Summary
To decipher the molecular basis for RET kinase activation and oncogenic deregulation, we defined the temporal sequence of RET autophosphorylation by label-free quantitative mass spectrometry. Early autophosphorylation sites map to regions flanking the kinase domain core, while sites within the activation loop only form at later time points. Comparison with oncogenic RET kinase revealed that late autophosphorylation sites become phosphorylated much earlier than wild-type RET, which is due to a combination of an enhanced enzymatic activity, increased ATP affinity, and surprisingly, by providing a better intermolecular substrate. Structural analysis of oncogenic M918T and wild-type RET kinase domains reveal a cis-inhibitory mechanism involving tethering contacts between the glycine-rich loop, activation loop, and αC-helix. Tether mutations only affected substrate presentation but perturbed the autophosphorylation trajectory similar to oncogenic mutations. This study reveals an unappreciated role for oncogenic RET kinase mutations in promoting intermolecular autophosphorylation by enhancing substrate presentation.
Graphical Abstract
Highlights
-
•
Kinetics of RET autophosphorylation identify early and late autophosphorylation sites
-
•
Evidence for a RET kinase domain cis-inhibitory tether is presented
-
•
Oncogenic RET subverts cis-inhibition and perturbs the autophosphorylation trajectory
-
•
Oncogenic RET kinase is overactive and a better trans-autophosphorylation substrate
Activation of receptor tyrosine kinases by oncogenic mutations frequently perturbs intrinsic cis-inhibitory mechanisms and promotes kinase activation loop phosphorylation. Here, Plaza-Menacho et al. reveal how a RET receptor tyrosine kinase oncogenic mutation alters its autophosphorylation trajectory not only by increasing nucleotide-affinity and kinase activity but also by producing a better substrate in trans.
Introduction
RET is a receptor tyrosine kinase (RTK) expressed and required during early development for the formation of neural crest-derived lineages and kidney organogenesis (Arighi et al., 2005; Plaza-Menacho et al., 2006). It has been reported, based on cell-based experimental models, that upon ligand and coreceptor binding to the extracellular domain, the catalytic and signaling activity of RET kinase are controlled by the autophosphorylation (autoP) of several tyrosine residues within the cytoplasmic domain (Airaksinen et al., 1999; Plaza-Menacho et al., 2006). For other RTK paradigms such as the insulin receptor and FGFR2, ligand-dependent activation leads to autoP of specific tyrosine residues to both relieve repressive cis-autoinhibitory interactions and to promote binding of PTB- and SH2-domain containing proteins that trigger downstream signaling (Chen et al., 2007; Hubbard, 1997). While the latter role for autoP has been demonstrated for RET, the effect of autoP on catalytic activation is less clear. In vitro, activation loop (AL) phosphorylation has little effect on catalytic activity (Knowles et al., 2006; Plaza-Menacho et al., 2011). A similar situation is found for the EGFR and nonreceptor tyrosine kinase ACK1 (Lougheed et al., 2004; Zhang et al., 2006). In these cases, allosteric mechanisms have been identified to stimulate receptor activation independent of AL phosphorylation status.
The canonical RET-AL between the DFG and APE motifs, common to RTKs, contains two tyrosine residues, Y900 and Y905, required for signaling in cells (Kawamoto et al., 2004). Many RTKs such as the FGFR1 and FGFR2 have two tyrosines in this region, whose autoP is carefully orchestrated leading to a precise and sequential order of tyrosine phosphorylation (Chen et al., 2007, 2008; Furdui et al., 2006; Lew et al., 2009). For FGFR1, a three-stage mechanism serves to initially stimulate the intrinsic tyrosine kinase activity and subsequently target tyrosine residues both within and outside of the kinase domain (KD) (Chen et al., 2008; Furdui et al., 2006; Lew et al., 2009). There are few, if any, studies examining the sequence of RET autoP (Coulpier et al., 2002). Exhaustive phosphopeptide mapping and western blots (WB) with antiphosphospecific tyrosine antibodies have identified many of the major autoP sites but crucially temporal information is lacking (Kawamoto et al., 2004). An alternative way to follow the order of RET autoP (without requiring a battery of phosphospecific antibodies) is to use label-free quantitative mass spectrometry (LFQMS). This approach can simultaneously monitor all tyrosine phosphorylation sites by employing more sensitive fast scanning mass spectrometers that acquire higher mass accuracy data in tandem with HPLC hardware and software now available (Cutillas et al., 2005; Steen et al., 2006). In label-free quantitation, peptide ion abundances are measured from extracted ion chromatograms and the relative abundance of integrated ion signals between independent mass spectometry (MS) runs is compared.
Germline missense activating point mutations in RET cause the cancer syndrome multiple endocrine neoplasia type 2 (MEN2). How these point mutations, scattered throughout the RET KD and elsewhere, impact on and subvert intrinsic activation mechanisms for RET is still not clear (Plaza-Menacho et al., 2006). Equally, it is not known whether such RET mutations impact on the sequential order of autoP as has been shown for pathogenic mutations in FGFR1 and FGFR2 (Chen et al., 2008; Lew et al., 2009).
Here, we combine quantitative proteomics, biochemical analysis, and X-ray crystallography to define the temporal sequence of RET autoP (trajectory) and identify intrinsic repressive interactions via a cluster of residues stabilizing an autoinhibited form of the kinase. In addition, perturbation of the autoP trajectory by oncogenic RET kinase mutants uncovered both an enhanced RET kinase substrate presentation in trans and ATP occupancy in cis as molecular determinants of oncogenic RET kinase activation.
Results
Early RET AutoP Sites Map to Flanking Regions outside the KD Core
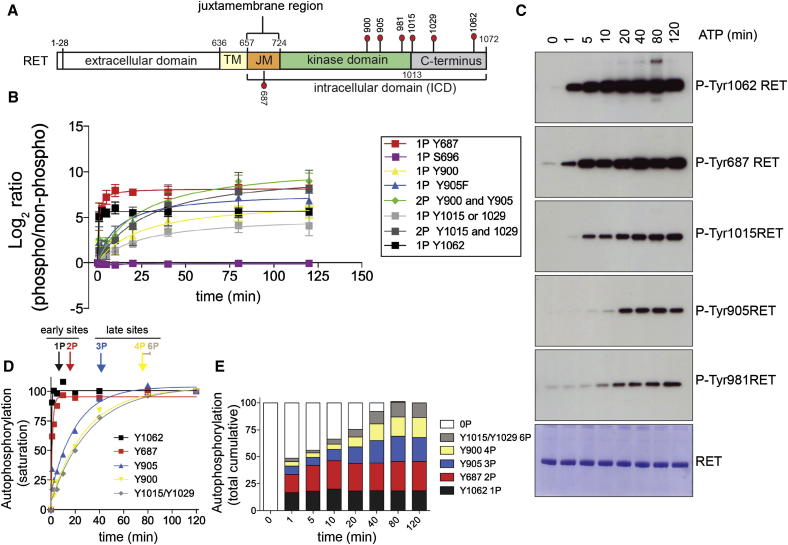
To define the precise temporal sequence of RET autoP, we used purified recombinant RET intracellular domain (ICD; residues 661–1072; Figure 1A) expressed in Sf9 insect cells and performed in vitro time course autoP assays using saturating concentrations of ATP (5 mM) and MgCl2 (10 mM) for 0–120 min. Monomeric status and kinase activity of purified soluble protein were confirmed by size exclusion chromatography and enzyme kinetics experiments using poly-E4Y peptide (Figure S1 available online). The temporal sequence of RET autoP was assessed by LFQMS (Figure 1B). This powerful technique has a higher dynamic range than isotope labeling methods and allows us to potentially compare multiple phosphorylation sites and samples (Cutillas et al., 2005; Steen et al., 2006). We identified seven tyrosines: Y687, Y826, Y900, Y905, Y1015, Y1029, and Y1062 that, upon KD activation, became efficiently phosphorylated in a time-dependent fashion (Figure 1B). Y826, which appeared to be weakly phosphorylated upon KD activation but did not change significantly with time was excluded from further analysis. Signal log2 ratios of phosphorylated peptides standardized to their nonphosphorylated counterparts were plotted relative to zero time point (Figure 1B). Global autoP analysis revealed that sites located at the flanking regions were phosphorylated first; in particular the C-terminal Y1062 followed by the juxtamembrane Y687 were phosphorylated more rapidly than sites located within the KD core such us AL Y900 and Y905 (Figures 1B and 1C). A closer examination of the temporal sequence of RET autoP showed that Y1062 and Y687 followed very fast saturation kinetics, taking between 1–5 min to reach a 25-fold and 50-fold increase in overall phosphorylation levels, respectively. This contrasted with AL Y900 and Y905 sites, which needed 40–80 min to reach saturation at ∼25-fold and 35-fold increased phosphorylation levels, respectively (Figures 1B and 1D). Interestingly, we observed that the double phosphorylated peptide containing both Y900 and Y905 resulted in higher phosphorylation levels (45-fold increase) together with faster saturation than the single phosphorylated peptides. Phosphorylation of Y1015 and Y1029 followed very similar kinetics to AL tyrosines, with the exception of the single phosphorylated peptides, which were slightly slower. Despite very rapid saturation kinetics for Y1062 and Y687 (Figure 1D), fully phosphorylated RET was found only at late time points, (95% phosphorylated by 40 min and 100% by 80 min), suggesting slow allosteric inputs in the autoP process (Figure 1E). We validated the results from the LFQMS by WB analysis using available phosphospecific antibodies (Figure 1C). Interestingly, another tyrosine site within the KD core Y981, which was not identified by LFQMS, showed the same tendency as Y905. We also tested a phosphospecific antibody against Y1015 and observed faster kinetics of autoP compared to the quantitative mass spectrometry data. An inspection of this particular peptide sequence analyzed by LFQMS indicated it also contained Y1029. We could not explain this discrepancy at this stage, as the LFQMS data would not discriminate between the two sites in the single phosphorylated peptide (see Figure 2). Overall, the definition of the temporal sequence of RET autoP (i.e., trajectory) identifies early sites flanking the RET KD core (e.g., Y1062 and Y687) and late sites within the AL (e.g., Y900 and Y905) and KD consistent with an autoP trajectory shown in Figure 1D.
Figure 1.
Early RET AutoP Sites Map to Regions outside the KD Core
(A) Schematic diagram of discrete RET functional domains together with the autoP sites analyzed in this study.
(B) Global temporal autoP analysis by LFQMS of recombinant purified RET ICD (2.5 μM) treated with saturating concentrations of ATP (5 mM) and MgCl2 (10 mM) for 0–120 min. Data represent the mean signal value (Log2 ratios of phosphorylated peptides standardized to the nonphosphorylated counterparts) ± SE of four independent experiments (n = 4) performed with different protein preparations.
(C) WB analyses of samples analyzed in (B) using the indicated antibodies.
(D) Phosphorylation status (saturation rate, percentage) for each LFQMS-identified autoP site and phosphorylation state (1P to 6P) from (B).
(E) Cumulative phosphorylation for each autoP site from (D).
See also Figure S1.
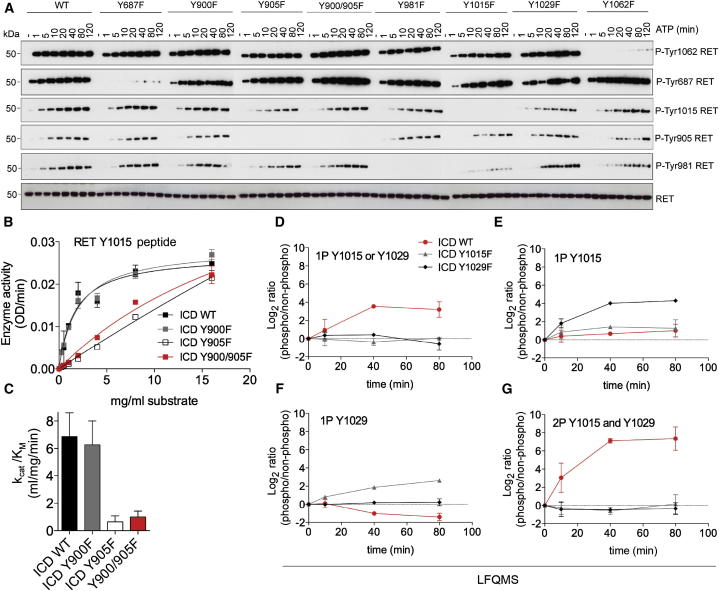
Figure 2.
Phosphoacceptor Site Mutants Display Minimal Changes in the Pattern of RET AutoP
(A) WB analyses of recombinant purified RET ICD (2.5 μM) WT and the indicated Y to F mutants treated with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min using the indicated antibodies.
(B) Enzymatic assay performed with RET ICD WT, Y900F, Y905F, or the double mutant Y900/905F incubated with increasing concentrations of the RET Y1015 KRRDYLDLAS peptide. Data shown represent the mean (OD/min) ± SE, n = 4.
(C) Catalytic efficiency constants (kcat/KM) of (B).
(D–G) Dissection of Y1015 and Y1029 autoP by LFQMS analysis of recombinant purified RET ICD WT and Y1015F or Y1029F mutants (2.5 μM) treated with ATP (5 mM) and MgCl2 (10 mM). Data represent the mean signal value (Log2 ratios of phosphorylated peptides standardized to the nonphosphorylated counterparts) ± SE, n = 4.
To confirm that RET autoP sites generated upon KD activation came from an intermolecular rather than intramolecular mechanism, time course autoP assays were performed with increasing concentrations of RET ICD (1 μM, 2.5 μM, and 5 μM, respectively). WB analysis using total and phospho-specific tyrosine antibodies demonstrated that the degree of autoP was directly proportional to the concentration of protein employed in the reaction indicating autoP in trans (Figure S1).
Primary Sequence Flanking the Phosphoacceptor Tyrosine Does Not Determine the Temporal Sequence of RET AutoP
We compared the consensus sequence of RET autoP sites obtained by LFQMS, including also Y981 (Figure S2), with that derived from an optimal RET KD peptide substrate (Plaza-Menacho et al., 2011). In addition, enzymatic assays were performed with RET ICD in both phosphorylated and nonphosphorylated states against peptides derived from the different autoP sites, and catalytic efficiency constants (kcat/KM) were determined (Figure S2). There was poor agreement between these data, from which we concluded that the precise sequence of RET autoP is not determined by primary sequence flanking the phosphoacceptor tyrosine. Instead, RET may recognize other structural features or conformations present within an intact RET receptor that may be distal to the phosphoacceptor tyrosine. Alternatively, accessibility of the autoP tyrosines may be controlled by conformational changes within RET ICD following KD activation.
RET AL Y900 and Y905 Are Not De Facto Activating Tyrosines
In the well-established RTK paradigm, autoP of tyrosine residues in the AL enhances catalytic activity and precedes the generation of further phosphotyrosines sites within and outside the KD core. These late autoP sites then serve as a platform for docking and effectors proteins to transmit downstream signaling (Furdui et al., 2006; Hubbard, 1997; Lew et al., 2009; McTigue et al., 1999). However, our LFQMS data demonstrate that Y900 and Y905 should not be considered “activating,” as their phosphorylation display delayed kinetics (Figure 1) and they are neither required for RET KD enzymatic activity (Knowles et al., 2006) nor autoP (Plaza-Menacho et al., 2011). To test this further, we analyzed the impact of single mutations Y900F and Y905F and the double mutant Y900/905F on the timing of RET ICD autoP (Figure 2A). Surprisingly, no significant effects on the levels and kinetics of autoP were observed for either individual point mutant or the double mutation compared to the wild-type (WT) protein (Figure 2A). This is an interesting finding when compared to a double mutant Y900/Y905 in the context of the RET KD core, which displayed severe defects in the autoP levels (Figure S3). We then conducted enzyme kinetic experiments using a RET peptide derived from the sequence containing the Y1015 autoP site (Figure 2B). Against this peptide substrate, Y905, but not Y900, was required for efficient RET kinase enzymatic activity; in contrast to the data from the autoP assay where neither residue was required for efficient RET autoP. These data suggest that (1) peptides substrates are poor surrogates to assess RET autoP, and (2) regions flanking the RET KD core influence kinase activity and AL conformation, and deletion of these regions exposes a dependency on AL phosphorylation lacking in the RET ICD (see Figure S3).
RET ICD Lacking Individual AutoP Sites Is Functional and Displays Small Changes in the Kinetics of AutoP
In order to assess the effect of each autoP site on the kinetics and levels of RET autoP, we also generated baculoviruses expressing RET ICD with the following mutants: Y687F, Y981F, Y1015F, Y1029F, and Y1062F. Time course autoP assays did not reveal significant changes in global terms of either specific or total levels of autoP (Figure 2A). As a control experiment, we tested a catalytically impaired RET ICD K758M mutant. This mutant could not undergo autoP upon ATP stimulation; however, when employed as a substrate, it could be efficiently phosphorylated (autoP “rescue”) by a shorter version of RET comprising the KD core (Figure S3).
Our initial LFQMS analysis did not discriminate between Y1015 or Y1029 phosphorylation in the monophosphorylated (1P) state of the peptide because of their proximity. The discrepancy between WB analysis using a phosphospecific Y1015 antibody and the mass spectrometry data, led us to investigate further the temporal sequence of autoP for these two specific sites. Using LFQMS we compared the kinetics of autoP of RET ICD WT, Y1015F and Y1029F (Figures 2D–2G). When the 1P phosphorylation status (regardless of which of the two sites) was analyzed, we observed that both sites are required to reach the optimal 1P state (Figure 2D). Both sites are also required to achieve optimal di-phosphorylation (2P) status (Figure 2G). Comparison of the kinetics and degree of autoP (Figures 2E and 2F) demonstrated that Y1015 phosphorylation was an earlier event (16-fold increased at 10 min) compared to Y1029 phosphorylation, which reached (16-fold increase between 40 and 80 min). These data demonstrate that Y1015 autoP precedes Y1029, despite both being required for full autoP. These data corroborate the results obtained by WB analysis where we observed phosphorylation of Y1015 at earlier stages than the LFQMS results predicted.
Oncogenic Mutations in the RET KD Perturb the AutoP Trajectory
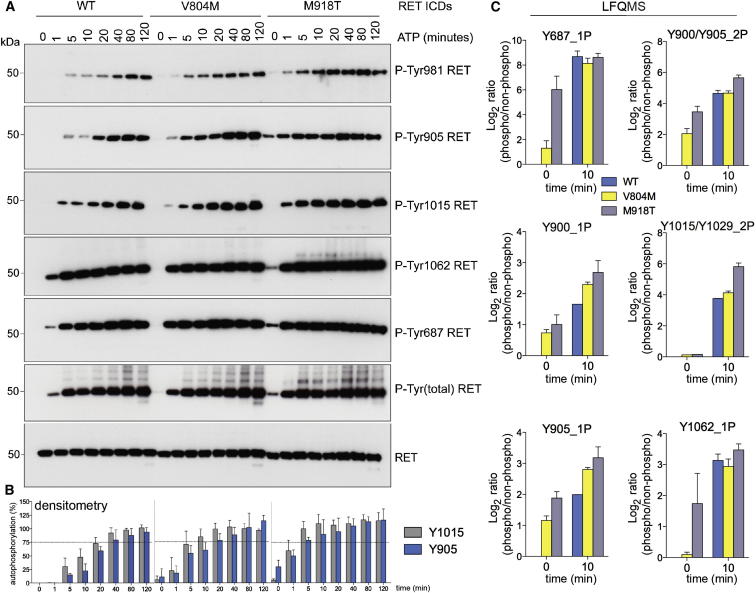
Having uncovered the precise temporal sequence of RET WT autoP, we hypothesized that oncogenic mutants targeting the RET KD, in particular M918T, may alter the kinetics of autoP by impacting on the conformation of the kinase. We therefore engineered baculoviruses expressing two oncogenic mutants targeting the KD, V804M a drug-resistant gatekeeper mutation associated with FMTC and MEN2A, (Gibelin et al., 2004; Plaza-Menacho et al., 2007a) and the most phenotypically aggressive MEN2B-associated M918T (Santoro et al., 1995). M918 maps to region beneath the AL referred to as the P+1 substrate-binding pocket (Hubbard, 1997). Time courses of RET autoP for the V804M and M918T mutants revealed much faster autoP kinetics for both oncogenic mutants compared to the native protein. In particular, late autoP sites within the KD core of RET WT (Y900, Y905, and Y981) become phosphorylated much earlier (Figures 3A and 3B). Likewise, Y1015 displayed faster kinetics and higher levels of autoP for the oncogenic mutants compared with the native protein. Early autoP sites Y1062 and Y687, displayed even faster kinetics of autoP compared to WT, consistent with an elevated enzymatic activity for both mutants (Figure 3A). Interestingly, RET M918T displayed some degree of phosphorylation at specific sites at zero time point, indicating that this oncogenic mutant overcomes endogenous insect cell tyrosine phosphatases. Alternatively, this mutant may overcome cis-inhibitory mechanisms of regulation or use a trans mechanism for oncogenic activation. These results were validated by LFQMS comparing phosphorylation status of WT versus oncogenic mutants at zero and 10 min time points, which gave sufficient temporal resolution to measure the rapid phosphorylation kinetics of oncogenic RET (Figure 3C).
Figure 3.
Oncogenic Mutations in the RET Tyrosine KD Perturb the AutoP Trajectory
(A) WB analysis of RET ICD (2.5 μM) WT and oncogenic mutants V804M and M918T stimulated with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min using the indicated antibodies.
(B) Quantitation by densitometry analysis of experiments depicted in (A). Data represent the mean signal value (percentage) ± SE, n = 6. For clarity, only Y905 and Y1015 are depicted.
(C) Temporal autoP analysis by LFQMS of recombinant purified RET ICD (2.5 μM) WT, V804M, or M918T treated with ATP (5 mM) and MgCl2 (10 mM) for 0–10 min. Data represent the mean signal value (Log2 ratios of phosphorylated peptides standardized to the nonphosphorylated counterparts) ± SE, n = 2.
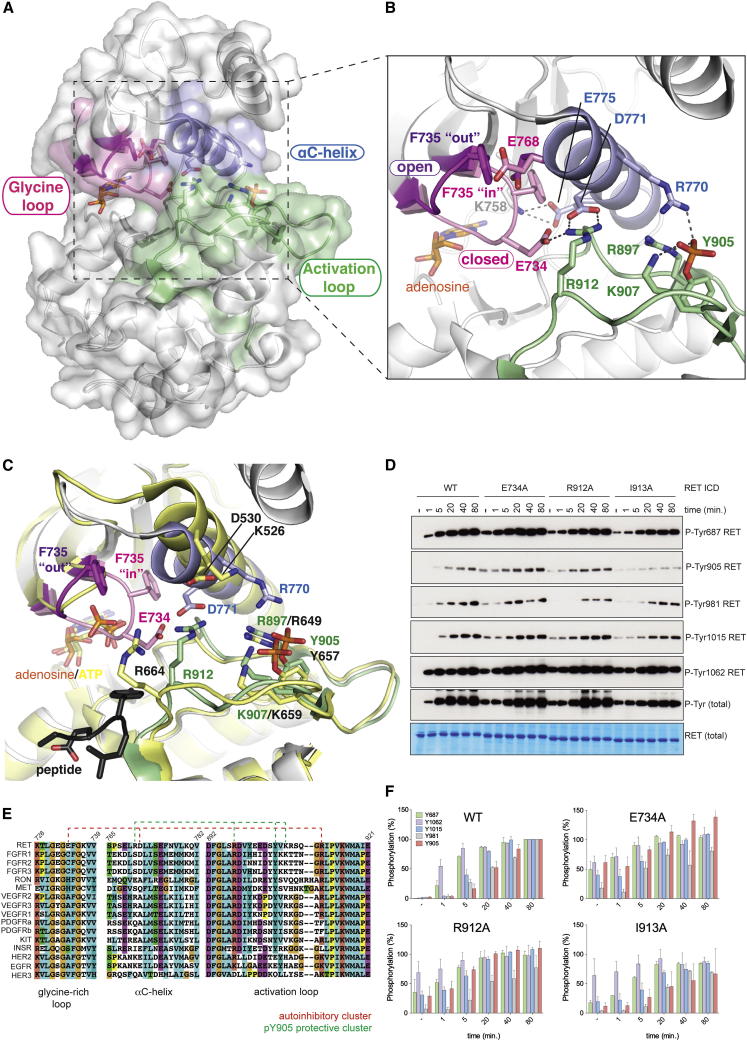
A Crystal Structure of Phosphorylated RET KD with Two Discrete Glycine-Rich Loop Conformations Reveals a Tethered Autoinhibited State
Despite several RET KD crystal structures solved with a number of inhibitors and AMP (Knowles et al., 2006; Mologni et al., 2010), the impact of ATP or ATP-analogs on RET KD conformation has been elusive. During efforts to trap a RET KD structure in complex with ATP, crystals took several months to grow. We determined the structure at 1.65 Å resolution and identified an adenosine moiety within the nucleotide cleft indicating a complete hydrolysis of the ATP (Figures 4A and 4B; Table 1; Supplemental Experimental Procedures). A striking feature of this RET KD structure is the presence of two distinct conformations of the glycine-rich loop (GRL) indicating the intrinsic flexibility of this region. In the “closed” conformation, the GRL is clamped over the kinase cleft with F735 side chain packed against the αC helix and the side chain of E734 from the GRL hydrogen bondings to the side chain of AL residue R912, which also interacted with side chain of D771 from the αC helix (Figure 4B). In the second “open” conformer the GRL pivots away from the AL and αC helix and has a solvent accessible F735 side chain. The two different GRL conformations are defined by the mutually exclusive conformation of two residues, E768 from the αC helix and F735 from the GRL (Figure 4B). F735 shifts by 5.4Å between these conformers, facing away from the nucleotide cleft in the open conformer and tucked into the cleft in the closed state. In the closed conformer, the E768 side chain points away from the cleft and forms hydrogen bonds with the main chain of S765 at the top of the αC helix.
Figure 4.
Crystal Structure of Phosphorylated RET KD with Two Distinct GRL Conformations Reveals an Autoinhibited Conformation
(A) Crystal structure of phosphorylated RET KD bound to adenosine at 1.65 Å (PDB ID code 4CKJ) displaying two distinct conformations of the GRL. Main secondary structure components involved in the GRL tether are depicted with discrete colors.
(B) Detailed view from (A), showing the two GRL conformations. Secondary structure components as well as the crucial residues are depicted.
(C) Superposition of phosphorylated (p) RET KD bound to adenosine structure (PDB ID code 4CKJ, color-coded) and ATP-analog and substrate-peptide bound pFGFR2 kinase domain structure (PDB ID code 2PVF, yellow). Peptide and main secondary structure components are depicted. Peptide (dark gray color) and homologous residues for both structures are shown (color-coded letters RET kinase, black letter FGFR2 kinase).
(D) WB analysis of RET ICD (2.5 μM) WT, E734A, R912A and I913A mutants stimulated with ATP (5 mM) and MgCl2 (10 mM) for 0–80 min using the indicated antibodies.
(E) Sequence alignment of RTKs showing secondary structure components and the two identified networks of residues interacting via hydrogen bonds defining two independent clusters. Numbering corresponds to RET sequence.
(F) Quantification by densitometry analysis (ImageJ) of (D). Data shown are the mean (percentage) ± SE, n = 2.
See also Figure S4.
Table 1.
Data Collection and Refinement Statistics
| RET M918T: Ade | RET WT: Ade | |
|---|---|---|
| Data collection | ||
| Space group | C 2 | C 2 |
| Cell Dimensions | ||
| a, b, c (Å) | 71.9, 70.6, 79.0 | 73.2, 69.1, 78.6 |
| α, β, γ (°) | 90.0, 101.1, 90.0 | 90.0, 101.7, 90.0 |
| Resolution (Å) | 40–2.11 | 40–1.65 |
| (Outer resolution shell) Å | (2.23–2.11) | (1.68–1.65) |
| Rsym (%) | 6.9 (18.0) | 7.4 (67.1) |
| 〈I/σI〉 | 7.4 (2.6) | 4.8 (1.2) |
| Completeness (%) | 99.0 (99.0) | 90.8 (91.0) |
| Redundancy | 5.1 (5.4) | 1.8 (1.9) |
| Refinement | ||
| Resolution (Å) | 40–2.1 | 40–1.65 |
| (Outer resolution shell) Å | (2.21–2.11) | (1.68–1.65) |
| No. unique reflections | 22,084 | 46,229 |
| Rwork | 15.6 (19.0) | 15.1 (26.3) |
| Rfreea | 19.0 (25.4) | 17.7 (32.3) |
| No. atoms | 2 710 | 2 571 |
| Average isotropic B-factors (Å2) | ||
| Protein | 32.1 | 33.4 |
| Ligand | 51.5 | 32.6 |
| Water | 37.9 | 47.4 |
| Rmsd | ||
| Bonds (Å) | 0.005 | 0.019 |
| Angles (°) | 0.90 | 1.82 |
| Ramachandran plot (%) (favored, allowed, disallowed) | 98.4/1.6/0.0 | 98.7/1.3/0.0 |
Ade, adenosine; Rmsd, root-mean-square deviation.
A total of 5% of the data were set aside to compute Rfree.
Superimposition of the RET KD-adenosine structure with the closely related FGFR2 bound to an ATP analog and substrate-peptide structure (Chen et al., 2007) reveals that the “closed” GRL conformation would clash with the β and γ phosphates from ATP. Therefore this conformer would reflect an apo-state of the enzyme, as it does resemble the apo-FGFR2 KD structure (Protein Data Bank [PDB] ID code 2PSQ), particularly residues 911–914 of the activation segment (equivalent to FGFR residues 663–666). In the presence of Mg-ATP and a phosphorylated AL, this region of FGFR2 KD is reorganized to accommodate a substrate peptide (PDB ID code 2PVF). The “open” GRL conformer, however, would readily accommodate ATP (Figure 4C), such a conformation is stabilized by hydrogen bonds with the loop that precedes the αC helix located above the GRL. These hydrogen bonds are formed between the main chain carbonyls of F735, E734 and G733 from the GRL and the side chain nitrogen of N763 and K761 and the main chain nitrogen of K761.
A closer look at the ATP-occluded conformer reveals a possible autoinhibitory mechanism (cis-inhibition) to block nucleotide binding and perturb substrate interaction, involving three key structural elements: the GRL, AL, and αC helix (Figure 4B). In the previously determined phosphorylated RET KD crystal structures (PDB ID codes 2IVT, 2IVV, and 2IVU), phosphorylated Y905 was stabilized via hydrogen bonds between the phosphate group and the side chains of R770 (αC helix) and AL R897 and K907, in contrast to the single tether found in other RTKs, e.g., R1155 in the activated IRK structure (that is structurally equivalent to R897 in RET). The closed GRL conformer also displays a second triad of tethered residues via salt bridge interaction involving hydrogen bonds between E734 from the GRL, R912 from AL and D771 from the αC helix as described above (Figures 4B and 4E).
To functionally validate these structural findings, we performed time course autoP assays with point mutants E734A, R912A, and I913A, which we anticipated would favor a GRL open conformation. We observed that mutation of these residues enhances the kinetics of autoP of sites located at the juxtamembrane, C-tail regions, and AL, with the exception of I913A (Figures 4D and 4F). Taken together, these structural and biochemical data indicate that the closed conformer of the GRL contribute to a cis autoinhibitory mechanism. Mutations that release this tether have a gain-of-function effect as indicated by the faster kinetics of RET autoP.
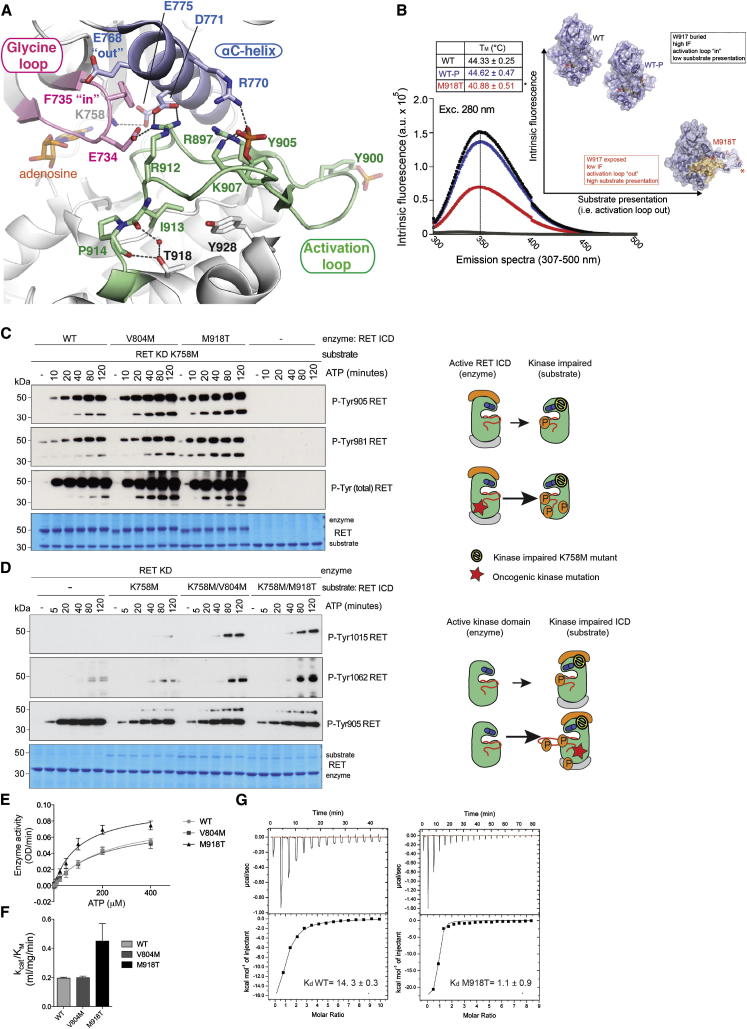
Structural and Biophysical Characterization of Oncogenic RET M918T KD
To understand the oncogenic mechanism of RET M918T activation, we also determined the crystal structure of the RET M918T KD bound to adenosine at 2.1 Å resolution (Figure S4; Table 1; Supplemental Experimental Procedures). The oncogenic RET M918T KD structure displayed several features not shared by previously solved structures of the native KD (Figure 5A). The AL was phosphorylated on both Y900 and Y905, compared to the singly phosphorylated AL on Y905 observed in RET WT KD structures (PDB ID codes 2IVV, 2IVT, and 2IVU). Furthermore, T918 forms a network of hydrogen bonds with a water molecule and main chain carbonyl from I913 and P914 (Figures 5 and S4). Paradoxically, we also observed the “closed” GRL conformer defined by the second cluster of tethered residues via hydrogen bonds involving E734 from the GRL, R912 from AL, and D771 from the αC helix (Figure 5A). Similar to native RET, this GRL conformer is not competent for ATP binding and despite its high activity in solution it appears in a cis-inhibited state in the crystal. A clue to this apparent paradox may come from the robust proteomic and biochemical characterization of RET M918T in solution (Figure 3), where we show that late autoP sites (i.e., AL tyrosines) become phosphorylated much earlier than RET WT, which in turn can affect the balance between enzymatic activity and substrate presentation in the process of autoP. We considered whether the M918T mutation could impact on the accessibility of the AL to be presented as a substrate to a second RET kinase molecule in trans. By destabilizing an “AL-in” conformer, the M918T oncogenic mutation would increase the exposure of phosphoacceptor tyrosine sites presented to the catalytic cleft of another RET kinase molecule (Figure 5B). Biophysical characterization based on intrinsic fluorescence (IF) and thermal denaturation assays were used to test the proposed “AL-out” model for enhanced substrate presentation in trans. Conformational changes of the AL can be measured in solution by IF differences taking advantage of a tryptophan (W917) situated next to M918 within the P+1 loop at the end of the activation segment (Figures 5B and S5). Indeed, RET M918T showed a marked difference in IF with a greatly decreased intrinsic tryptophan fluorescence compared to RET WT (Figure 5B). We interpret this large difference as showing that the W917 environment is perturbed by M918T. Hence, W917 would be buried within RET WT kinase (AL-in) and accessible within M918T (AL-out). This would be in agreement with an enhanced intermolecular substrate presentation by oncogenic RET M918T. This is also consistent with a reduced melting temperature (∼4°C lower) for this mutant measured by thermal shift assay when compared to RET WT indicating a significantly less stable protein (Figure 5B).
Figure 5.
Structural, Biophysical, and Biochemical Characterization of Oncogenic RET M918T
(A) Crystal structure of oncogenic RET M918T KD at 2.1Å resolution (PDB ID code 4CKI). Closed-up view shows the GRL in the closed conformation. Main secondary structure components, ligand adenosine, and important residues are depicted with discrete colors.
(B) Biophysical characterization of RET ICD WT and oncogenic M918T. IF (arbitrary units [a.u.]) emission spectra (307–500 nm, excitation at 280 nm) of RET ICD WT, phosphorylated RET ICD WT, and RET M918T. Left inset: shows the melting temperatures for the indicated proteins, showing a significantly lowered melting temperature for oncogenic RET M918T. Right inset: depicts our proposed model of enhanced substrate presentation in trans by an “AL-out” conformer. Asterisk (∗) denotes W917.
(C) WB analysis of autoP rescue experiments in trans of catalytically impaired RET K758M KD (substrate, 2 μM) by active RET ICD (2.5 μM) WT, V804M, and M918T using the indicated antibodies. Total levels of RET (both enzyme and RET K758M substrate) were monitored by Coomassie staining. Time course with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min, n = 2.
(D) WB analysis of autoP rescue experiments in trans of catalytically impaired RET K758M, RET K758M/V804M, or K758M/M918T ICD (1 μM) by RET KD (2.5 μM) using the indicated antibodies. Total levels of RET were monitored by Coomassie staining. Time course with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min, n = 3.
(E) Enzymatic assay performed with purified recombinant RET ICD WT, V804M, or M918T (2.5 μM) incubated with increasing concentrations of ATP (0–400 μM) using 8 mg/ml ABL peptide (EAYAAPFAKKK). Data represent the mean (OD/min) ± SE, n = 4.
(F) Catalytic efficiency constants (kcat/KM) of (E).
(G) Isothermal titration calorimetry profiles of ATP for RET ICD WT and M918T at pH 7.65 at 20°C. The plot of the heat released (kcal) per mol of ATP added, against the molar radio of ATP to protein is depicted. Note that a minor proportion of ATP may be converted to ADP via autoP. The data were fitted to a single sequential-binding site(s) model, and the solid lines represent the best fit.
See also Figures S4, S5, and Table S1.
Oncogenic Mutations in the RET KD Enhance Both Enzymatic and Substrate Presentation Properties
To assess whether oncogenic mutations in the RET KD enhance the enzymatic properties of the receptor, we performed autoP rescue experiments in trans using a catalytically impaired RET K758M KD as substrate. In this experiment, comparison between active RET ICD WT, V804M, and M918T indicated that oncogenic mutations in the KD enhance the catalytic properties of RET (Figure 5C). These data recapitulate the results of the autoP process in situ (Figure 3). Next, to investigate whether oncogenic mutations in the KD also enhance the substrate presentation properties of the receptor, the reciprocal experiment was performed. Here, catalytically impaired RET K758M ICD with or without the V804M or M918T mutations were used as substrates. AutoP rescue experiments in trans by RET KD revealed the unexpected finding that oncogenic mutations in the KD also enhance the substrate presentation of the receptor (Figure 5D). Next, we calculated the enzyme kinetic parameters of the oncogenic mutants and native kinases for ATP (Figures 5E and 5F). Enzyme activity assessed upon increasing concentrations of ATP revealed that only RET M918T displayed increased enzyme kinetics parameters as indicated by the higher catalytic efficiency constant (kcat/KM). To test if the increased enzymatic constants observed for M918T correlated with increased affinity for ATP, we directly measured ATP binding by isothermal titration calorimetry (ITC) to establish the apparent dissociation constants (Kd). We found the Kd values for RET M918T were an order of magnitude lower than RET WT (1.1 ± 0.9 and 14.3 ± 0.3 μM, respectively) indicating a significantly tighter ATP affinity (Figure 5G; Table S1). The basis for this tighter binding is unclear, but we note that several hydrophobic core side chains adopt distinct rotamers from those in RET WT, which together with the lower apparent melting temperature for M918T, may suggest a greater dependence on ATP-binding to stabilize the C-spine of the kinase (Figure S4). Taken together, these data imply that oncogenic mutations in the RET KD perturb the autoP trajectory by enhanced substrate presentation in trans and increased nucleotide pocket occupancy for ATP in cis.
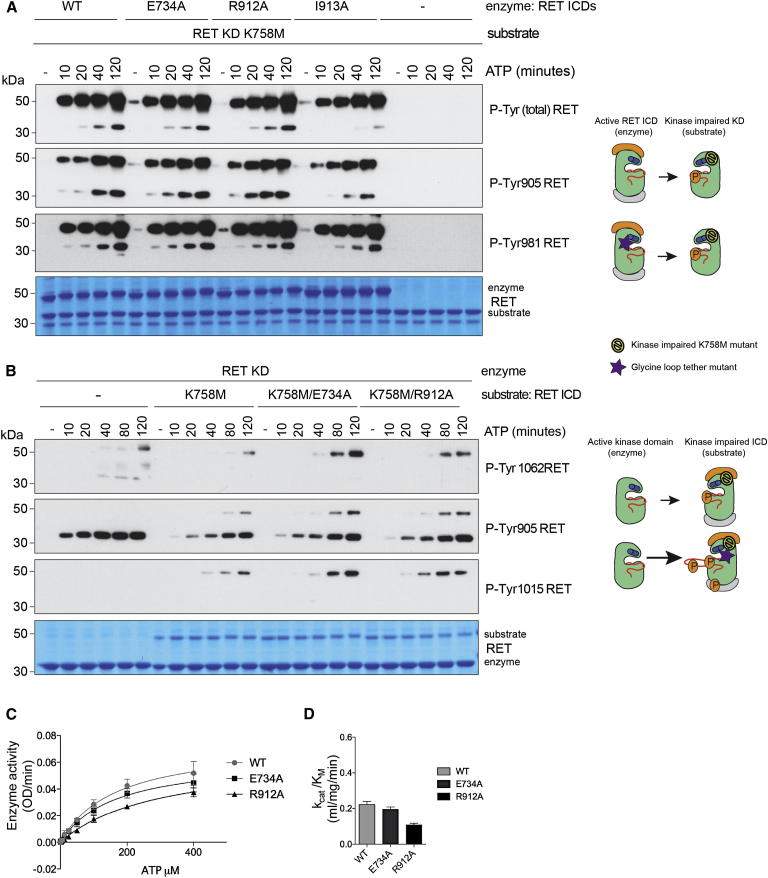
Mutations Disrupting the GRL Tether Do Not Affect the Enzymatic Activity But Do Enhance the Substrate Presentation Properties of RET
To uncover whether mutations disrupting the GRL tether enhance the enzymatic properties of the receptor, we performed autoP rescue experiments in trans using catalytically impaired RET K758M KD as a substrate. In this experiment, quantitative comparison between active RET ICD WT, E734A, R912A, and I913A revealed no significantly enhanced enzymatic activity (Figures 6A and S6). Next, we tested whether these mutations enhanced the substrate presentation properties of the receptor; again the reciprocal experiment was performed. Catalytically impaired RET K758M ICD with or without the E734A or R912A mutations were used as substrates. AutoP rescue experiments in trans by RET KD revealed that contrary to the effect on enzymatic activity, tether mutations do enhance the substrate presentation properties of RET, presumably by releasing the AL for presentation in trans (Figure 6B). To test this hypothesis, IF and thermal denaturation assays were performed (Figure S5). GRL tether mutants exhibited an intermediate IF emission pattern between RET WT (ordered AL, W917 buried, and high IF) and oncogenic M918T (unfolded AL conformation, W917 exposed, and low IF). These data seemed also independent of the binding of ATP, as indicated by the use of a kinase-impaired version in analogous experiments (Figure S5). The stability of these mutants by thermal shift assay was also measured and found no significant differences compared with RET WT. These data indicate that tether mutations can impact on AL conformation to enhanced substrate presentation, without affecting the stability of the protein. Next we calculated the enzyme kinetic parameters for ATP for the described mutants and native kinases (Figures 6C and 6D; Table S1). Enzyme activity assessed upon increasing concentrations of ATP revealed no significant increase in the enzymatic parameters as indicated by their catalytic efficiency constants. To measure the impact of these mutations on the affinity for ATP we used ITC, which showed that none of these mutants, contrary to what we anticipated, significantly affected the affinity for ATP as indicated by their Kd constant values (Table S1). These data show that tether mutations within the GRL or AL enable nucleotide binding without affecting the enzymatic activity of the kinase but rather by enhancing RET substrate presentation in trans. However, contrary to oncogenic mutations, they do not alter affinity for ATP (occupancy). These data support and emphasize the unexpected enhanced autoP substrate explanation for oncogenic RET kinase activation.
Figure 6.
Mutations Disrupting the GRL Tether Do Not Affect the Enzymatic Activity but Enhance RET Kinase Substrate Presentation In trans
(A) WB analysis of autoP rescue experiments in trans of catalytically impaired RET K758M KD (substrate, 2 μM) by active RET ICD (2.5 μM) WT, E734A, R912A, and I913A using the indicated antibodies. Total levels of RET were monitored by Coomassie staining. Time course with ATP (5 mM) and MgCl2 (10 mm) for 0–120 min, n = 2.
(B) WB analysis of autoP rescue experiments in trans of catalytically impaired RET ICD K758M, K758M/E734A, or K758M/R912A as substrates (1.5 μM) by active RET KD (2.5 μM) using the indicated antibodies. Total levels of RET were monitored by Coomassie staining. Time course with ATP (5 mM) and MgCl2 (10 mM) for 0–120 min, n = 3.
(C) Enzymatic assay performed with purified recombinant RET ICD WT, E734A, or R912A (2.5 μM) incubated with increasing concentrations of ATP (0–400 μM) using 8 mg/ml ABL peptide (EAYAAPFAKKK). Data represent the mean (OD/min) ± SE, n = 4.
(D) Catalytic efficiency constants (kcat/KM) of (C).
See also Figures S5, S6, and Table S1.
Discussion
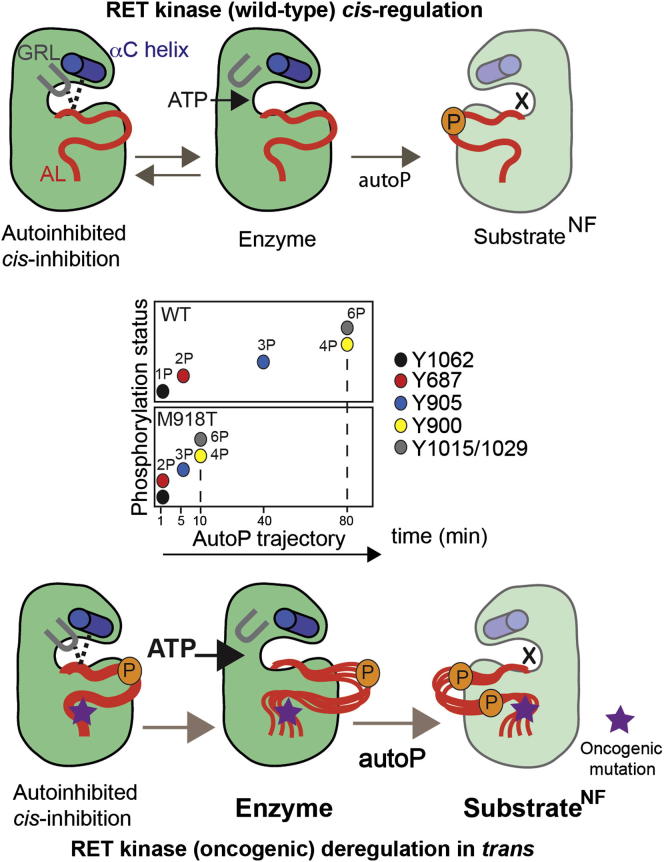
Despite the crucial biological and pathological implications of RET in early development and neoplastic processes, the precise molecular mechanisms for RET KD activation and oncogenic deregulation have so far eluded definition. The current paradigm for RTK activation is that binding of extracellular ligand promotes rapid phosphorylation of the canonical AL to boost catalytic activity, remove cis-inhibitory conformations and generate effector-docking sites (Artim et al., 2012). Examples include the IRK, FGFR2, TrkA, and Ror (Artim et al., 2012; Hubbard, 1997). In this study, we show that contrary to many RTKs the earliest detectable autoP sites generated following RET activation map to regions flanking the KD core while those sites within the AL of the RET KD only form at much later time points (Figures 1 and 7). One interpretation of these results is that the flanking regions restrict access to AL tyrosine sites, as part of a repressive allosteric interaction (Chan et al., 2003; Hubbard, 2004). In such a scenario, juxtamembrane and C-tail tyrosine autoP would precede AL phosphorylation as part of a concerted ordered activation trajectory for RET. Arguing against such a concerted mechanism controlling access to AL autoP sites is the observation that mutation of individual autoP sites does not impact dramatically on kinase activation or perturb the overall pattern of autoP. For instance, Y1062F and Y687F mutants do not significantly affect the kinetics of Y905 autoP (Figure 2A). We cannot exclude the existence of compensatory mechanisms between the juxtamembrane and C-terminal regions, or redundancy of adjacent autoP sites. A plausible alternative is a more stochastic activation mechanism where sites might be preferentially phosphorylated depending on primary sequences surrounding the phosphoacceptor site or spatially-close determinants. However, we show that the phosphorylation efficiency of autoP sites-based peptide substrates does not recapitulate the sequence of autoP using an intact RET ICD or KD substrate (Figure S2). This argues that determinants influencing how rapidly sites are targeted are not local to the tyrosine autoP site but appear opportunistic relating to tyrosine site accessibility and location within the kinase structure consistent with a highly active artificially “forced” RET dimer (Figure S6). Our data point toward kinase regulation independent of AL phosphorylation but determined by inputs from flanking regions and other structural and functional components (e.g., GRL, αC helix) independently of the autoP sites, because these would play a key role in docking and downstream signaling rather than being catalytically required.
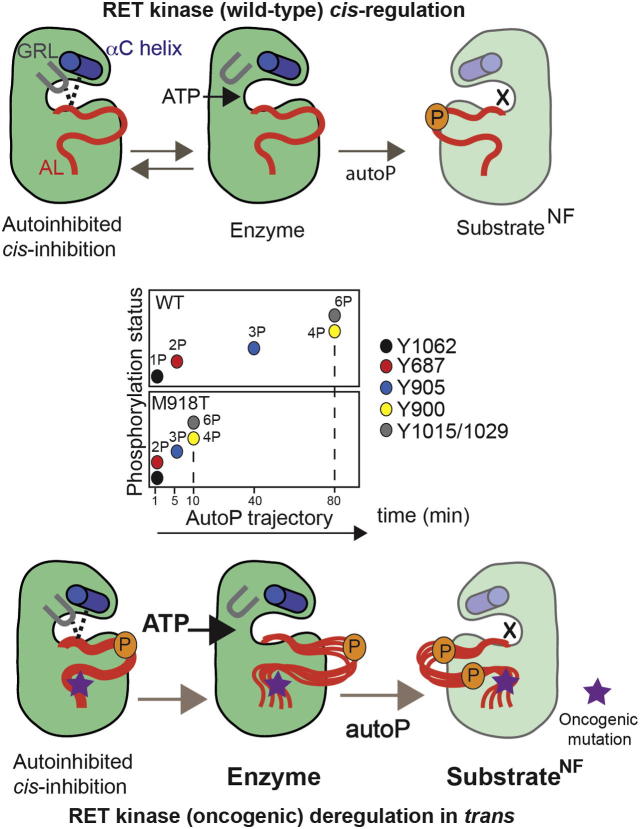
Figure 7.
A Proposed Model for RET (WT) Kinase cis-Regulation and Oncogenic Perturbation of the AutoP Trajectory In trans
Inset: summary of changes to the autoP trajectory by oncogenic RET M918T (quantified by LFQMS based on saturation rates). This is caused by an enhanced enzymatic activity and higher affinity for ATP and the production of a better intermolecular RET substrate. RET WT KD cis-regulation by a tether affecting GRL and AL conformation leading to autoinhibition. An ATP-bound conformer disrupts the GRL tether between E734 (GRL), R912 (AL), and D771 (αC helix) allowing ATP to access the active site (enzyme, active kinase) and releasing the AL. This leads to phosphorylation in trans (autoP) of a RET kinase presented as a substrate. Oncogenic RET M918T kinase is autoinhibited in cis by the GRL closed conformer despite displaying enhanced phosphorylation status. Enhanced ATP affinity favors an open GRL conformer and a more active enzyme, which also displays enhanced substrate presentation by promoting more flexible “AL-out” conformers (X denotes in a nucleotide-free state of a catalytically impaired RET kinase presented as a surrogate substrate for autophosphorylation in trans).
Our data confirm earlier reports (Knowles et al., 2006; Plaza-Menacho et al., 2011) showing that RET-AL phosphorylation is not a prerequisite for kinase activation. Precedents exist for AL phosphorylation-independent RTKs such as the EGFR, c-Kit, and ACK1 kinases (DiNitto et al., 2010; Lougheed et al., 2004; Stamos et al., 2002; Zhang et al., 2006). In the case of EGFR, activation of the KD is achieved independently of AL phosphorylation by both overcoming steric constraints in the extracellular region and promoting the association of transmembrane and intracellular regions into a ligand-dependent asymmetric EGFR dimer (Endres et al., 2013). Whether similar mechanisms operate for RET activation independent of AL phosphorylation requires further study, but it is suggested by our results.
The structural data presented here uncover a possible cis-inhibitory mechanism connecting three crucial structural components of the RET KD. In the closed GRL conformer, ATP access to the nucleotide-binding pocket is restricted by the GRL being tethered to the AL (Figures 4 and 7). The tether couples side chains of E734 (GRL), R912 (AL), and D771 (αC helix) through hydrogen bonds, a structural feature that was not seen in previous phosphorylated RET KD structures (Knowles et al., 2006). These interactions explain a role for a GRL glutamate (i.e., RET E734), an unusual feature in RTKs (Figure 4E). Mutations disrupting the GRL tether also perturb the autoP trajectory of RET in a gain-of-function manner, in contrast to single Y/F mutants targeting the autoP sites, which did not have a significant effect (Figures 2 and 4). ATP-binding-site occlusion by unphosphorylated activation segments has been previously observed (Artim et al., 2012). We propose that a tethered GRL to the AL and αC helix conformation provides a distinct means to couple substrate interaction and ATP cleft access reminiscent of, but distinct from, the FGFR2 autoinhibitory “molecular brake” promoted by the crosstalk between the kinase hinge region, αC helix and AL (Chen et al., 2007).
A major focus of this study was to understand how oncogenic RET V804M and M918T deregulate kinase activity compared with RET WT. Comparison of oncogenic V804M and M918T RET kinase mutants in autoP assays revealed that late autoP sites become phosphorylated more rapidly than for RET WT (Figures 3 and 7). We hypothesized that the perturbation of the autoP trajectory could arise either by increasing RET catalytic activity or by enhancing the presentation of RET as a substrate in trans, or both. In addition, it could also affect the enzyme kinetics parameters and affinity of the mutant kinase for ATP. We verified that oncogenic RET M918T increases enzymatic activity and ATP affinity analogous to the T790M mutation of the EGFR (Yun et al., 2008). The structure of oncogenic RET M918T KD however also exhibited a tethered GRL conformer indicating an occluded nucleotide-binding pocket, or cis-inhibited state (Figure 5A). This led us to question whether in solution, perturbation of the autoP trajectory by RET M918T could arise through presentation of a better intermolecular substrate by favoring an “AL-out” conformer presenting Y900 or Y905 as a substrate to an active RET kinase in trans. The striking differences in the IF properties between RET WT and M918T suggest that in solution the conformational dynamics and flexibility of the AL are increased by the oncogenic mutation. Similar studies for FGFR2 and Eph RTKs have shown that kinase activation involves an increase in the conformational dynamics of their respective ICDs (Chen et al., 2013; Wiesner et al., 2006). Our biochemical evidence suggests that the impact of M918T in a kinase-impaired substrate is to generate a better substrate for phosphorylation in trans. This unexpected facet of oncogenic RET M918T is in addition to increasing RET enzymatic activity and ATP affinity (Figures 5 and 7). We note that M918T also has increased activity toward exogenous substrates and is itself phosphorylated to a greater degree by exogenous kinases such as FAK (Figure S7). How M918T mutation perturbs the GRL cis-inhibition in solution and increases ATP affinity is unclear from the DFG-in conformer crystal structure. Solution data indicate a change in stability and conformational dynamics, which could affect the catalytic spine and thus ATP affinity (Figure S4). Whether M918T could disrupt a DFG-out conformer in RET remains to be shown. However, disruption of the GRL tether by mutation selectively alters the autoP trajectory and increases substrate presentation in trans without altering catalytic activity or ATP occupancy (Figure 6).
In summary, our findings indicate that RET is a member of a select group of RTKs whose kinase activity is regulated by regions flanking the KD core rather than by AL phosphorylation. The slow time course for AL Y900/Y905 phosphorylation may be crucial for signaling because of a role in effector recruitment rather than as an activation event. AutoP in trans appears more complex than a simple recognition of primary sequence motifs containing an accessible phospho-acceptor site. Our data show that oncogenic RET mutations promote autoP by both increasing catalytic activity and generating a better intermolecular substrate. At the same time they introduce greater flexibility, lower stability, and higher ATP affinity for the kinase domain with important implications for oncogenic RET chemical inhibition. This illustrates the subtlety and complex effects a single point mutation can exert to perturb RTK activation mechanisms.
Experimental Procedures
Full experimental details are found in the Supplemental Information.
Expression and Purification of Recombinant Protein
Protein expression was carried out using Sf9 insect cells infected with recombinant Baculoviruses as previously described (Knowles et al., 2006). Human RET9 ICD (residues 661–1,072) and KD core (705–1,013) WT and the indicated mutants proteins were purified as previously described (Knowles et al., 2006).
Mass Spectrometric Label-Free Quantitation
For label-free quantitation, three to five biological replicates and three technical replicates were analyzed per condition. Data was acquired using the Agilent’s LC-Chip Cube-6520 QTOF in MS-only mode. MS data was recorded with the settings required in profile mode with the exception of recording one spectrum per second. Data was acquired randomly as determined by list randomizer (http://www.random.org/lists/). Files were imported into Progenesis LC-MS version 4.0.4573.30654 (Nonlinear Dynamics) for LC-MS run alignment and MS peak extraction for label-free quantitation after ion peak identification by MASCOT. Peaks of interest were analyzed and validated using Excel and Graph Pad.
AutoP Assays, SDS-Page, and WB
Unless otherwise indicated, time course autoP assays were performed with recombinant purified RET ICD and/or KD core (2.5 μM) in the presence of saturating concentrations of ATP (5 mM) and MgCl2 (10 mM) for the indicated time points. Reactions were stopped by the addition of 4× loading sample buffer (Invitrogen) with 10% β-mercaptoethanol and boiled for 5 min after which samples were loaded on gel. Samples were run in a NuPAGE Invitrogen 4%–12% Bis-Tris precast gels, and WB was performed with the indicated antibodies as previously described (Plaza Menacho et al., 2005, 2007a, 2007b). Details of antibodies used are in the Supplemental Information.
Phosphospecific RET Y687 Antibody Generation
Polyclonal antisera against phosphorylated RET Y687 was generated by immunizing rabbits with a synthetic peptide (AFPVSpYSSSGA) KLH-conjugated via glutaraldehyde following a standard protocol (Pettingill Technology).
Enzymatic Kinase Assays
Enzyme kinetic experiments were performed as previously described (Knowles et al., 2006; Plaza-Menacho et al., 2011).
Acknowledgments
We thank to the following London Research Institute (Cancer Research UK) colleagues: Sara Kisakye Nambozo, Roger George, and Svend Kjaer (Protein Production Facility) for their technical assistance in the production of baculoviruses; Nicola O’Reilly and the Peptide Production Facility for peptides supply; Andrew Purkiss (Structural Biology Laboratory) for helping with data collection and structure determination; Diamond Light Source Synchrotron (Oxford, UK) for access to I24 beam line. This work was supported by core funding from Cancer Research UK to N.Q.M.
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/3.0/).
Contributor Information
Iván Plaza-Menacho, Email: ivan.plaza-menacho@cancer.org.uk.
Neil Q. McDonald, Email: neil.mcdonald@cancer.org.uk.
Accession Numbers
The crystallographic coordinates and structure factors have been deposited in the PDB (http://www.pdb.org). PDB ID codes for the RET WT and RET M918T KD crystal structures are 4CKJ and 4CKI, respectively.
Supplemental Information
References
- Airaksinen M.S., Titievsky A., Saarma M. GDNF family neurotrophic factor signaling: four masters, one servant? Mol. Cell. Neurosci. 1999;13:313–325. doi: 10.1006/mcne.1999.0754. [DOI] [PubMed] [Google Scholar]
- Arighi E., Borrello M.G., Sariola H. RET tyrosine kinase signaling in development and cancer. Cytokine Growth Factor Rev. 2005;16:441–467. doi: 10.1016/j.cytogfr.2005.05.010. [DOI] [PubMed] [Google Scholar]
- Artim S.C., Mendrola J.M., Lemmon M.A. Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family. Biochem. J. 2012;448:213–220. doi: 10.1042/BJ20121365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.M., Ilangumaran S., La Rose J., Chakrabartty A., Rottapel R. Autoinhibition of the kit receptor tyrosine kinase by the cytosolic juxtamembrane region. Mol. Cell. Biol. 2003;23:3067–3078. doi: 10.1128/MCB.23.9.3067-3078.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Ma J., Li W., Eliseenkova A.V., Xu C., Neubert T.A., Miller W.T., Mohammadi M. A molecular brake in the kinase hinge region regulates the activity of receptor tyrosine kinases. Mol. Cell. 2007;27:717–730. doi: 10.1016/j.molcel.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Xu C.F., Ma J., Eliseenkova A.V., Li W., Pollock P.M., Pitteloud N., Miller W.T., Neubert T.A., Mohammadi M. A crystallographic snapshot of tyrosine trans-phosphorylation in action. Proc. Natl. Acad. Sci. USA. 2008;105:19660–19665. doi: 10.1073/pnas.0807752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Huang Z., Dutta K., Blais S., Neubert T.A., Li X., Cowburn D., Traaseth N.J., Mohammadi M. Cracking the molecular origin of intrinsic tyrosine kinase activity through analysis of pathogenic gain-of-function mutations. Cell Rep. 2013;4:376–384. doi: 10.1016/j.celrep.2013.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulpier M., Anders J., Ibáñez C.F. Coordinated activation of autophosphorylation sites in the RET receptor tyrosine kinase: importance of tyrosine 1062 for GDNF mediated neuronal differentiation and survival. J. Biol. Chem. 2002;277:1991–1999. doi: 10.1074/jbc.M107992200. [DOI] [PubMed] [Google Scholar]
- Cutillas P.R., Geering B., Waterfield M.D., Vanhaesebroeck B. Quantification of gel-separated proteins and their phosphorylation sites by LC-MS using unlabeled internal standards: analysis of phosphoprotein dynamics in a B cell lymphoma cell line. Mol. Cell. Proteomics. 2005;4:1038–1051. doi: 10.1074/mcp.M500078-MCP200. [DOI] [PubMed] [Google Scholar]
- DiNitto J.P., Deshmukh G.D., Zhang Y., Jacques S.L., Coli R., Worrall J.W., Diehl W., English J.M., Wu J.C. Function of activation loop tyrosine phosphorylation in the mechanism of c-Kit auto-activation and its implication in sunitinib resistance. J. Biochem. 2010;147:601–609. doi: 10.1093/jb/mvq015. [DOI] [PubMed] [Google Scholar]
- Endres N.F., Das R., Smith A.W., Arkhipov A., Kovacs E., Huang Y., Pelton J.G., Shan Y., Shaw D.E., Wemmer D.E. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152:543–556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furdui C.M., Lew E.D., Schlessinger J., Anderson K.S. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell. 2006;21:711–717. doi: 10.1016/j.molcel.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Gibelin H., Bezieau S., Misso C., Bouin-Pineau M.H., Maréchaud R., Kraimps J.L. Germline RET V804M mutation associated with multiple endocrine neoplasia type 2A. Br. J. Surg. 2004;91:1458–1459. doi: 10.1002/bjs.4718. [DOI] [PubMed] [Google Scholar]
- Hubbard S.R. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard S.R. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat. Rev. Mol. Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Kawamoto Y., Takeda K., Okuno Y., Yamakawa Y., Ito Y., Taguchi R., Kato M., Suzuki H., Takahashi M., Nakashima I. Identification of RET autophosphorylation sites by mass spectrometry. J. Biol. Chem. 2004;279:14213–14224. doi: 10.1074/jbc.M312600200. [DOI] [PubMed] [Google Scholar]
- Knowles P.P., Murray-Rust J., Kjaer S., Scott R.P., Hanrahan S., Santoro M., Ibáñez C.F., McDonald N.Q. Structure and chemical inhibition of the RET tyrosine kinase domain. J. Biol. Chem. 2006;281:33577–33587. doi: 10.1074/jbc.M605604200. [DOI] [PubMed] [Google Scholar]
- Lew E.D., Furdui C.M., Anderson K.S., Schlessinger J. The precise sequence of FGF receptor autophosphorylation is kinetically driven and is disrupted by oncogenic mutations. Sci. Signal. 2009;2:ra6. doi: 10.1126/scisignal.2000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougheed J.C., Chen R.H., Mak P., Stout T.J. Crystal structures of the phosphorylated and unphosphorylated kinase domains of the Cdc42-associated tyrosine kinase ACK1. J. Biol. Chem. 2004;279:44039–44045. doi: 10.1074/jbc.M406703200. [DOI] [PubMed] [Google Scholar]
- McTigue M.A., Wickersham J.A., Pinko C., Showalter R.E., Parast C.V., Tempczyk-Russell A., Gehring M.R., Mroczkowski B., Kan C.C., Villafranca J.E., Appelt K. Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis. Structure. 1999;7:319–330. doi: 10.1016/s0969-2126(99)80042-2. [DOI] [PubMed] [Google Scholar]
- Mologni L., Rostagno R., Brussolo S., Knowles P.P., Kjaer S., Murray-Rust J., Rosso E., Zambon A., Scapozza L., McDonald N.Q. Synthesis, structure-activity relationship and crystallographic studies of 3-substituted indolin-2-one RET inhibitors. Bioorg. Med. Chem. 2010;18:1482–1496. doi: 10.1016/j.bmc.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Plaza Menacho I., Koster R., van der Sloot A.M., Quax W.J., Osinga J., van der Sluis T., Hollema H., Burzynski G.M., Gimm O., Buys C.H. RET-familial medullary thyroid carcinoma mutants Y791F and S891A activate a Src/JAK/STAT3 pathway, independent of glial cell line-derived neurotrophic factor. Cancer Res. 2005;65:1729–1737. doi: 10.1158/0008-5472.CAN-04-2363. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho I., Burzynski G.M., de Groot J.W., Eggen B.J., Hofstra R.M. Current concepts in RET-related genetics, signaling and therapeutics. Trends Genet. 2006;22:627–636. doi: 10.1016/j.tig.2006.09.005. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho I., Mologni L., Sala E., Gambacorti-Passerini C., Magee A.I., Links T.P., Hofstra R.M., Barford D., Isacke C.M. Sorafenib functions to potently suppress RET tyrosine kinase activity by direct enzymatic inhibition and promoting RET lysosomal degradation independent of proteasomal targeting. J. Biol. Chem. 2007;282:29230–29240. doi: 10.1074/jbc.M703461200. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho I., van der Sluis T., Hollema H., Gimm O., Buys C.H., Magee A.I., Isacke C.M., Hofstra R.M., Eggen B.J. Ras/ERK1/2-mediated STAT3 Ser727 phosphorylation by familial medullary thyroid carcinoma-associated RET mutants induces full activation of STAT3 and is required for c-fos promoter activation, cell mitogenicity, and transformation. J. Biol. Chem. 2007;282:6415–6424. doi: 10.1074/jbc.M608952200. [DOI] [PubMed] [Google Scholar]
- Plaza-Menacho I., Morandi A., Mologni L., Boender P., Gambacorti-Passerini C., Magee A.I., Hofstra R.M., Knowles P., McDonald N.Q., Isacke C.M. Focal adhesion kinase (FAK) binds RET kinase via its FERM domain, priming a direct and reciprocal RET-FAK transactivation mechanism. J. Biol. Chem. 2011;286:17292–17302. doi: 10.1074/jbc.M110.168500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M., Carlomagno F., Romano A., Bottaro D.P., Dathan N.A., Grieco M., Fusco A., Vecchio G., Matoskova B., Kraus M.H. Activation of RET as a dominant transforming gene by germline mutations of MEN2A and MEN2B. Science. 1995;267:381–383. doi: 10.1126/science.7824936. [DOI] [PubMed] [Google Scholar]
- Stamos J., Sliwkowski M.X., Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J. Biol. Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- Steen H., Jebanathirajah J.A., Rush J., Morrice N., Kirschner M.W. Phosphorylation analysis by mass spectrometry: myths, facts, and the consequences for qualitative and quantitative measurements. Mol. Cell. Proteomics. 2006;5:172–181. doi: 10.1074/mcp.M500135-MCP200. [DOI] [PubMed] [Google Scholar]
- Wiesner S., Wybenga-Groot L.E., Warner N., Lin H., Pawson T., Forman-Kay J.D., Sicheri F. A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J. 2006;25:4686–4696. doi: 10.1038/sj.emboj.7601315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C.H., Mengwasser K.E., Toms A.V., Woo M.S., Greulich H., Wong K.K., Meyerson M., Eck M.J. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc. Natl. Acad. Sci. USA. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Gureasko J., Shen K., Cole P.A., Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.