Summary
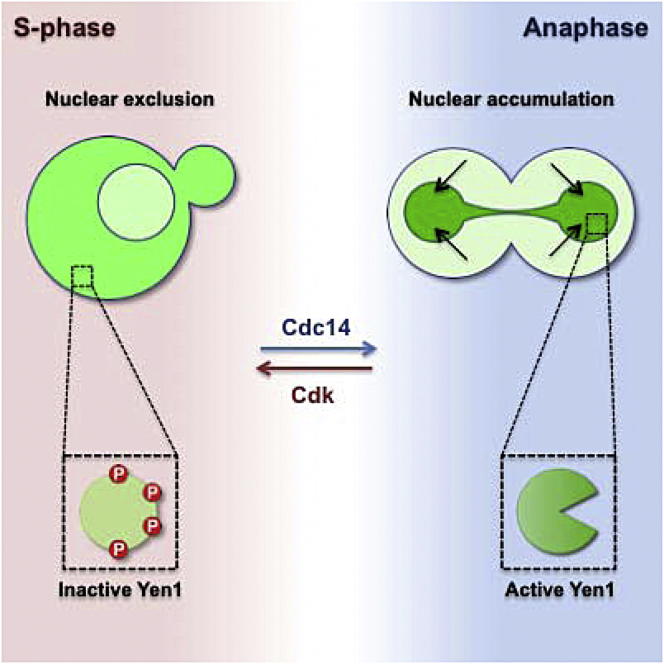
The careful orchestration of cellular events such as DNA replication, repair, and segregation is essential for equal distribution of the duplicated genome into two daughter cells. To ensure that persistent recombination intermediates are resolved prior to cell division, the Yen1 Holliday junction resolvase is activated at anaphase. Here, we show that the master cell-cycle regulators, cyclin-dependent kinase (Cdk) and Cdc14 phosphatase, control the actions of Yen1. During S phase, Cdk-mediated phosphorylation of Yen1 promotes its nuclear exclusion and inhibits catalytic activity by reducing the efficiency of DNA binding. Later in the cell cycle, at anaphase, Cdc14 drives Yen1 dephosphorylation, leading to its nuclear relocalization and enzymatic activation. Using a constitutively activated form of Yen1, we show that uncontrolled Yen1 activity is detrimental to the cell: spatial and temporal restriction of Yen1 protects against genotoxic stress and, by avoiding competition with the noncrossover-promoting repair pathways, prevents loss of heterozygosity.
Graphical Abstract
Highlights
-
•
Yen1 undergoes a dual mode of regulation: activity and subcellular localization
-
•
Cdk phosphorylation inhibits Yen1 at S phase by reducing its DNA binding affinity
-
•
Yen1 activation at anaphase is driven by the Cdc14 phosphatase
-
•
Premature activation of Yen1 leads to loss of heterozygosity and genome instability
The complete elimination of DNA recombination intermediates is essential for chromosome segregation. Blanco et al. show that the master cell-cycle regulators Cdk and Cdc14 control both the localization and nuclease activation of the Holliday junction resolvase Yen1. Cdk/Cdc14 therefore control the final wave of joint molecule resolution at anaphase.
Introduction
Before every cell division, our genetic material needs to be duplicated and assembled into two equal packages that can be faithfully transmitted to the next generation. The temporal coordination of molecular events such as DNA replication, repair, and chromosome segregation is therefore critical for the maintenance of genome integrity. Homologous recombination (HR), although essential for the repair of DNA breaks and the restoration of damaged replication forks, can occasionally create a block to chromosome segregation by generating covalently linked DNA intermediates such as Holliday junctions (HJs) (Branzei and Foiani, 2008; Holliday, 1964; West, 2003). These joint molecules (JMs) constitute a physical connection between sister chromatids or homologous chromosomes, and their timely removal is needed to avoid any interference with normal chromosome segregation.
Cells possess a battery of helicases and nucleases that can deal with HR intermediates. For example, in S. cerevisiae there are two major pathways for the processing of late HR intermediates. The first, mediated by the Sgs1-Top3-Rmi1 (STR) complex, specializes in the elimination of double HJs (dHJs) that arise from HR-mediated strand break repair (Bzymek et al., 2010; Cejka et al., 2010). For this, the RecQ-family helicase Sgs1 catalyzes the convergent branch migration of two HJs, resulting in a hemicatenate structure that is decatenated by the type I topoisomerase Top3. A related mechanism of double HJ “dissolution” occurs in human cells, driven by BLM-TopoIIIα-RMI1-RMI2 (BTR complex) and is important for the avoidance of sister chromatid exchanges and loss of heterozygosity (Luo et al., 2000; Wu and Hickson, 2003). Indeed, STR/BTR play an important role in limiting crossover (CO) formation by directing the products of recombination to noncrossovers (NCOs). The second pathway for the processing of late HR intermediates involves two structure-selective endonucleases: in yeast, these are the XPF-family endonuclease Mus81-Mms4 (Boddy et al., 2001; Ehmsen and Heyer, 2008; Interthal and Heyer, 2000; Kaliraman et al., 2001) and the XPG-family member Yen1 (Blanco et al., 2010; Ip et al., 2008). Again, there is a good analogy with human systems, where MUS81-EME1 and GEN1 are important for promoting Holliday junction “resolution” (Castor et al., 2013; Chen et al., 2001; Garner et al., 2013; Ip et al., 2008; Wechsler et al., 2011; Wyatt et al., 2013). These reactions, unlike STR/BTR, have the potential to promote both the formation of NCO and CO products. Therefore, the choice of pathway for the removal of late HR intermediates directs the outcome of repair, with NCO pathways being the preferred choice in mitotic cells (Ira et al., 2003).
How the processing of recombination intermediates is biased toward NCOs in mitosis has been the focus of extensive research. In sgs1Δ mutants, the elimination of JMs is delayed until G2/M, becomes dependent on Mus81-Mms4, and results in increased CO formation (Dayani et al., 2011; Ira et al., 2003). Consistent with these observations, sgs1Δ mus81Δ double mutants are synthetically lethal due to the accumulation of toxic HR intermediates (Fabre et al., 2002), indicating that Mus81-Mms4 resolves HR intermediates that escape the actions of the STR complex at earlier stages of the cell cycle. The DNA repair deficiency of mus81Δ and mms4Δ mutants is exacerbated by further deletion of YEN1, and mus81Δ yen1Δ double mutants display both aberrant chromosome segregation and reduced mitotic CO formation (Blanco et al., 2010; Ho et al., 2010). Thus, Yen1 provides an additional mechanism for JM resolution.
The apparent hierarchy among the STR complex, Mus81-Mms4, and Yen1 for DNA damage repair is supported by observations showing that Mus81-Mms4 and Yen1 are sequentially activated in two waves that occur late in the cell cycle. Cdk/Cdc5-mediated phosphorylation of Mms4 drives the hyperactivation of Mus81-Mms4 at the G2/M transition, whereas S phase phosphorylation of Yen1 holds this protein in an inactive state until it is activated at anaphase (Gallo-Fernández et al., 2012; Matos et al., 2011, 2013; Szakal and Branzei, 2013). Intuitively, such a regulatory mechanism creates a window during S/G2 in which the NCO-promoting pathways have preferential access to the majority of repair intermediates. Any persistent dHJs, and single HJs that might arise from replication fork processing, would then be captured by Mus81-Mms4 in G2/M or by Yen1 in anaphase, thus ensuring the segregation of DNA. The temporal restriction of Mus81-Mms4 and Yen1 activities to late stages of the cell cycle may also be important to protect against the unscheduled cleavage of DNA replication intermediates that arise during S phase.
Although the biochemical properties of Mus81-Mms4 have been studied extensively, the biochemical properties of Yen1 (759 amino acids) or its human ortholog GEN1 (908 amino acids) have yet to be determined. Indeed, the analysis of Yen1 has been restricted to immunoprecipitates, whereas most studies of GEN1 were carried out with a purified N-terminal fragment (GEN11-527) of the protein (Ip et al., 2008; Matos et al., 2011; Rass et al., 2010). Like other members of the XPG family of 5′-flap endonucleases, Yen1/GEN1 exhibit 5′-flap and fork cleavage activities, but, unique to this nuclease family, Yen1 and GEN1 cleave HJs by the introduction of symmetrically related nicks across the junction to produce ligatable nicked duplex products.
Yen1 is a target of the S phase cyclin-dependent kinase Cdk, and its subcellular localization is cell-cycle-regulated in a Cdk-dependent manner (Kosugi et al., 2009; Loog and Morgan, 2005; Ubersax et al., 2003). However, we know very little about the mechanism of Yen1 inhibition during S phase, how it is activated at anaphase, or how these dynamic cycles of inactivation/activation are coupled with cell-cycle progression. Here, we investigate the mechanism of Yen1 regulation, in terms of its biochemical activation and subcellular localization, and show how both are linked to cell-cycle control. Importantly, we demonstrate that Cdk and the mitotic exit phosphatase Cdc14 (Stegmeier and Amon, 2004) constitute the “off” and “on” switches for Yen1 function, respectively. Employing Yen1 mutants that are refractory to Cdk-dependent inhibition and are constitutively active throughout the cell cycle, we show that premature activation of Yen1 results in DNA damage sensitivity and increased loss of heterozygosity. The purification of full-length Yen1, in both the phosphorylated and nonphosphorylated states, revealed that the mechanistic basis of inhibition lies in modulation of the DNA binding activity of the protein. As such, Yen1 is uniquely regulated at two distinct levels, by both biochemical inhibition and by nuclear exclusion, until anaphase when the protein is activated and undergoes nuclear entry in response to Cdc14-mediated dephosphorylation.
Results
Generation of a Constitutively Active Form of Yen1
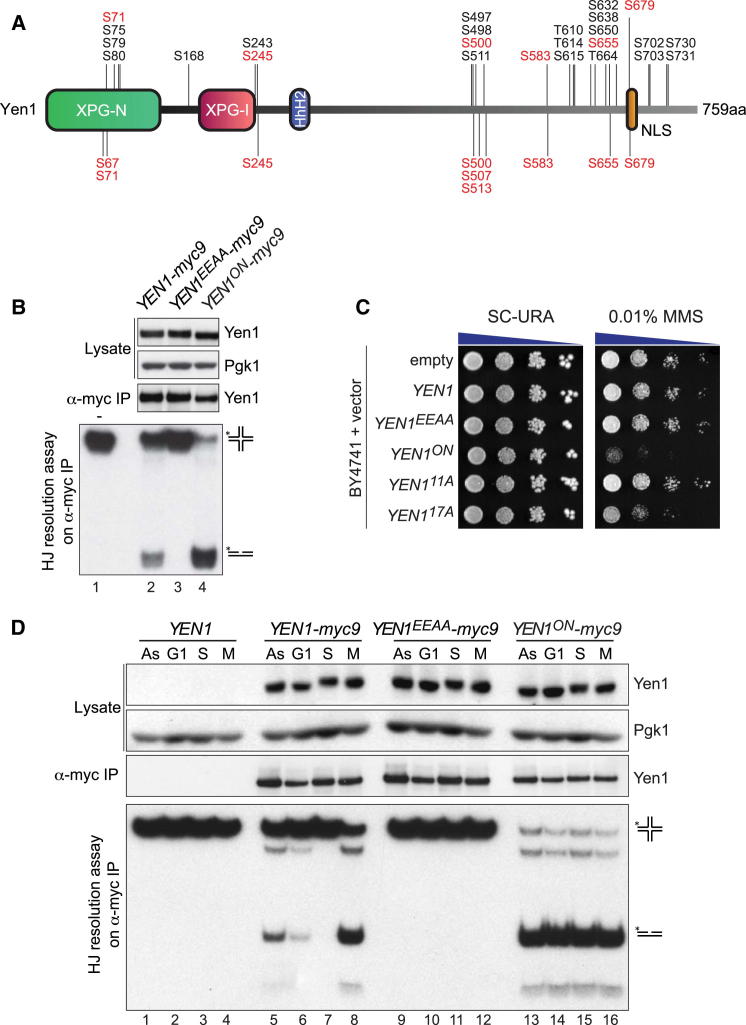
Yen1 regulation involves cycles of phosphorylation and dephosphorylation that control both its activity and cellular localization (Kosugi et al., 2009; Matos et al., 2011). To determine the sites of phosphorylation, affinity-purified Yen1 was analyzed by mass spectrometry, revealing a total of 25 different phosphorylated residues (Figure 1A). Several phosphorylated serines and threonines were located in consensus target sequences for cell-cycle control and DNA damage response kinases, including Cdk (S/T-P; S71, S245, S500, S583, S655, S679), Mec1/Tel1 (S/T-Q; S80, S498, T664), and Cdc5 (D/E-X-S/T-ϕ-X-D/E; S614). Although our peptide coverage for Yen1 was incomplete, it is significant that phosphorylation was detected in six of the nine predicted Cdk-consensus sites.
Figure 1.
Yen1ON Is a Constitutively Active Version of Yen1
(A) Schematic representation of Yen1, indicating the 25 phosphoresidues identified by mass spectrometry (above) and the nine predicted Cdk consensus target sites (below). Cdk sites are highlighted in red.
(B) Analysis of the nuclease activity of Yen1ON. Soluble lysates were prepared from asynchronous cultures of strains in which YEN1-myc9, nuclease-dead YEN1EEAA-myc9, and YEN1ON-myc9 alleles replaced endogenous YEN1. Following α-myc immunoaffinity purification, the IPs were analyzed by western blotting and for nuclease activity using 32P-labeled Holliday junction DNA. Upper panel: western blots of the lysate and IP, with Pgk1 as a loading control. Lower panel: resolution assays. (−), untreated substrate.
(C) Yen1ON overexpression causes increased MMS sensitivity. Ten-fold dilutions of wild-type strain BY4741 transformed with centromeric plasmids carrying different YEN1 alleles (see Figure S1) were plated on SC-URA with or without MMS and imaged after 3 days.
(D) Yen1ON activity escapes cell-cycle control. The activity profiles of Yen1-myc9, Yen1EEAA-myc9 and Yen1ON-myc9 were analyzed as in (B) following their immunoprecipitation from synchronized cultures. As, asynchronous culture; G1, α-factor treated culture; S, S and G2 cells 25 min after α-factor release; M, G2/M cells 60 min after α-factor release.
See also Figure S1.
Using this information, and the fact that Yen1 is a known target of Cdk, we generated a collection of YEN1 alleles with mutations in the identified/predicted phosphoresidues (Figure S1 available online). One allele, named YEN1ON, in which the serines in all nine Cdk consensus sites were mutated to alanine, was of particular interest because immunoprecipitates (IPs) of Yen1ON-myc9 displayed a greater HJ resolvase activity than Yen1-myc9, as measured by the conversion of a 32P-labeled HJ into nicked duplex products (Figure 1B, compare lanes 2 and 4). Moreover, we found that constitutive expression of Yen1ON from a plasmid resulted in increased cellular sensitivity to the DNA-damaging agent methyl methanesulfonate (MMS) (Figure 1C), whereas expression of the wild-type protein or a catalytically inactive form of Yen1 (Yen1EEAA, in which residues E193 and E195 were converted to alanine) did not affect MMS sensitivity. In contrast to Yen1, which exhibited little HJ resolvase activity in S phase but was activated at anaphase, Yen1ON was constitutively active throughout the entire cell cycle (Figure 1D, compare lanes 6–8 with 14–16), explaining the high levels of activity observed in asynchronous cultures (Figure 1B, lane 4).
Among the different phosphomutants generated, containing between 1 and 17 amino acid changes (Figure S1), YEN1ON exhibited constitutive nuclease activity and the strongest sensitivity to DNA-damaging agents, coupled with the lowest number of amino acid substitutions (Figure 1C; data not shown). It was therefore selected for further study. Analysis of the other mutants, however, showed that Yen1 inhibition during S phase was primarily dependent on the phosphorylation status of S500, S507, S513, and S583, located in the central region of Yen1 (compare Yen14A and Yen15A in Figure S1).
Deregulated Yen1 Resolves HR Intermediates Leading to Increased CO Frequency and Defective DNA Repair
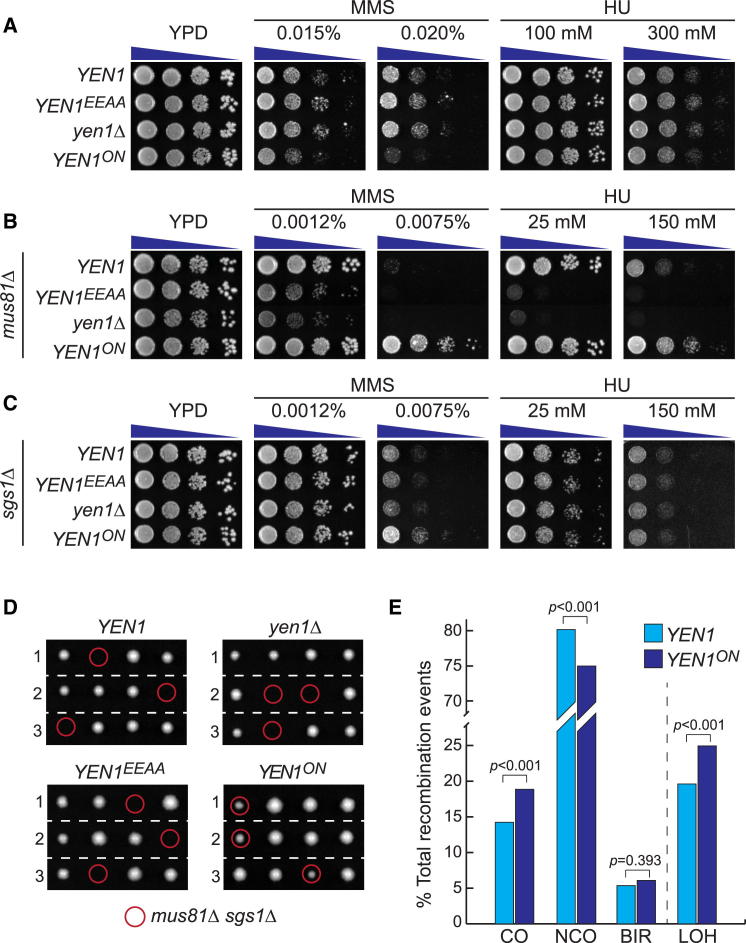
To determine the consequences of Yen1 deregulation in more detail, we constructed strains in which the endogenous YEN1 allele was replaced with YEN1ON. Cells harboring YEN1ON displayed increased sensitivity to high doses of MMS, but not hydroxyurea (HU) (Figure 2A). Remarkably, YEN1ON fully suppressed the DNA damage sensitivity of mus81Δ strains, which are deficient for the catalytic subunit of the structure-selective endonuclease Mus81-Mms4 (Figure 2B). YEN1ON could also partially suppress the MMS sensitivity of sgs1Δ mutants, whereas no effect was observed for HU (Figure 2C). Finally, we found that YEN1ON rescued the synthetic lethality of sgs1Δ mus81Δ double mutants (Figure 2D). These results show that prematurely activated Yen1 is capable of acting upon recombination intermediates that are normally processed by Mus81-Mms4 or Sgs1.
Figure 2.
Premature Activation of Yen1 Causes Increased DNA Damage Sensitivity and Loss of Heterozygosity
(A) Expression of Yen1ON at physiological levels causes MMS sensitivity. Assays were carried out as described in Figure 1C, using strains in which endogenous YEN1 was replaced by the indicated YEN1 alleles. Cells were plated on YPD containing the indicated amount of drugs.
(B) YEN1ON suppresses the DNA damage sensitivity of mus81Δ mutants. As in (A), but employing strains deleted for MUS81.
(C) Mild suppression of the MMS sensitivity of sgs1Δ mutants by YEN1ON.
(D) YEN1ON suppresses the synthetic lethality of mus81Δ sgs1Δ double mutants. Diploid strains homozygous for the indicated YEN1 alleles and heterozygous for MUS81/mus81Δ and sgs1Δ/SGS1 were sporulated and analyzed by tetrad dissection. Images were taken after 3 days incubation at 30°C. The expected position of each mus81Δ sgs1Δ colony is indicated (red circles).
(E) YEN1ON promotes crossover formation and loss of heterozygosity during mitotic recombination. The percentage of different recombination events in YEN1 and YEN1ON strains is indicated. CO, crossovers; NCO, noncrossovers; BIR, break-induced replication. LOH indicates the total loss of heterozygosity (the sum of CO and BIR events). p values were calculated using a chi-square test with Pearson’s correction.
See also Table S1.
The action of STR complex early in the cell cycle generates noncrossovers (NCOs) and limits the formation of crossovers (COs) that could lead to loss of heterozygosity (LOH). We therefore reasoned that premature activation of Yen1 would increase CO formation by competing with the NCO-promoting pathways. To test this, we utilized a genetic assay for the detection of unselected products of mitotic recombination after the formation of a single I-SceI-mediated double-strand break at the ade2-I locus on chromosome XV (Ho et al., 2010). When we scored the recombination events in a wild-type YEN1 background, we found 14.2% COs and 80.5% NCOs (Figure 2E; Table S1). In strains carrying YEN1ON, however, the CO frequency increased significantly to 18.9% at the expense of NCO products, with little effect on the frequency of break-induced replication (BIR) events. These results show that restriction of Yen1 nuclease activity until mitosis is important for efficient DNA repair and to avoid LOH through the formation of excess mitotic COs.
Phosphorylation-Dependent Control of Yen1 Localization
Yen1 has been identified as a protein that undergoes cell-cycle-dependent nucleocytoplasmic relocalization (Kosugi et al., 2009), and it has been suggested that the subcellular localization of Yen1 is determined by the phosphorylation status of the Cdk consensus site (S679) that overlaps the nuclear localization signal (NLS; 679-SPIKKSRTT-687). Indeed, overexpressed Yen1 is predominantly nuclear in G1-arrested cells and becomes cytoplasmic after release into S phase in a Cdk-dependent manner. Substitution of S679 with alanine, which abrogates the phosphorylation of the NLS-overlapping Cdk consensus site, promotes a greater extent of nuclear localization throughout the cell cycle (Kosugi et al., 2009).
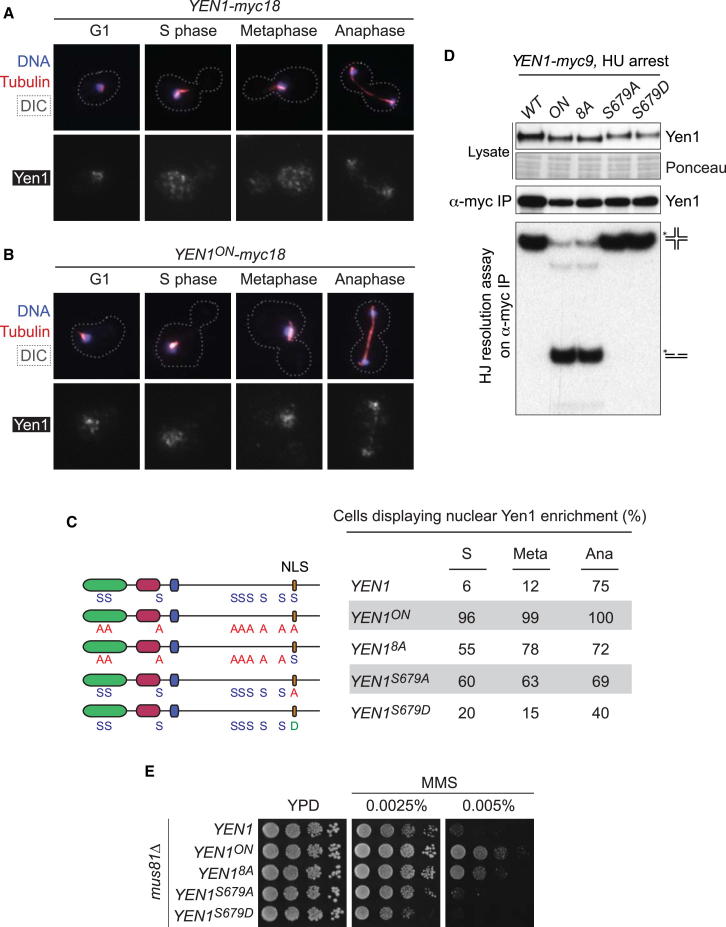
S679 is one of the residues mutated to alanine in Yen1ON (Figure 1B). We therefore used immunofluorescence analyses to determine how this mutational change affected the localization of Yen1ON-myc18 (expressed from the endogenous YEN1 locus) throughout the cell cycle. Control experiments showed that wild-type Yen1 was enriched in the nucleus during G1 but became diffuse throughout the cell during S phase. Yen1 remained cytoplasmic in metaphase, but strikingly relocalized into the nucleus at anaphase (Figure 3A). In contrast, we did not observe similar dynamics with Yen1ON, which was prominently nuclear throughout the cell cycle (Figure 3B). These results show that Yen1ON is not only deregulated in terms of its biochemical activity, but also in its subcellular compartmentalization.
Figure 3.
Biochemical Activation and Subcellular Localization Are Independent Layers of Yen1 Regulation
(A) Nuclear localization of Yen1 throughout the cell cycle. Yen1-myc18 at different stages of the cell cycle was analyzed by immunofluorescence. Spindle morphology and DNA were visualized using anti-tubulin antibodies and DAPI. The contours of the cells, determined from differential interference contrast (DIC) images, are depicted by a dotted line.
(B) As in (A), but with Yen1ON.
(C) Subcellular localization of Yen1 phosphomutants. Nuclear enrichment (as opposed to diffuse pan-cellular staining) of the indicated Yen1 mutants was analyzed as in (A) and quantified (200 cells per condition). The schematic indicates the mutations at the Cdk sites in each allele. Protein domains are colored as in Figure 1A.
(D) Analysis of the biochemical activities of the Yen1 mutants used in (C). Myc9-tagged proteins were immunoprecipitated from S phase-arrested cells, and their nuclease activities were assayed.
(E) Suppression of the mus81Δ phenotype depends on both the biochemical activation and nuclear localization of Yen1. mus81Δ strains carrying the indicated YEN1 alleles were analyzed as in Figure 2B.
It was expected that the nuclear enrichment of Yen1ON would be a consequence of the mutational change at S679A. However, we found that Yen1 carrying a single change at this site, Yen1S679A, failed to fully recapitulate the localization pattern of Yen1ON (Figure 3C). These results indicate that additional phosphorylation events contribute to the compartmentalization of Yen1, and this concept was supported by observations with a Yen18A mutant (identical to Yen1ON, except for position S679). This mutant displayed a profile of nuclear enrichment remarkably similar to Yen1S679A, confirming that phosphorylation events at Cdk sites other than S679 contribute to the exclusion of Yen1 from the nucleus prior to the onset of anaphase.
Knowledge of the compartmentalization patterns of these mutants also allowed us to investigate whether biochemical activation and localization were interdependent events. IPs of myc-tagged Yen1, Yen1ON, Yen18A, Yen1S679A, or Yen1S679D from S phase cells revealed that Yen18A, but not Yen1S679A, exhibited deregulated HJ resolvase activity (Figure 3D), although both proteins showed similar levels of nuclear enrichment (Figure 3C). These results show that the biochemical activation of Yen1 is separable from nuclear relocalization. In accord with this proposal, only the activity-deregulated versions of Yen1 (Yen1ON and Yen18A) were effective in reducing the MMS sensitivity of mus81Δ mutants, as the Yen1S679A mutant failed to suppress the mus81Δ phenotype despite its nuclear enrichment (Figure 3E).
The “Yin” Cdk and “Yang” Cdc14 of Yen1 Regulation
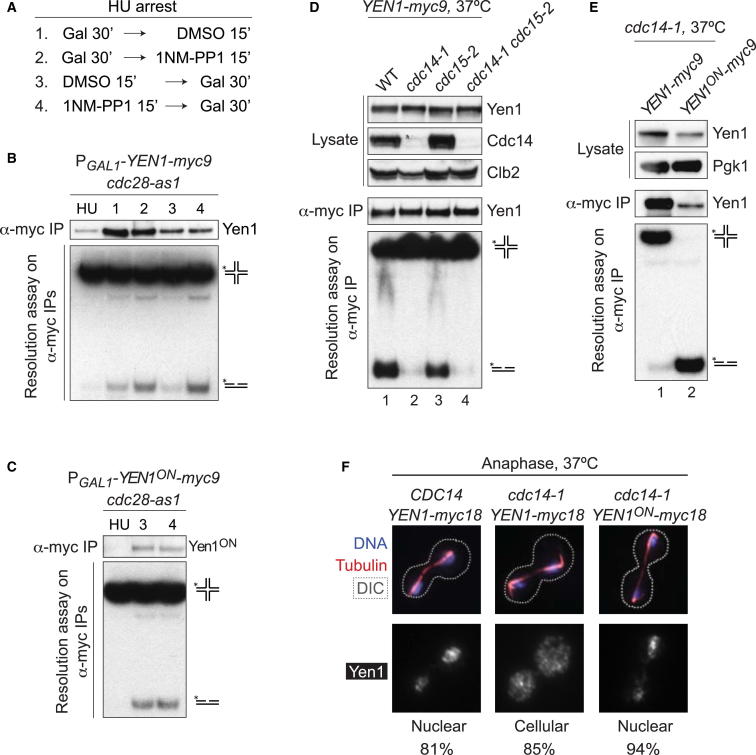
We have shown that the cycles of Yen1 activation/inactivation depend on its phosphorylation status and that its nuclease functions are inhibited at S phase (Matos et al., 2011). Yen1 is known to be a target of the cyclin-dependent kinase Cdk (Cdc28), and in particular of the S phase Cdk complex, Cdc28-Clb5 (Loog and Morgan, 2005; Ubersax et al., 2003). Because Yen1 localization also depends on Cdk phosphorylation, we determined whether Cdk-mediated phosphorylation was directly responsible for the inhibition of Yen1 activity during S phase. To do this, Yen1-myc9 was expressed under the control of a galactose-inducible promoter (PGAL1) in cells arrested in S phase that carried the cdc28-as1 allele, in which Cdk can be specifically inhibited by the addition of ATP-analog 1NM-PP1 (Bishop et al., 2000) (Figure 4A). When Yen1 expression was induced after Cdk inhibition, we observed significantly more HJ resolvase activity than in the absence of inhibitor (Figure 4B, compare lanes 3 and 4). A similar, although less efficient, boost in activity was observed when Yen1 was induced before Cdk inhibition (lanes 1 and 2). These results demonstrate that Cdk activity is responsible for the inhibition of Yen1 activity in S phase. Consistent with this notion, the activity of Yen1ON could not be further enhanced by Cdk inhibition (Figure 4C).
Figure 4.
Cdk and Cdc14 Phosphatase Regulate the Nuclease Activity and Subcellular Localization of Yen1
(A) Schematic representation of the experimental conditions employed in the Cdk inhibition experiments (B and C).
(B) Cdk inhibition in S phase cells increases Yen1 activity. cdc28-as1 cells carrying galactose-inducible YEN1-myc9 were treated with HU. To one-half of the culture, galactose was added before DMSO (lane 1) or the Cdc28-as1 kinase inhibitor 1NM-PP1 (lane 2). The other half was split, and galactose was added after DMSO (lane 3) or 1NM-PP1 (lane 4). Yen1-myc9 was immunoprecipitated and assayed for HJ resolution.
(C) Yen1ON activity is refractory to Cdk inhibition. As in (B), except that YEN1ON-myc9 was expressed after addition of DMSO (3) or 1NM-PP1 (4). The amounts of immunoprecipitated protein were reduced compared with (B) in order to limit DNA cleavage.
(D) cdc14-1 mutants fail to activate Yen1 at restrictive temperature. Yen1-myc9 IPs from the indicated strains (grown at 25°C and then shifted to 37°C for 2 hr) were tested for nuclease activity.
(E) Yen1ON activity is refractory to Cdc14 inactivation. As in (D), but employing cdc14-1 strains carrying either YEN1-myc9 or YEN1ON-myc9.
(F) Cdc14 is required for nuclear enrichment of Yen1 at anaphase. The immunofluorescence analysis of Yen1-myc18 or Yen1ON-myc18 during anaphase in CDC14 or cdc14-1 strains at the restrictive temperature was carried out as in Figure 3. The percentage of the cells displaying the most representative type of Yen1 distribution (nuclear or cellular) is shown. The contours of the cells, determined from DIC images, are depicted by a dotted line.
We next wanted to determine the identity of the phosphatase that reverses inhibitory phosphorylation and activates Yen1 at anaphase. Cdc14 phosphatase appeared to be a prime suspect for this reaction for several reasons: (1) Cdc14 is released from the nucleolus at the onset of anaphase (Shou et al., 1999; Visintin et al., 1999), at a time that coincides with Yen1 activation; (2) Cdc14 is responsible for the dephosphorylation of several Cdk substrates to allow exit from mitosis (Visintin et al., 1998); and (3) Cdc14 displays a preference for targets with phosphoserine over phosphothreonine in the Cdk consensus phosphorylation sites (9:0 for Yen1), and therefore the S-P to T-P ratio is, to some extent, predictive of Cdc14 targets (Bremmer et al., 2012). We therefore determined whether the absence of Cdc14 would result in a defect in the activation of Yen1. Because CDC14 is an essential gene, strains carrying the temperature sensitive allele cdc14-1 were generated. When switched to 37°C, the restrictive temperature at which Cdc14-1 is unstable, we observed a complete lack of activation of Yen1 (Figure 4D, compare lanes 1 and 2). However, analysis of DNA content by fluorescence-activated cell sorting (data not shown) and the abundance of Clb2 cyclin indicated that we were comparing a cycling population of wild-type cells with a population arrested in anaphase in cdc14-1 mutants. To eliminate the possibility of cell-cycle-dependent effects, the same experiment was carried out in a cdc15-2 background. CDC15 encodes an essential kinase and a component of the mitosis exit network (Bardin and Amon, 2001) such that cdc15-2 mutants arrest in late anaphase/telophase when switched to the restrictive temperature of 37°C. Under these conditions Yen1 exhibited a robust nuclease activity, whereas little Yen1 activity was observed in cdc14-1 cdc15-2 double mutants (Figure 4D, lanes 3 and 4). These results confirm that Cdc14 is responsible for the activation of Yen1 nuclease activity in anaphase. Moreover, in agreement with a direct role of Cdc14 in the dephosphorylation and activation of Yen1, we found that Yen1ON remained active when cdc14-1 mutants were switched to 37°C (Figure 4E).
Because both inactivation and nuclear exclusion of Yen1 are driven by Cdk activity, whereas its biochemical activation relies on Cdc14, we next determined whether the nuclear relocalization of Yen1 at anaphase also requires Cdc14. For this purpose, we analyzed the subcellular localization of Yen1 during anaphase both in CDC14 and cdc14-1 cells at the restrictive temperature of 37°C (Figure 4F). In a wild-type (CDC14) strain, more than 80% of anaphase cells presented a clear enrichment of Yen1 in the nucleus, whereas Yen1 staining was pan-cellular in most anaphases of cdc14-1 mutants (85%). Yen1ON was found to bypass the requirement for Cdc14 in re-establishing its nuclear localization, with 94% of the cells displaying nuclear enrichment of the protein after Cdc14 inactivation. These results show that Cdc14 promotes not only the activation of Yen1, but also its nuclear enrichment at anaphase.
Ectopic Expression of Cdc14 in S Phase Is Sufficient to Activate Yen1
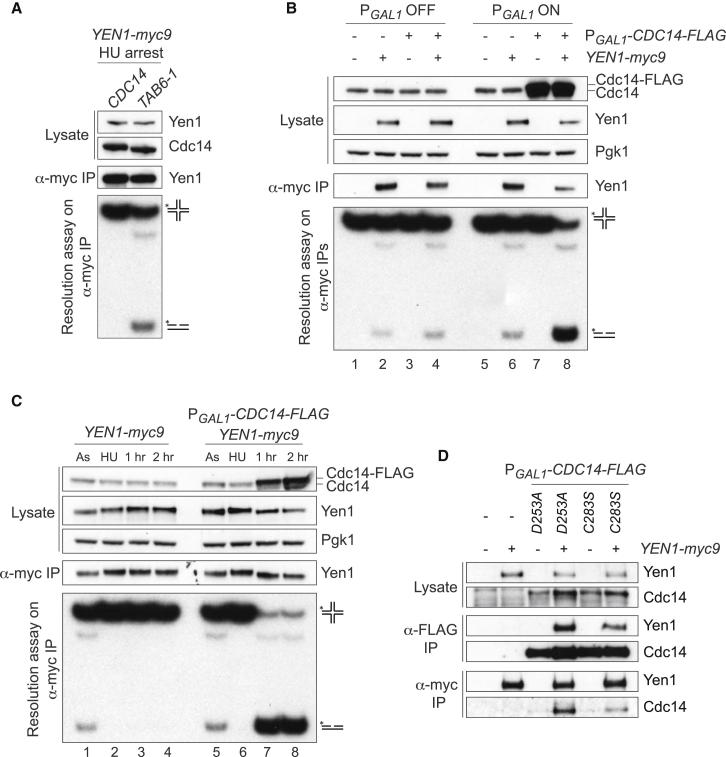
Because our results have shown that Cdc14 is required for Yen1 activation at anaphase, we determined whether inappropriate expression of Cdc14 at other stages of the cell cycle would suffice to activate Yen1. To do this, strains were employed in which the endogenous CDC14 locus was replaced by the TAB6-1 allele. This mutant displays reduced nucleolar sequestration of Cdc14 due to a mutation (Cdc14P116L) that impairs its interaction with Net1 (Shou et al., 2001). Our prediction was that if more Cdc14 could be made available in the nucleus, then the activity of Yen1 should also be increased. When we analyzed Yen1 in immunoprecipitates from CDC14 and TAB6-1 cells arrested in S phase (Figure 5A), we found that, whereas Yen1 activity was undetectable in wild-type cells, Yen1 from TAB6-1 mutants displayed elevated activity.
Figure 5.
Direct Activation of Yen1 by Cdc14 Phosphatase
(A) Analysis of Yen1 activity in TAB6-1 (CDC14P116L) mutants. Yen1-myc9 IPs from CDC14 and TAB6-1 strains treated with HU (enriched at the G1/S phase transition) were tested for nuclease activity.
(B) Overexpression of Cdc14 in cycling cells results in Yen1 activation. Cycling cultures of the indicated strains were supplemented with 2% galactose for 2 hr to induce Cdc14 expression. As controls, similar cultures were left uninduced. Lysates and IPs were analyzed as described in Figure 1A.
(C) Dephosphorylation and activation of Yen1 by ectopic overexpression of Cdc14 in HU-treated cells. The indicated strains were blocked at G1/S and Cdc14 expression was induced by adding 2% galactose for 2 hr. As, asynchronous culture.
(D) Physical interactions between Yen1 and Cdc14. PGAL1-CDC14 mutants were induced by addition of 0.5% galactose for 90 min. For each culture, lysates were prepared and split into two, and proteins were immunoprecipitated with either α-FLAG or α-myc. Proteins were analyzed by western blotting.
Further evidence to support our observations that Cdc14 is the phosphatase that activates Yen1 was provided by experiments in which Cdc14 was overexpressed from the GAL1 promoter. We found that Cdc14 overexpression resulted in the dephosphorylation of Yen1 leading to high levels of activity (Figure 5B, lane 8). However, because it is known that CDC14 overexpression results in a G1 arrest (Visintin et al., 1998), it was necessary to rule out cell-cycle-related effects. To do this, we overexpressed Cdc14 in S phase-arrested cells and found that it was sufficient to trigger a robust activation of Yen1 outside of anaphase (Figure 5C, lanes 7 and 8).
Finally, to determine whether Yen1 physically interacts with its activator, Cdc14, we took advantage of the Ccd14D253A and Cdc14C283S mutations, which inhibit the catalytic activity of Cdc14, but retain the ability to interact with its targets, effectively behaving as “substrate trapping” mutants (Bloom et al., 2011). We found that IPs of both Cdc14D253A and Cdc14C283S contained Yen1. Similarly, an IP of Yen1 contained both Cdc14D253A and Cdc14C283S (Figure 5D). These results demonstrate that physical interactions occur between Cdc14 and Yen1.
Mechanistic Basis of Yen1 Activation/Inactivation: Phosphorylation Modulates DNA Binding
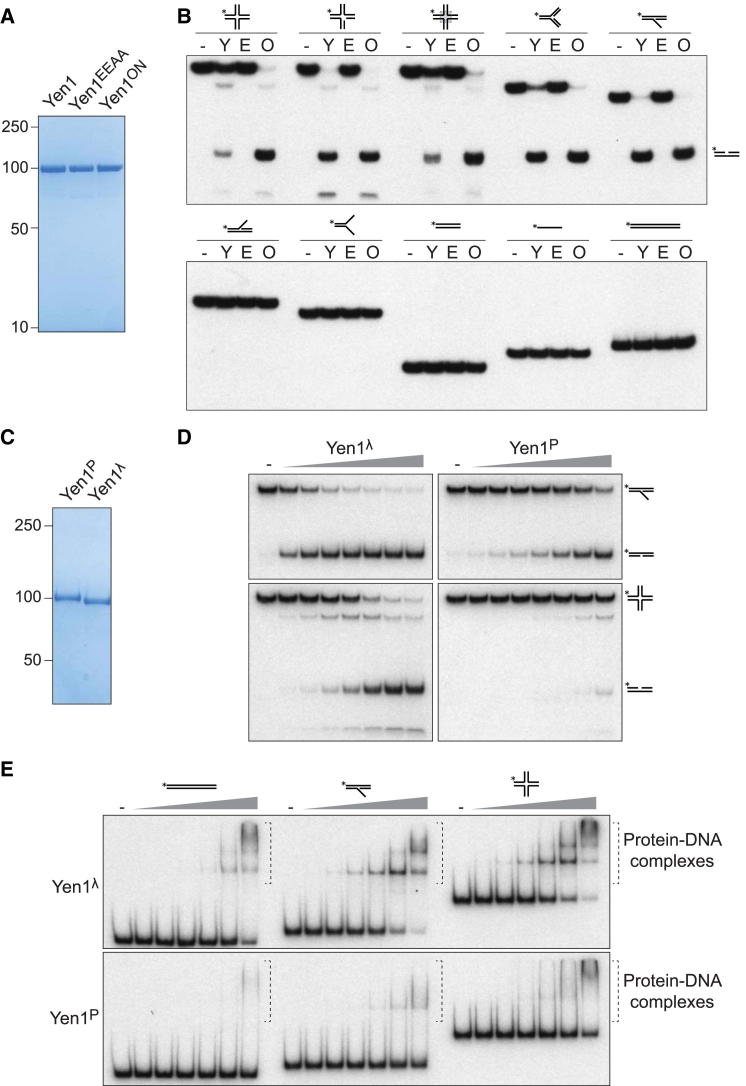
To understand how changes in the phosphorylation status of Yen1 modulate its nuclease activity, three versions of Yen1 were purified to homogeneity: wild-type Yen1 protein, the catalytically inactive Yen1EEAA mutant, and the phosphomutant Yen1ON (Figure 6A). Because all three proteins were purified from asynchronous cultures, they would contain a mixture of phosphorylated and nonphosphorylated protein. When equimolar amounts of Yen1 were compared with Yen1ON, we observed that Yen1ON exhibited much greater activity (Figure 6B). We did not, however, observe any changes to the substrate specificity of these two nucleases: both were active upon Holliday junctions (static, mobile, or nicked), replication fork structures, and 5′ flaps. The catalytic-dead mutant Yen1EEAA did not process any of these substrates. Neither Yen1 nor Yen1ON showed any activity toward 3′ flaps, splayed arm, double-stranded, or single-stranded DNA substrates, in agreement with the expected specificity of other members of the XPG family of nucleases (Lieber, 1997).
Figure 6.
Phosphorylation of Yen1 Modulates DNA Binding
(A) FTH-tagged Yen1, Yen1EEAA, and Yen1ON were purified from asynchronous cells and analyzed by SDS-PAGE and Coomassie staining (1 μg each).
(B) Yen1ON exhibits elevated nuclease activity. FTH-tagged Yen1 (Y), Yen1EEAA (E), and Yen1ON (O) (10 nM) were analyzed for nuclease activity using a static Holliday junction X0, nicked HJ X0, mobile HJ X26, replication fork, 5′-flap (upper panel); 3′-flap, splayed arm, double-stranded DNA (dsDNA) (60 bp), oligo dT60, and dsDNA (120 bp). (−): no protein.
(C) SDS-PAGE of purified Yen1P and Yen1λ (1 μg each). Yen1-FTH was expressed in cdc14-1 cells arrested at 37°C and purified without (Yen1P) or with (Yen1λ) λ-phosphatase treatment.
(D) Phosphorylated Yen1 exhibits reduced nuclease activity. Upper panels: increasing amounts of Yen1λ or Yen1P (0, 0.25, 0.5, 1, 2, 4, 8, 16 nM) were incubated with 5′-flap DNA. Lower panels: Yen1λ or Yen1P (0, 1, 2, 4, 8, 16, 32, 64 nM) with HJ X0.
(E) Yen1P exhibits reduced ability to bind DNA. DNA binding assays were carried out using dsDNA, 5′-flap, and X0 HJ DNA, with either Yen1λ (upper panel) or Yen1P (lower panel). Proteins were used at 0, 10, 20, 40, 80, 160, and 320 nM.
Additionally, Yen1 was purified in both fully phosphorylated (Yen1P) and dephosphorylated forms (Yen1λ), by employing cdc14-1 strains shifted to 37°C, in which half of the preparation was subjected to an additional λ phosphatase treatment step (Figure 6C). When increasing amounts of these two proteins were incubated with 5′-flap and HJ DNA (Figure 6D), we observed that the dephosphorylated version of Yen1 was approximately 8-fold more active than Yen1P. These results demonstrate that phosphorylation has a direct influence on the catalytic activity of Yen1.
To investigate the molecular basis for this difference in activity, we compared the ability of Yen1λ and Yen1P to bind DNA. Using electrophoretic mobility shift assays, carried out with 32P-labeled double-stranded, 5′-flap, and HJ DNAs, we observed that Yen1λ exhibited a significantly greater ability to bind DNA than phosphorylated Yen1P (Figure 6E, compare upper and lower panels). These results indicate that phosphorylation reduces the general affinity of Yen1 for its DNA substrates, which directly affects its catalytic activity.
Discussion
The careful regulation of activities that can process recombination intermediates is essential for proper chromosome segregation and the avoidance of genome instability. The primary mechanism to remove double Holliday junctions involves the STR complex, which promotes dissolution reactions leading to NCO formation. To ensure that this pathway is favored in S phase, the activities of the Holliday junction resolving nucleases, Mus81-Mms4 and Yen1, are held in check by cycles of phosphorylation and dephosphorylation. Recent work defined the mechanism by which Mus81-Mms4 is hyperactivated at G2/M by Cdk/Cdc5-mediated phosphorylation, to provide a first wave of Holliday junction resolution activity that captures and processes persistent junctions that have escaped the attention of STR (Dehé et al., 2013; Gallo-Fernández et al., 2012; Matos et al., 2011, 2013; Schwartz et al., 2012; Szakal and Branzei, 2013). Until now, however, little was known about the mechanism by which the second wave of HJ resolution, mediated by Yen1, might be regulated. Here, we have shown that Yen1 undergoes a dual mode of regulation, in which both its subcellular localization and nuclease activation are carefully controlled by changes to its phosphorylation status.
First, we found that phosphorylation drives the exclusion of Yen1 from the nucleus in S phase. Consistent with previous observations (Kosugi et al., 2009), we observed that phosphorylation of S679, a residue that overlaps the nuclear localization sequence, plays an important role in ensuring the cytoplasmic localization of Yen1 by impairing nuclear import. However, phosphoregulation of the NLS alone does not fully account for the localization of Yen1 because a single mutation at this site (Yen1S679A) was insufficient to promote nuclear localization to the same extent as Yen1ON (in which nine phosphorylation sites were mutated). In this regard, it is interesting to note that wild-type Yen1 shows strong nuclear enrichment in null mutants lacking the β-karyopherin Msn5 protein (Kosugi et al., 2009). As reported for other Msn5 cargos, phosphorylation might play an important role in enhancing the nuclear export of Yen1 (Kaffman et al., 1998). At anaphase, dephosphorylation of Yen1 S679 and other phosphoresidues restores NLS functionality and potentially impairs nuclear export, resulting in the rapid accumulation of Yen1 in the nucleus.
Second, we found that phosphorylation has a direct inhibitory effect on Yen1 nuclease activity. By purifying Yen1 proteins in their phosphorylated and nonphosphorylated states, we directly compared their DNA binding and nuclease activities and found that phosphorylation hinders the ability of Yen1 to process its substrates by a strikingly simple mechanism: inhibition of DNA binding, which directly impacts upon its catalytic activity. Although the precise molecular basis for the reduction of DNA binding imposed by phosphorylation is presently unknown, our observations indicate that Cdk sites located in the central region of Yen1 are important for S phase inactivation (Figure S1). We therefore propose two possible hypotheses: (1) that the central region might undergo a phosphorylation-dependent conformational change that influences DNA binding activity, or (2) that the central region itself is directly required for DNA interaction, or stability of the protein-DNA complex, and that its substrate affinity is reduced by the increased negative charge caused by phosphorylation.
The direct regulation of a nuclease activity by sequential cycles of phosphorylation and dephosphorylation is somewhat unusual. There are, however, suggestions that other members of the XPG-family of structure-selective nucleases, to which Yen1 belongs, are also negatively regulated by phosphorylation events. For instance, phosphorylation of yeast Exo1 and human EXO1 by the DNA damage checkpoint kinases downregulates their ability to promote DNA resection and leads to proteasomal degradation (El-Shemerly et al., 2005, 2008; Morin et al., 2008). The 5′-flap endonuclease FEN1 is also subject to phosphorylation, which affects its localization, degradation, nuclease activity, and ability to interact with PCNA (Guo et al., 2008, 2012; Henneke et al., 2003). Phosphorylation, however, does not affect FEN1’s affinity for DNA. Yen1 provides an example for the regulation of a structure-selective nuclease by the direct modification of its DNA binding properties through changes to its phosphorylation status.
So how are the two separate layers of control over Yen1 function enforced and coordinated with cell-cycle progression? Yen1 contains nine consensus Cdk sites and is known to be one of many Cdk substrates (Ubersax et al., 2003). Previous work and our own results indicate that the changes in Yen1 localization are dependent on Cdk-mediated phosphorylation (Kosugi et al., 2009). Moreover, we found that Cdk-mediated phosphorylation inhibits Yen1 nuclease activity in S phase. Because Yen1 is a specific target of the S phase Cdk complex, Cdc28-Clb5 (Loog and Morgan, 2005), the spatial and temporal inhibition of Yen1 activity might be one of the earliest events to occur as cells enter S phase.
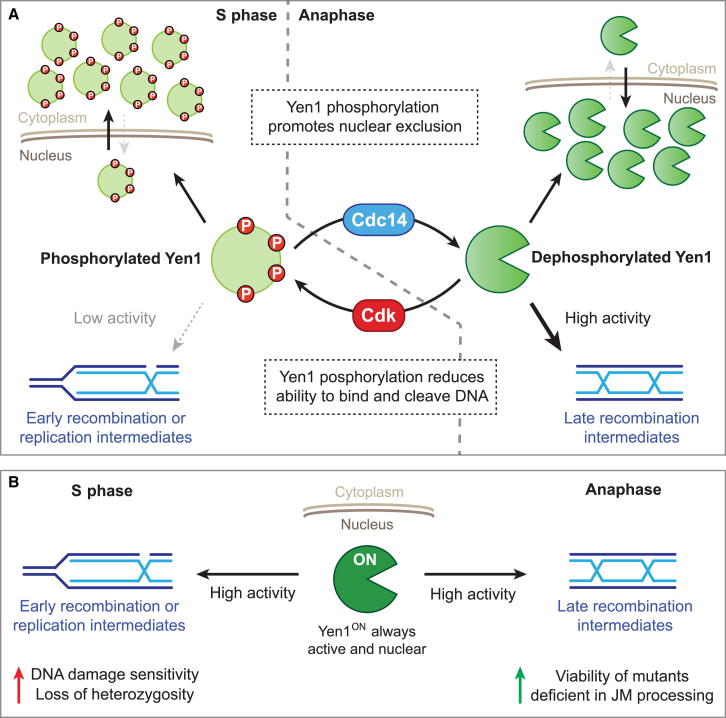
The dual inhibitory actions of Cdk-mediated phosphorylation of Yen1 are reversed by the mitotic exit phosphatase Cdc14, which is sequestered in the nucleolus throughout most of the cell cycle, but is released in two bursts at anaphase (Shou et al., 1999; Visintin et al., 1999). Cdc14 associates with Yen1 and promotes its dephosphorylation, triggering both its nuclear enrichment and increased DNA binding affinity. Therefore, whereas the two levels of control on Yen1 function are to some extent separable, they are interconnected through their common dependence on the master cell-cycle regulators Cdk and Cdc14, which determine the accurate timing of the second wave of joint molecule resolution. A model for the bimodal regulation of Yen1 by Cdk and Cdc14 is shown in Figure 7A.
Figure 7.
Regulatory Control of Yen1 by Cdk and Cdc14
(A) Cdk and Cdc14 control the phosphorylation status of Yen1. In S phase, Cdk phosphorylates Yen1 to (1) promote its nuclear exclusion by impairing its NLS function and, potentially, enhance Msn5-dependent export, and (2) reduce its DNA binding affinity, avoiding the cleavage of S phase-specific DNA structures such as replication or early recombination intermediates. At anaphase, Cdc14 dephosphorylates Yen1 to reinstate a fully functional NLS and potentially block Msn5-mediated export, leading to its nuclear accumulation. Additionally, the removal of phosphate groups increases the DNA binding affinity and catalytic activity of Yen1, allowing it to resolve HR intermediates that have persisted until anaphase. Red circles (P) depict phosphate groups.
(B) Yen1ON cannot be turned off or shuttled to the cytoplasm by Cdk. This may lead to the unscheduled and detrimental cleavage of replication or early recombination intermediates. In mutants that accumulate HR intermediates, the constitutive activation of Yen1ON provides an alternative way to process these potentially toxic DNA structures.
But why is it so critical to limit the actions of Yen1 to anaphase? The answer to this question was provided by our analysis of cells carrying constitutively active Yen1ON (Figure 7B). The nuclear exclusion and inhibition of Yen1 activity in S/G2 has two purposes. First, it restricts its HJ resolvase activity in order to facilitate dHJ processing by the STR complex, thus favoring NCO formation. In this regard, our results demonstrated that a deregulated form of Yen1 could compete for HR intermediates that are normally channeled toward NCOs and instead redirect them to COs, with a concomitant increase in LOH, despite the presence of the STR pathway. Interestingly, similar increases in CO frequency and LOH were observed following the deregulation of Mus81-Mms4 (Matos et al., 2013; Szakal and Branzei, 2013). Second, it ensures that the 5′ flap/fork endonuclease activity of Yen1 will not have access to DNA structures such as replication forks and/or early recombination intermediates that may arise in S/G2, because they may be susceptible to nucleolytic cleavage. In contrast to the deregulation of Mus81-Mms4, however, we found that constitutive activation of Yen1 led to a hypersensitivity to genotoxic stress. One possibility for this difference is the structure selectivity of Yen1 compared with Mus81-Mms4. For example, exposed replication forks might be cleaved on the leading strand by Mus81-Mms4, whereas Yen1 could promote lagging strand cleavage, with potentially different outcomes for repair.
Taken together, our results indicate that cells reserve the efficient, but potentially dangerous, activity of Yen1 until anaphase, when it is unleashed as a last resort to remove persistent HR intermediates that could interfere with chromosome segregation. Cdk and Cdc14 enforce this deliberate delay of the second wave of JM resolution in order to prevent the inappropriate actions of Yen1 in S phase, which could lead to genome instability.
Experimental Procedures
All experimental procedures are described in detail in Supplemental Information.
Yeast Strains and Cultures
All strains employed are described in Table S2. Synchronous release of mitotic cultures and DNA damage sensitivity assays were carried out as described (Blanco et al., 2010). S phase arrests were performed by addition of HU to 150 mM, followed by 2–2.5 hr incubation.
Protein Analysis and Nuclease Assays on Beads
TCA lysates, soluble lysates, and affinity-purified proteins were analyzed by SDS-PAGE followed by western blotting. Myc-tagged Yen1 was detected using an α-myc antibody. For nuclease assays on beads, Yen1-myc9 was immunoaffinity purified, and the beads were washed extensively and supplemented with 10 μl of resolution buffer (50 mM Tris-HCl [pH 7.5], 1 mM MgCl2) containing ∼0.5 nM 5′-32P-end-labeled synthetic Holliday junction (Ip et al., 2008). Reactions were typically incubated for 15–20 min at 30°C and stopped by SDS/proteinase K deproteinization. Radiolabeled products were separated by PAGE and analyzed by autoradiography or phosphorimaging.
Genetic Analysis of Recombination
Analysis of recombination outcome during mitotic DSB repair was performed as described (Ho et al., 2010).
Protein Purification and Biochemical Assays
Yen1-FTH (Yen1-3xFLAG-2xTEV-10xHis) proteins were purified from yeast lysates by two consecutive rounds of affinity chromatography. For the generation of dephosphorylated Yen1-FTH, protein preps were treated with λ phosphatase as part of the purification scheme. Nuclease assays were carried out using purified Yen1 proteins and 10 nM unlabeled DNA (supplemented with ∼0.2 nM 5′-32P-end-labeled DNA) in 50 mM Tris-HCl (pH 7.5), 1 mM MgCl2, 50 mM NaCl, and 2% glycerol. DNA binding assays were carried out on ice by mixing Yen1 with DNA in binding buffer (50 mM Tris-HCl [pH 7.5], 125 mM NaCl, 5 mM EDTA, 1 mM DTT, 100 μg/ml BSA, 11% glycerol, and 2 ng/μl duplex poly dIdC). Protein-DNA complexes were separated by 5% PAGE, and radiolabeled DNAs were visualized. Radiolabeled products were analyzed by autoradiography.
Acknowledgments
We thank John Diffley, David Morgan, Pedro San-Segundo, Francesca Storici, Michael Resnick, Lorraine Symington, and Wolfgang Zachariae for yeast strains, plasmids, and antibodies; members of the West lab, and Fernando Monje-Casas for helpful comments and discussions; John Diffley for codon optimization of YEN1; and Sarah Maslen and Mark Skehel for the phosphomapping of Yen1. We are also grateful to Mark Hall for communication of data prior to publication. This work was supported by Cancer Research UK, the European Research Council, the Louis-Jeantet Foundation, the Swiss Bridge Foundation, and the Breast Cancer Campaign. J.M. was a recipient of a Fellowship from the Human Frontiers Science Program.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Supplemental Information
References
- Bardin A.J., Amon A. Men and sin: what’s the difference? Nat. Rev. Mol. Cell Biol. 2001;2:815–826. doi: 10.1038/35099020. [DOI] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Blanco M.G., Matos J., Rass U., Ip S.C.Y., West S.C. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair (Amst.) 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Bloom J., Cristea I.M., Procko A.L., Lubkov V., Chait B.T., Snyder M., Cross F.R. Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J. Biol. Chem. 2011;286:5434–5445. doi: 10.1074/jbc.M110.205054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddy M.N., Gaillard P.H.L., McDonald W.H., Shanahan P., Yates J.R., 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- Bremmer S.C., Hall H., Martinez J.S., Eissler C.L., Hinrichsen T.H., Rossie S., Parker L.L., Hall M.C., Charbonneau H. Cdc14 phosphatases preferentially dephosphorylate a subset of cyclin-dependent kinase (Cdk) sites containing phosphoserine. J. Biol. Chem. 2012;287:1662–1669. doi: 10.1074/jbc.M111.281105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bzymek M., Thayer N.H., Oh S.D., Kleckner N., Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castor D., Nair N., Déclais A.C., Lachaud C., Toth R., Macartney T.J., Lilley D.M.J., Arthur J.S., Rouse J. Cooperative control of holliday junction resolution and DNA repair by the SLX1 and MUS81-EME1 nucleases. Mol. Cell. 2013;52:221–233. doi: 10.1016/j.molcel.2013.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P., Plank J.L., Bachrati C.Z., Hickson I.D., Kowalczykowski S.C. Rmi1 stimulates decatenation of double Holliday junctions during dissolution by Sgs1-Top3. Nat. Struct. Mol. Biol. 2010;17:1377–1382. doi: 10.1038/nsmb.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.B., Melchionna R., Denis C.M., Gaillard P.H.L., Blasina A., Van de Weyer I., Boddy M.N., Russell P., Vialard J., McGowan C.H. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- Dayani Y., Simchen G., Lichten M. Meiotic recombination intermediates are resolved with minimal crossover formation during return-to-growth, an analogue of the mitotic cell cycle. PLoS Genet. 2011;7:e1002083. doi: 10.1371/journal.pgen.1002083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehé P.-M., Coulon S., Scaglione S., Shanahan P., Takedachi A., Wohlschlegel J.A., Yates J.R., 3rd, Llorente B., Russell P., Gaillard P.-H. Regulation of Mus81-Eme1 Holliday junction resolvase in response to DNA damage. Nat. Struct. Mol. Biol. 2013;20:598–603. doi: 10.1038/nsmb.2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen K.T., Heyer W.D. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2008;36:2182–2195. doi: 10.1093/nar/gkm1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shemerly M., Janscak P., Hess D., Jiricny J., Ferrari S.R. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 2005;65:3604–3609. doi: 10.1158/0008-5472.CAN-04-4069. [DOI] [PubMed] [Google Scholar]
- El-Shemerly M., Hess D., Pyakurel A.K., Moselhy S., Ferrari S.R. ATR-dependent pathways control hEXO1 stability in response to stalled forks. Nucleic Acids Res. 2008;36:511–519. doi: 10.1093/nar/gkm1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F., Chan A., Heyer W.D., Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc. Natl. Acad. Sci. USA. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo-Fernández M., Saugar I., Ortiz-Bazán M.A., Vázquez M.V., Tercero J.A. Cell cycle-dependent regulation of the nuclease activity of Mus81-Eme1/Mms4. Nucleic Acids Res. 2012;40:8325–8335. doi: 10.1093/nar/gks599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner E., Kim Y., Lach F.P., Kottemann M.C., Smogorzewska A. Human GEN1 and the SLX4-associated nucleases MUS81 and SLX1 are essential for the resolution of replication-induced Holliday junctions. Cell Rep. 2013;5:207–215. doi: 10.1016/j.celrep.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Qian L.M., Liu R., Dai H.F., Zhou M., Zheng L., Shen B. Nucleolar localization and dynamic roles of flap endonuclease 1 in ribosomal DNA replication and damage repair. Mol. Cell. Biol. 2008;28:4310–4319. doi: 10.1128/MCB.00200-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Kanjanapangka J., Liu N., Liu S., Liu C., Wu Z., Wang Y., Loh T., Kowolik C., Jamsen J. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol. Cell. 2012;47:444–456. doi: 10.1016/j.molcel.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke G., Friedrich-Heineken E., Hübscher U. Flap endonuclease 1: a novel tumour suppresser protein. Trends Biochem. Sci. 2003;28:384–390. doi: 10.1016/S0968-0004(03)00138-5. [DOI] [PubMed] [Google Scholar]
- Ho C.K., Mazón G., Lam A.F., Symington L.S. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Interthal H., Heyer W.D. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Ip S.C.Y., Rass U., Blanco M.G., Flynn H.R., Skehel J.M., West S.C. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- Ira G., Malkova A., Liberi G., Foiani M., Haber J.E. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Rank N.M., O’Neill E.M., Huang L.S., O’Shea E.K. The receptor Msn5 exports the phosphorylated transcription factor Pho4 out of the nucleus. Nature. 1998;396:482–486. doi: 10.1038/24898. [DOI] [PubMed] [Google Scholar]
- Kaliraman V., Mullen J.R., Fricke W.M., Bastin-Shanower S.A., Brill S.J. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Tomita M., Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA. 2009;106:10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber M.R. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- Loog M., Morgan D.O. Cyclin specificity in the phosphorylation of cyclin-dependent kinase substrates. Nature. 2005;434:104–108. doi: 10.1038/nature03329. [DOI] [PubMed] [Google Scholar]
- Luo G.B., Santoro I.M., McDaniel L.D., Nishijima I., Mills M., Youssoufian H., Vogel H., Schultz R.A., Bradley A. Cancer predisposition caused by elevated mitotic recombination in Bloom mice. Nat. Genet. 2000;26:424–429. doi: 10.1038/82548. [DOI] [PubMed] [Google Scholar]
- Matos J., Blanco M.G., Maslen S.L., Skehel J.M., West S.C. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell. 2011;147:158–172. doi: 10.1016/j.cell.2011.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matos J., Blanco M.G., West S.C. Cell-cycle kinases coordinate the resolution of recombination intermediates with chromosome segregation. Cell Rep. 2013;4:76–86. doi: 10.1016/j.celrep.2013.05.039. [DOI] [PubMed] [Google Scholar]
- Morin I., Ngo H.P., Greenall A., Zubko M.K., Morrice N., Lydall D. Checkpoint-dependent phosphorylation of Exo1 modulates the DNA damage response. EMBO J. 2008;27:2400–2410. doi: 10.1038/emboj.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U., Compton S.A., Matos J., Singleton M.R., Ip S.C.Y., Blanco M.G., Griffith J.D., West S.C. Mechanism of Holliday junction resolution by the human GEN1 protein. Genes Dev. 2010;24:1559–1569. doi: 10.1101/gad.585310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz E.K., Wright W.D., Ehmsen K.T., Evans J.E., Stahlberg H., Heyer W.D. Mus81-Mms4 functions as a single heterodimer to cleave nicked intermediates in recombinational DNA repair. Mol. Cell. Biol. 2012;32:3065–3080. doi: 10.1128/MCB.00547-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W.S., Jang J., Shevchenko A., Charbonneau H., Deshaies R.J. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Shou W., Sakamoto K.M., Keener J., Morimoto K.W., Traverso E.E., Azzam R., Hoppe G.J., Feldman R.M., DeModena J., Moazed D. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit. Mol. Cell. 2001;8:45–55. doi: 10.1016/s1097-2765(01)00291-x. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Szakal B., Branzei D. Premature Cdk1/Cdc5/Mus81 pathway activation induces aberrant replication and deleterious crossover. EMBO J. 2013;32:1155–1167. doi: 10.1038/emboj.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., Morgan D.O. Targets of the cyclin-dependent kinase Cdk1. Nature. 2003;425:859–864. doi: 10.1038/nature02062. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M.D., Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Wechsler T., Newman S., West S.C. Aberrant chromosome morphology in human cells defective for Holliday junction resolution. Nature. 2011;471:642–646. doi: 10.1038/nature09790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S.C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Wyatt H.D.M., Sarbajna S., Matos J., West S.C. Coordinated actions of SLX1-SLX4 and MUS81-EME1 for Holliday junction resolution in human cells. Mol. Cell. 2013;52:234–247. doi: 10.1016/j.molcel.2013.08.035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.