCP genes. The maximal bar length corresponds to a NLS frequency of 30%. Placement of ASW and CEU individuals in sub-Saharan Africa and Western Europe, respectively, reflects their approximate historical geographical origins rather than their present location.
In agreement with previous observations10, the genomes of contemporary humans of European and Asian descent showed greater similarities to the Neanderthal genome than did the genomes of the three populations of purely African descent. On average, the NLS frequency was 6.1±0.2% for contemporary humans of European and Asian descent, thus indicating a substantial excess of NLS in contemporary out-of-Africa populations (Fig. 1b,c, blue bars; Supplementary Tables 1–3). This D-statistic estimate is similar to the ones reported by other studies (4.8±0.2%)10, with the higher values obtained in our study potentially arising from the additional filtering of genomic sites polymorphic in Neanderthals. Further, in agreement with other studies10, there was no substantial difference in the genome-wide frequencies of NLS between European and Asian populations, with a slight tendency for higher frequencies in Asians: 5.9±0.08 and 6.2±0.06%, respectively18,19.
For each pair of human populations, we searched for the presence of functional groups of genes showing an unusual excess, or paucity, of NLS. This analysis, based on gene groups compiled according to gene ontology terms20, and conducted using the gene set enrichment analysis (GSEA) algorithm21, yielded an unexpected observation; we indeed find significant clustering of NLS in specific functional groups, but these functional groups differed substantially between contemporary European and Asian populations (Supplementary Data 1). Functional groups showing NLS enrichment in Asian populations mainly represent immune and haematopoietic pathways. The strongest signal of NLS enrichment was, however, observed in contemporary Europeans and included two functional groups: the lipid catabolic process (LCP) and its nested term—cellular LCP (GSEA, permutations P<0.01, significance score >3; Fig. 2a). Specifically, genes in the LCP term had the greatest excess of NLS in populations of European descent, with an average NLS frequency of 20.8±2.6% versus 5.9±0.08% genome wide (two-sided t-test, P<0.0001, n=379 Europeans and n=246 Africans; Fig. 1b,c, red bars; Supplementary Table 4; Supplementary Fig. 1). Further, among examined out-of-Africa human populations, the excess of NLS in LCP genes was only observed in individuals of European descent: the average NLS frequency in Asians is 6.7±0.7% in LCP genes versus 6.2±0.06% genome wide (Supplementary Table 4).
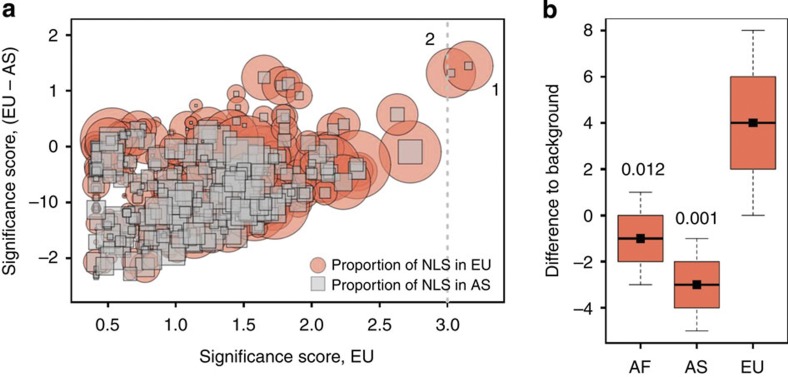
Figure 2. Outstanding genetic features of lipid catabolism genes.
(a) Clustering of NLS in functional categories in the genomes of contemporary humans of European (EU) and Asian (AS) descent. The significance scores are proportional to the NES and inversely proportional to the false discovery rates calculated for each gene ontology (GO) term in the biological process category using the GSEA algorithm. Sizes of circles and squares represent NLS fractions in Europeans and Asians, respectively, for each GO term. Numbers near the circles mark the top two GO terms with a functional enrichment significance score greater than three, based on the distribution of Neanderthal-like genomic sites in Europeans: (1) LCP, (2) cellular LCP. (b) Positive selection signals in LCP gene regions estimated using CMS scores. Black squares represent the frequency of sites with elevated CMS scores (>1), potentially indicating genomic regions under recent positive selection in LCP gene regions, normalized by the frequency of such sites in all annotated genes within the same population. The boxplots show the variation of normalized site frequency estimates obtained by 1,000 bootstraps over LCP gene regions (n=38). The boxes show quartiles and the median of the data, the whiskers extend to the minimum and maximum data values located within 0.5 interquartile range from the box. The numbers above the boxplots show the proportion of bootstrap values where the normalized site frequency of elevated CMS scores in Africans (AF) or Asians (AS) was greater than, or equal to, the normalized site frequency in Europeans (EU).
The excess of NLS observed in LCP genes for populations of European descent was based on a large number of sites (n=498), and was robust to bootstrapping across sites (P<0.01, 1,000 bootstraps, Supplementary Table 5; Supplementary Fig. 2). Notably, NLS were located in 23 independent genomic regions. Among the remaining 15 LCP genes that did not contain NLS, 8 did not contain sites showing divergence between Neanderthals and chimpanzees and the remaining 7 contained only a few such divergent sites (Supplementary Table 6). It is furthermore robust to the potential effects of DNA damage characteristic of ancient DNA samples, as excluding the C/T and A/G substitutions that may stem from deamination of cytosine residues in ancient DNA22,23 did not affect the results (Supplementary Table 4). Repeating the analysis using the genome sequences of African (ASW, LWK, YRI), European (CEU, FIN, GBR, IBS) and East Asian (CHB, JPT) individuals, which were sequenced to deeper coverage at the pilot stage of the 1,000 genomes project, as well as the high-coverage genome sequences of African (ASW, LWK, YRI, MKK—Maasai in Kinyawa, Kenya), European (CEU, TSI) and East Asian (CHB, JPT) individuals provided by the Complete Genomics human diversity set24, confirmed our observations (Supplementary Tables 7 and 8). Finally, repeating the analysis using the high-coverage (Altai) and the low-coverage (Vindjia) Neanderthal genomes, separately, resulted in similar findings (Supplementary Table 9).
Adaptive signature of Neanderthal sequences in LCP genes
The excess of NLS in LCP genes in the genomes of contemporary Europeans may be due to a rapid spread of Neanderthal alleles in European ancestors because of their adaptive significance. Specifically, one may hypothesize that, over time, Neanderthals acquired changes to lipid catabolism, which were beneficial for survival in the environmental conditions of prehistoric Europe and Central Asia. These adaptive variants may then have been acquired by the modern humans through introgression and rapidly brought to high frequency by positive selection. To test this hypothesis, we searched for signatures of positive selection in the genomes of contemporary humans of European, Asian and African decent using composite of multiple signals (CMS) scores25. High CMS values indicate genomic regions under recent positive selection based on three distinct signatures of selection: long-range haplotypes, differentiated alleles and high-frequency-derived alleles. We indeed found a significant excess of high CMS scores in the LCP gene regions of contemporary Europeans but not Asians or Africans (Fig. 2b). This effect was robust at different CMS score cutoffs and was specific to LCP: no significant excess of high CMS scores in individuals of European descent was observed in comparable genomic regions containing other metabolic genes (Supplementary Fig. 3). Furthermore, within the LCP term, high CMS scores found in contemporary Europeans were associated with genes containing the excess of NLS, but were not associated with other LCP genes (two-sample Wilcoxon test, P=0.0003; n=45 and 20; Supplementary Fig. 4).
Metabolic changes associated with Neanderthal ancestry
The observed signatures of positive selection suggest that genetic variants shared with Neanderthals resulted in adaptive changes in lipid catabolism in Europeans, but not in Asians. At a physiological level, these adaptive changes should affect the concentrations of LCP metabolites, and the expression levels of the corresponding metabolic enzymes, in a manner specific to Europeans. To test this, we analysed the lipid composition of prefrontal cortex (PFC) tissue in 14 adult humans of European, African and Asian descent, as well as 14 adult chimpanzees (Supplementary Data 2) using C8-reversed phase liquid chromatography coupled to high-resolution mass spectrometry26,27. Out of 4,243 detected mass spectrometric peaks, 1,314 could be computationally matched to lipid compounds belonging to 63 metabolic categories using metabolite annotation databases28,29. After elimination of low confidence matches supported by less than 5 mass spectrometric peaks, 16 metabolic categories, containing 1,253 peaks, remained and were used in further analyses (Fig. 3a; Supplementary Data 2).
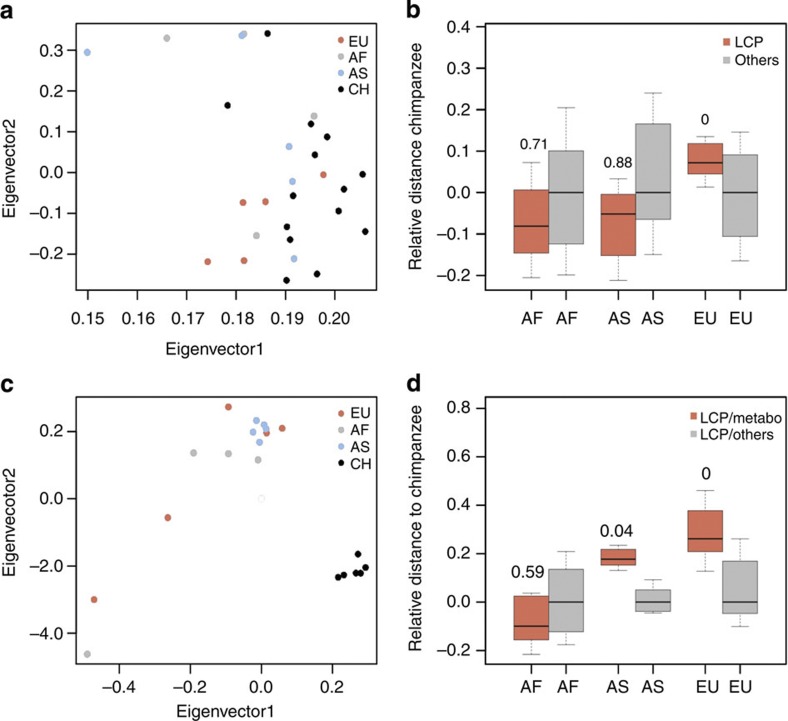
Figure 3. Lipid concentration and gene expression divergence in LCP and other metabolic pathways.
(a) Principal component analysis based on normalized intensities of 1,314 annotated mass spectrometric peaks corresponding to 63 metabolic categories. Each circle represents an individual: blue, Asians; grey, Africans; red, Europeans; black, chimpanzees. (b) The distribution of lipid concentration divergence estimates measured between chimpanzees and humans of African (AF, n=4 individuals), Asian (AS, n=5 individuals) and European (EU, n=5 individuals) descent for metabolic categories directly linked to LCP genes (red, n=1,090 mass spectrometric peaks) and metabolites in other metabolic pathways (grey, n=163 mass spectrometric peaks). To minimize the influence of environmental differences among populations, metabolic divergence in the LCP term was normalized to the divergence of all other metabolic pathways within the same population. The numbers above the red boxplots show the proportion of values from the LCP divergence distribution obtained by 1,000 bootstraps over individuals within populations that were smaller than, or equal to, the divergence values calculated based on other metabolic pathways represented by the grey boxplots. All boxes in this and the other panels show quartiles and the median of the data, the whiskers extend to the minimum and maximum data values located within 0.5 interquartile range from the box. (c) Principal component analysis based on the expression levels of 25,813 genes. Each circle represents an individual; colours are as in panel a. (d) The distribution of gene expression divergence estimates measured between chimpanzees and human populations for LCP genes directly linked to seven metabolic categories shown in panel b (red, n=6 expressed genes) and other LCP genes (grey, n=26 expressed genes). Normalization procedure and significance estimation were conducted the same way as for metabolite data presented in panel b.
Seven of these 16 metabolic categories were directly linked with genes in the LCP term (Supplementary Table 10) based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway annotation30. In Europeans, the concentrations of lipids within these seven metabolic categories linked with LCP were more diverged from chimpanzees than the concentrations of lipids in the other nine metabolic categories not linked with LCP (q<0.0001; q value shows the proportion of the LCP divergence values that were smaller than, or equal to, the divergence values in other metabolic categories). By contrast, in contemporary Africans or Asians there were no differences between the concentration divergence of lipids associated with LCP and the lipids associated with other metabolic categories (q>0.1; Fig. 3b). This result was not driven by one or several metabolites, but represented a general property of this metabolite group, as shown by bootstrap analysis (q<0.0001) Further, this result was not caused by differences among population samples with respect to age, sex, tissue preservation or postmortem delay (Supplementary Table 11; Supplementary Fig. 5). We note that, while we cannot control for environmental differences among populations, our analysis is based on the relative divergence of seven metabolic categories associated to LCP and was normalized to the divergence of the other nine metabolic categories not associated with LCP within the same population. This normalization removes the influence of environmental factors affecting LCP and non-LCP metabolites to the same extent. Furthermore, our study design provides no indications that environmental effects should be particular to Europeans: all individuals of European and African descent used for lipidome analysis came from the same region within the United States, while all individuals of Asian descent came from central China.
Expression changes associated with Neanderthal ancestry
We next asked whether the greater concentration divergence of LCP metabolites observed in Europeans could be linked to a similarly accelerated expression level divergence of the corresponding enzymes. To test this, we measured gene expression levels in the 14 human PFC samples used in the lipid analysis, as well as 6 out of 14 chimpanzee PFC samples, using high-throughput RNA sequencing (RNA-seq). For each sample, we obtained an average of 15 million reads, 85% of which could be mapped31 uniquely to the corresponding genome (Fig. 3c; Supplementary Table 12; Supplementary Fig. 6). Using KEGG pathway annotation, six genes annotated in the LCP term and detected in the RNA-seq data could be directly linked to the seven LCP metabolic categories (Supplementary Table 10). For these six genes the expression divergence from chimpanzees, relative to the expression divergence of other LCP genes expressed in PFC, was the largest in Europeans (q<0.0001; q value shows the proportion of the divergence values that were smaller than, or equal to, the divergence values in other LCP genes), intermediate in Asians (q=0.04) and absent in Africans (q=0.59; Fig. 3d; Supplementary Fig. 6). This result was robust, as shown by bootstrap analysis, and was not caused by differences in age, sex, RNA quality or postmortem delay among the three human populations (Supplementary Fig. 7). Thus, accelerated metabolic divergence in the LCP term found in Europeans appears to be linked to an accelerated expression level divergence of the corresponding metabolic enzymes.
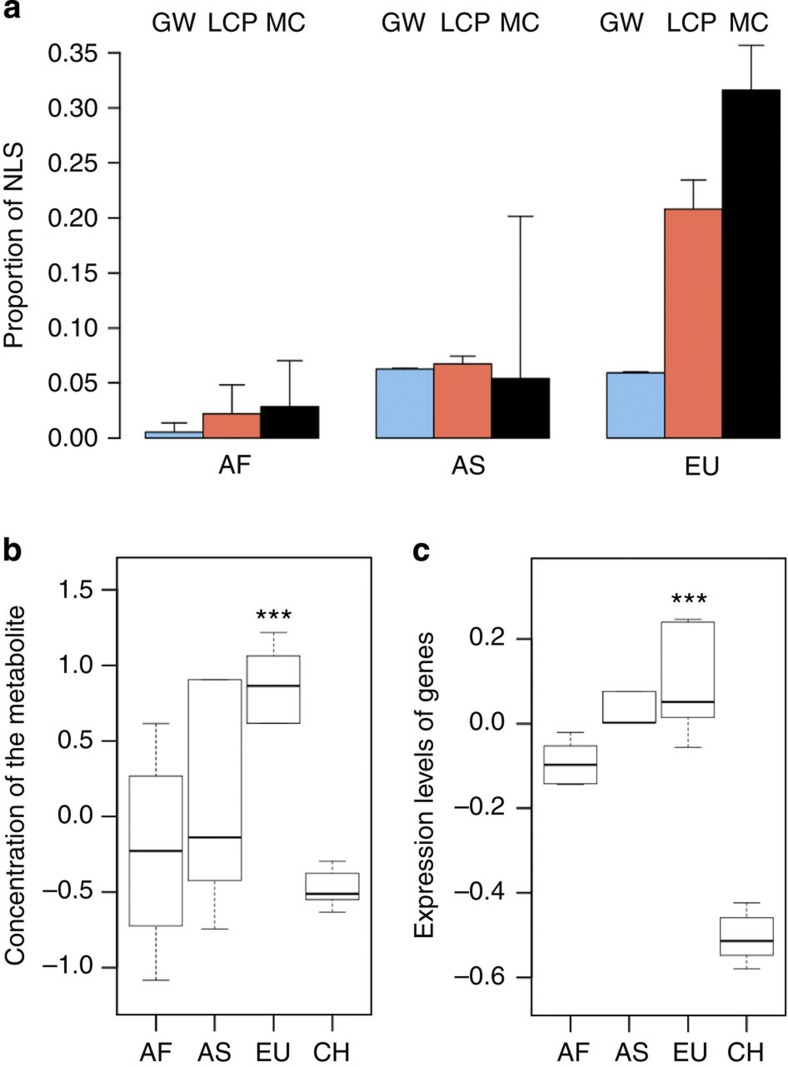
Notably, the gene regions of the six LCP enzymes linked to European-specific metabolic changes contained an even higher proportion of NLS in Europeans (31.6±4.1%) than all LCP genes (20.8±2.6%). Furthermore, these NLS were not distributed uniformly within the gene regions, but clustered in the vicinity of transcription start sites, suggesting that they may have a role in causing gene expression level changes seen in Europeans (two-sample Wilcoxon test, P=0.048; n=14 and 27; Supplementary Fig. 8). By contrast, individuals of Asian and African descent did not show any significant excess of NLS within the same gene regions (Fig. 4a).
Figure 4. Potential regulatory effects of NLS in LCP pathway.
(a) Average proportions of NLS in contemporary African (AF), European (EU) and Asian (AS) populations calculated based on sequence data from the 1,000 genomes project13; blue, genome wide (GW, n=1,158,559 sites); red, all LCP genes (n=498 sites); black, LCP genes connected to European-specific metabolic changes (MC, n=114 sites). The error bars show the s.d. of the NLS proportion estimates. (b) Relative concentration levels of 2-lysophosphatidylcholine, in three contemporary human populations and in chimpanzees (CH). The boxplots represent the median and the variation of normalized, z-transformed metabolite concentrations in each sample group calculated by 1,000 bootstraps over individuals within populations (n=11 mass spectrometric peaks). The *** indicates significance of 2-lysophosphatidylcholine concentration difference between European (n=5) and chimpanzee (n=14) individuals (P<0.001) estimated by the bootstrapping procedure. (c) Gene expression levels of genes directly linked with 2-lysophosphatidylcholine according to KEGG annotation, in the three human populations and chimpanzees. The boxplots are as in panel b. The *** indicates significance of expression difference between European (n=5) and chimpanzee (n=6) individuals (P<0.001) for genes directly linked with 2-lysophosphatidylcholine (n=21) estimated by the bootstrapping procedure.
Possible functional implications of changes in LCP
While we find changes in lipid catabolism particular to Europeans at the metabolite concentration and enzyme expression levels, the significance of these changes at the organismal level remains to be investigated. Still, the changes observed at the molecular level provide some clues. Among the seven metabolic categories associated with LCP, 2-lysophosphatidylcholine has been implicated in a number of functions, including reactive oxygen species generation, apoptotic and non-apoptotic death, as well as glucose-dependent insulin secretion32 (Fig. 4b,c). Furthermore, genetic variants linked to obesity (DAVID33, Fisher’s exact test, P<0.01 after multiple testing correction, n=38) hypertriglyceridemia and coronary heart disease, as well as triglycerides and cholesterol levels (DAVID, Fisher’s exact test, P<0.01 after multiple testing correction, n=38) in genome-wide association studies34 show a significant enrichment of LPC genes containing an excess of NLS (Supplementary Table 13). Notably, frequencies of these diseases have been shown to differ between individuals of European descent and other human populations35. These observations support a contribution of Neanderthal genetic variants to the phenotype of contemporary Europeans.
Discussion
Our results show that NLS—genetic variants shared between modern humans and Neanderthals, but distinct from chimpanzees—are specifically enriched in genes involved in lipid catabolism in contemporary humans of European descent. Signatures of recent positive selection associated with lipid catabolism genes containing NLS in contemporary Europeans further indicate that these genetic variants may have been swept to high frequency by positive selection. This notion presumes the presence of functional changes in the LCP associated with NLS ancestry. Our analysis of lipid concentration and enzyme expression levels in brain samples of chimpanzees and contemporary humans of African, East Asian and European decent supports this assumption. Specifically, we show an increased lipid concentration and enzyme expression divergence between Europeans and the ancestral state exemplified by chimpanzees in lipid catabolism pathways compared with other metabolic pathways. Finally, we show that lipid catabolism genes showing increased expression divergence in Europeans contain an even higher proportion of NLS than other lipid catabolism genes, and these NLS are located in closer proximity to the genes’ transcriptional start sites than would be expected by chance.
It is appealing to speculate that genetic variants affecting lipid catabolism in modern Europeans were acquired by modern human ancestors through genetic flow from Neanderthals, and then spread rapidly though the ancestral population by means of positive selection. Action of positive selection could be further explained by the adaptive significance of these variants in the geographic environment where Neanderthals had evolved and where Neanderthals and archaic Europeans later coexisted. It is noteworthy, however, that our observations are compatible with both introgression and incomplete lineage sorting hypotheses explaining the excess of genetic variants shared between contemporary humans and Neanderthals in out-of-Africa populations. Furthermore, while the excess of NLS in lipid catabolism genes is not observed in East Asian populations, we cannot assume that it is specific to Europeans. Complete genome sequences from a larger spectrum of human populations, especially from geographical regions coinciding with the Neanderthal living range, are needed to determine the full geographical spectrum of this effect. This is particularly true, given the fact that the Neanderthal area included Asian regions, such as the Altai Mountains. Still, the absence of NLS frequency increase in lipid catabolism genes in East Asian populations accompanied by no increase in the lipid concentration and enzyme expression divergence in the corresponding pathways indicates the geographical specificity of this phenomenon.
One further argument indirectly supporting geographical specificity and local adaptive significance of the lipid catabolism changes potentially induced by Neanderthal variants comes from a comparison with the genome of another archaic human species—Denisovans36. Analysis of lipid catabolism gene sequences with the Denisova genome revealed a higher frequency of sites shared between Neanderthals and moderns humans, derived respective to both the chimpanzees and Denisovans, in contemporary Europeans than in East Asians (Supplementary Table 14). Notably, no such difference was observed in the genome-wide analysis. This result shows that changes in lipid catabolism genes shared between Neanderthals and contemporary Europeans were not fully present in Denisovans. Given that the presumed geographical area of Denisovans includes most of Asia36, this result indirectly supports specificity of observed lipid catabolism change to the European part of the Eurasian continent.
We further note that a high frequency of NLS in lipid catabolism genes of contemporary Europeans does not require introgression, but is compatible with alternative scenarios. For instance, an alternative explanation of the general increase in NLS frequency in humans outside Africa, postulating the existence of a complex population structure within the African continent at the time of human and Neanderthal lineage divergence, has been hypothesized4. This hypothesis explains the presence of Neanderthal variants in non-African human populations by shared ancestry specific to ancestral human populations that left the African continent. If the lipid catabolism gene variants we find in Neanderthals and contemporary Europeans were already present in the ancestors of Neanderthals and out-of-Africa human populations, they may have independently increased in frequency in Neanderthals and humans situated in the European region. This scenario is probable if these genetic variants provided an adaptive advantage to both Neanderthal and human populations in the conditions of prehistoric Europe. While the presence of a recent positive selection signal in lipid catabolism gene variants containing NLS in modern Europeans supports such an adaptive scenario, the environmental pressures or functional mechanisms of this possible adaptive change remain elusive. Further studies of LCP conducted across multiple tissues and multiple contemporary human populations are needed to more fully assess the potential functional effects of this event.
Methods
D-statistic calculation
The genome sequences of individuals in 11 contemporary human populations, ASW, CEU, CHB, CHS, FIN, GBR, IBS, JPT, LWK, TSI and YRI were obtained from the 1,000 genomes project (release 2011.05.21)13. The genome sequence reads from 10 chimpanzee individuals were obtained from ref. 16 and mapped to the panTro2 reference genome. The sites polymorphic among chimpanzees were identified using GATK program17 with filtering parameters DP<30, DP>180, MQ0>40, (MQ0/(1.0*DP))>0.40, AN<16, MQ<25. This procedure identified 323,948 polymorphic sites that were removed from further analyses. The non-overlapping genome sequence contigs representing the Vindija Neanderthal genomes were obtained from the UCSC genome browser. The Altai Neanderthal genome contigs were obtained from http://www.eva.mpg.de/neandertal/index.html15. For both genomes, 4,360,373 sites were removed following the filtering procedure described in ref. 10. Coordinates of the chimpanzee and Vindija Neanderthal sequences were mapped to the human reference genome hg19 using the UCSC liftover tool with default parameters. A total of 411,783 polymorphic sites identified in a comparison between Vindija and Altai Neanderthal sequences were removed from further analysis.
To estimate similarity between the Neanderthal genome and the genomes of contemporary humans, we applied the following procedure to all possible pairs of contemporary human populations. The following procedure was applied to each pair of populations (Pop1 and Pop2). We randomly selected two individuals from populations Pop1 and Pop2—Ind1 and Ind2, respectively. Only sites with different genotypes in chimpanzee (A) and Neanderthal (B) were considered. We calculated #BABA as the number of sites where Ind1=B, Ind2=A (and neand.=B, chimp.=A) and #ABBA as the number of sites where Ind1=A, Ind2=B (and neand.=B, chimp.=A). Then, D score was calculated as following: (#ABBA−#BABA)/(#ABBA+#BABA). To estimate the proportions of NLS for populations Pop1 and Pop2, D score was calculated for all possible pairs of individuals Ind1 and Ind2, and then averaged across these pairs.
Functional analysis of gene groups
The GSEA program21 was applied to find functional gene groups with a significant enrichment or depletion of NLS in out-of-Africa human populations. All out-of-Africa human populations were compared with a purely African population YRI. We calculated #ABBA and #BABA for each pair of individuals (one from out-of-Africa population, another from YRI). The GSEA program ranks genes according to a difference between two values (#ABBA and #BABA), and each value can have several replicates. We treated each pair of individuals as a replicate, which was reasonable because our pairs of individuals were independent. To make the pairs of individuals independent (and also to lower their number and save the calculation time), pairs of individuals were composed in such a way that one individual could participate in one pair only.
We used two key statistics that GSEA reported for the gene set enrichment analysis: normalized enrichment score (NES) and false discovery rate to calculate the GSEA significance score. NES is the actual enrichment score divided by the mean of enrichment scores against all permutations of the data set to account for the differences in gene set size. False discovery rate is the estimated probability that a gene set with a given NES represents a false-positive finding. In all GSEA analyses, we used MSigDB collection21 of 825 gene ontology biological process gene sets ( http://www.broadinstitute.org/gsea/msigdb/download_file.jsp?filePath=/resources/msigdb/3.0/c5.bp.v3.0.symbols.gmt) as an annotation database.
Samples used for metabolite and RNA-seq experiments
Chimpanzee samples were obtained from the Anthropological Institute and Museum of the University of Zürich-Irchel, Switzerland, the Yerkes Primate Center, GA, USA and the Biomedical Primate Research Centre, the Netherlands. All chimpanzees used in this study suffered sudden deaths for reasons other than their participation in this study and without any relation to the tissue used. Human samples were obtained from the NICHD Brain and Tissue Bank for Developmental Disorders at the University of Maryland and the Chinese Brain Bank Center. All subjects suffered sudden death with no prolonged agonal state. They were defined as healthy controls by forensic pathologists at the corresponding tissue bank. Specific permission for brain autopsy and use of the brain tissue for research purpose was given by the donors or their relatives. Use of human autopsy tissue is considered non-human-subject research and is institutional review board exempt under the National Institutes of Health guidelines.
The samples were dissected from frozen postmortem tissue on dry ice. The frontal part of the superior frontal gyrus, a cortical region approximately corresponding to Brodmann area 10, was used for dissection. The tissue samples were first powdered using a mortar and pestle cooled by liquid nitrogen and next separated in two parts for metabolite and RNA extraction procedures. Each sample was ~100 mg in weight.
Metabolite sample preparation and measurement
Metabolites were extracted from the frozen brain tissue powder by methanol: methyl-tert-butyl-ether (1:3 (v/v)) extraction26,27. In brief, 50 mg of frozen powdered PFC tissue was re-suspended in 1 ml extraction solution containing two internal standards (0.5 μg of corticosterone and 1.5 μg of 1,2-diheptadecanoyl-sn-glycero-3-phosphocholine (PC 34:0)). The samples were incubated for 10 min at 4 °C on an orbital shaker, before subjecting them to ultrasonication for 10 min in an ice-cooled bath-type sonicator. The insoluble tissue material (including proteins) was pelleted by a centrifugation step (5 min; 14,000g) and the supernatant was transferred to a fresh 2 ml Eppendorf tube. To separate the organic from the aqueous phase, 500 μl of an H2O:methanol mixture (3:1(v/v)) was added to the supernatant, mixed by vortexing and centrifuged (5 min; 14,000g). Five hundred microlitres of the upper methyl-tert-butyl-ether-phase was transferred to a 1.5-ml Eppendorf tube, concentrated in a speed vacuum and re-suspended in 100 μl of an acetonitrile:isopropanol mixture (7:3 (v/v)) before liquid chromatography–mass spectrometry analysis. For the analysis, 5 μl of lipid extract was injected onto the ultra performance liquid chromatography C8-reversed phase column (BEH C8, Waters), connected to an Orbitrap Exactive mass spectrometer26,27.
Metabolite data pre-processing and analysis
Mass spectrometric peaks that had zero values within one of the sample groups (European, Chinese, African American or chimpanzee) were directly removed from the resulting matrix because of inconsistency between the replicates. Lipid metabolite data were then normalized for the internal standard (PC 34:0), log scaled and then quantile normalization was applied. Each peak intensity value was z-transformed. The resulting 4,243 spectrometric peaks were preliminarily annotated using database searches against the HMDB and LipidMaps databases27,28,29. These searches, which were based on pure mass to charge (m/z) ratio matches of the mass that was spectrometrically measured and the theoretical masses of the lipids in the above-mentioned databases, were performed with an m/z tolerance of 10 p.p.m., allowing different ionization adducts27. A total of 2,111 mass spectrometric peaks could thus be assigned to known metabolites in the HMDB and LipidMaps database and 1,314 of these could be linked to the KEGG database via the corresponding compound ID. Principal component analysis was performed using the ‘prcomp’ R function for all peaks linked to the KEGG database using normalized data before z-transformation.
If several peaks could be assigned to the same metabolic category, a median across these peaks was used as the metabolite category concentration value. Metabolic categories that could be assigned fewer than five peaks were removed from further analysis. For each metabolite, a median value was calculated within European, Chinese, African American and chimpanzee sample groups. The metabolite divergence between European, Chinese, and African American populations and chimpanzees was calculated as an absolute difference between the chimpanzee and human values.
To test the robustness of metabolite divergence estimates, we bootstrapped the individuals within populations 1,000 times. As a result, we obtained two divergence distributions: (i) for metabolic categories directly linked to the LCP pathway according to KEGG annotation (KEGG metabolism pathways only were used) and (ii) for all other metabolites detected in the PFC. One-sided P values were calculated as the proportion of values from the distribution (i) that were smaller than, or equal to, the actual value for the distribution (ii).
RNA-seq sample preparation and measurement
Total RNA was isolated using Trizol reagent (catalogue number 15596-026, Invitrogen). The libraries were constructed from the isolated total RNA according to the ‘TruSeq RNA Sample Preparation Kit’ protocol ( http://www.illumina.com) without modification. As the protocol includes the procedure of enrichment for messenger RNA, an additional polyA selection was not necessary. Samples were sequenced on the Illumina HiSeq 2,000 system, using the 100-bp single-end sequencing protocol.
RNA-seq data pre-processing and analysis
Human reads were mapped using Tophat31 (parameters were set to -a 8 -m 2 no-coverage-search microexon-search segment-mismatches 3 segment-length 25) to the hg19 reference genome. Chimpanzee reads were mapped to the panTro3 reference genome using the same procedure. To match human and chimpanzee gene annotations, we used a reciprocal liftover to map the human annotation to the chimpanzee genome (Ensembl annotation v. 69). Only exons that could be matched between the two genomes by reciprocal liftover were used to calculate gene expression intensities (RPKM). Gene expression values were further normalized by quantile normalization and z-transformation across all samples. Principal component analysis was performed using the ‘prcomp’ R function for all genes using normalized data before z-transformation.
For gene expression divergence calculations, a median value was calculated within European, Chinese, African American and chimpanzee sample groups. The distance from European, Chinese and African American populations to chimpanzee was calculated as an absolute difference between chimpanzee and human values. To test the robustness of gene expression divergence measurements, we bootstrapped individuals within populations 1,000 times, and obtained two distributions: (i) for genes directly linked to LCP metabolites according to KEGG annotation (KEGG metabolism pathways only were used); and (ii) for all other LCP genes expressed in the PFC. One-sided P values were calculated as the proportion of values from the distribution (i) that were smaller than, or equal to, the actual value for the distribution (ii).
Author contributions
P.K., M.L., S.P. and P.G. conceived and designed the experiments; Z.Y., X.J. and P.G. performed the experiments; E.E.K., K.B., L.H., Y.W., P.G. and K.T. analysed the data; P.K., E.E.K., M.L., M.S.G., P.G. and S.P. wrote the manuscript.
Additional information
Accession Codes: All human and chimpanzee RNA-seq data were uploaded to the Gene Expression Omnibus ( http://www.ncbi.nlm.nih.gov/geo/) under accession code GSE45263.
How to cite this article: Khrameeva, E. E. et al. Neanderthal ancestry drives evolution of lipid catabolism in contemporary Europeans. Nat. Commun. 5:3584 doi: 10.1038/ncomms4584 (2014).
Supplementary Material
Supplementary Figures 1-8 and Supplementary Tables 1-14
Enrichment of NLS in specific functional categories within the genomes of 10 contemporary human populations conducted using Gene Set Enrichment Analysis (GSEA).
Lipidomics data. The file contains intensities of 4,243 mass spectrometry peaks in 14 human and 14 chimpanzee prefrontal cortex samples.
Acknowledgments
We thank Mehmet Somel for helpful discussions and Jerome Boyd-Kirkup for help in manuscript preparation. This study was supported by the National Natural Science Foundation of China (Grant numbers 91331203, 31171232 and 31250110071); Ministry of Science and Technology of China (Grant number 2012DFG31940); Russian Academy of Sciences (program ‘Molecular and Cellular Biology'); and the Russian Foundation of Basic Research (Grant numbers 14-04-91342, 12-04-31789 and 12-04-91332). P.K. was supported by the Foreign Expert 1,000 Talents Plan program; E.E.K. and M.L. were supported by Chinese Academy of Sciences fellowships (Grant numbers 2011Y2SB13 and 2009S1-23).
References
- Krings M. et al. Neandertal DNA sequences and the origin of modern humans. Cell 90, 19–30 (1997). [DOI] [PubMed] [Google Scholar]
- Serre D. et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2, e57 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E. et al. Analysis of one million base pairs of Neanderthal DNA. Nature 444, 330–336 (2006). [DOI] [PubMed] [Google Scholar]
- Green R. E. et al. A complete Neandertal mitochondrial genome sequence determined by high-throughput sequencing. Cell 134, 416–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noonan J. P. et al. Sequencing and analysis of Neanderthal genomic DNA. Science 314, 1113–1118 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause J. et al. Neanderthals in central Asia and Siberia. Nature 449, 902–904 (2007). [DOI] [PubMed] [Google Scholar]
- Mellars P. Neanderthals and the modern human colonization of Europe. Nature 432, 461–465 (2004). [DOI] [PubMed] [Google Scholar]
- Finlayson C. et al. Late survival of Neanderthals at the southernmost extreme of Europe. Nature 443, 850–853 (2006). [DOI] [PubMed] [Google Scholar]
- Wall J. D., Lohmueller K. E. & Plagnol V. Detecting ancient admixture and estimating demographic parameters in multiple human populations. Mol. Biol. Evol. 26, 1823–1827 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R. E. et al. A draft sequence of the Neandertal genome. Science 328, 710–722 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. D., Mekel-Bobrov N., Vallender E. J., Hudson R. R. & Lahn B. T. Evidence that the adaptive allele of the brain size gene microcephalin introgressed into Homo sapiens from an archaic Homo lineage. Proc. Natl Acad. Sci. USA 103, 18178–18183 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D. & Hammer M. F. Archaic admixture in the human genome. Curr. Opin. Genet. Dev. 16, 606–610 (2006). [DOI] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mednikova M. B. A proximal pedal phalanx of a Paleolithic hominin from denisova cave, Altai. Archaeol. Ethnol. Anthropol. Eurasia 39, 129–138 (2011). [Google Scholar]
- Prüfer K. et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505, 43–49 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auton A. et al. A fine-scale chimpanzee genetic map from population sequencing. Science 336, 193–198 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall J. D. et al. Higher levels of Neanderthal ancestry in East Asians than in Europeans. Genetics 194, 199–209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. et al. Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M., Jaenicke V., Serre D., von Haeseler A. & Paabo S. DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Res. 29, 4793–4799 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs A. W. et al. Patterns of damage in genomic DNA sequences from a Neandertal. Proc. Natl Acad. Sci. USA 104, 14616–14621 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drmanac R. et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science 327, 78–81 (2010). [DOI] [PubMed] [Google Scholar]
- Grossman S. et al. Identifying recent adaptations in large-scale genomic data. Cell 152, 703–713 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel J. et al. Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front. Plant Sci. 2, 54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giavalisco P. et al. Elemental formula annotation of polar and lipophilic metabolites using 13C, 15N and 34S isotope labelling, in combination with high-resolution mass spectrometry. Plant J. 68, 364–376 (2011). [DOI] [PubMed] [Google Scholar]
- Wishart D. S. et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 41, d801–d807 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahy E., Sud M., Cotter D. & Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 35, w606–w612 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Sato Y., Furumichi M. & Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 40, d109–d114 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Pachter L. & Salzberg S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzelczyk A. & Gendaszewska-Darmach E. Novel bioactive glycerol-based lysophospholipids: new data—new insight into their function. Biochimie 95, 667–679 (2013). [DOI] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. & Lempicki R. A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K. G., Barnes K. C., Bright T. J. & Wang S. A. The genetic association database. Nat Genet. 36, 431–432 (2004). [DOI] [PubMed] [Google Scholar]
- Yu S. S., Castillo D. C., Courville A. B. & Sumner A. E. The triglyceride paradox in people of African descent. Metab. Syndr. Relat. Disord. 10, 77–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M. et al. A high-coverage genome sequence from an archaic Denisovan individual. Science 338, 222–226 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1-8 and Supplementary Tables 1-14
Enrichment of NLS in specific functional categories within the genomes of 10 contemporary human populations conducted using Gene Set Enrichment Analysis (GSEA).
Lipidomics data. The file contains intensities of 4,243 mass spectrometry peaks in 14 human and 14 chimpanzee prefrontal cortex samples.