Abstract
Fitting of atomic components into electron cryo-microscopy (cryoEM) density maps is routinely used to understand the structure and function of macromolecular machines. Many fitting methods have been developed, but a standard protocol for successful fitting and assessment of fitted models has yet to be agreed upon among the experts in the field. Here, we created and tested a protocol that highlights important issues related to homology modelling, density map segmentation, rigid and flexible fitting, as well as the assessment of fits. As part of it, we use two different flexible fitting methods (Flex-EM and iMODfit) and demonstrate how combining the analysis of multiple fits and model assessment could result in an improved model. The protocol is applied to the case of the mature and empty capsids of Coxsackievirus A7 (CAV7) by flexibly fitting homology models into the corresponding cryoEM density maps at 8.2 and 6.1 Å resolution. As a result, and due to the improved homology models (derived from recently solved crystal structures of a close homolog – EV71 capsid – in mature and empty forms), the final models present an improvement over previously published models. In close agreement with the capsid expansion observed in the EV71 structures, the new CAV7 models reveal that the expansion is accompanied by ∼5° counterclockwise rotation of the asymmetric unit, predominantly contributed by the capsid protein VP1. The protocol could be applied not only to viral capsids but also to many other complexes characterised by a combination of atomic structure modelling and cryoEM density fitting.
Keywords: Coxsackievirus A7, Picornaviridae, Flexible fitting, Electron cryo-microscopy, Model assessment
1. Introduction
In recent years, electron cryo-microscopy (cryoEM) has become one of the most prominent techniques for visualising macromolecular assemblies (Orlova and Saibil, 2011; Sali et al., 2003). However, the vast majority of density maps resulting from the various cryoEM reconstruction techniques are not of atomic or near-atomic resolution (even for icosahedral viruses) but rather belong to the so-called intermediate resolution zone (∼5–20 Å) (Baker et al., 1999; Beck et al., 2011), where a detailed interpretation of the map can only be achieved by docking (or fitting) into it an atomic model. Docking of atomic models (from X-ray crystallography, NMR or structure prediction methods) into EM maps has become common practice with a rapidly increasing number of atomic models associated with EM maps deposited in the PDB (currently over 460) (Lawson et al., 2011; Patwardhan et al., 2012).
Due to the differences between the conformations of the atomic model being fitted and the EM map, modifying the conformation of the atomic structure during the fitting process, referred to as flexible fitting, is often needed (Beck et al., 2011). The variety of flexible fitting approaches is currently large. Common to all is the limited sampling of conformational degrees of freedom. Therefore, they are usually applied to components that are first placed into the density map by rigid fitting, whereby a global search of the fit is performed on the atomic model as a single component in six translation/rotation degrees of freedom (Ahmed et al., 2012; Beck et al., 2011). Both rigid and flexible fitting result in a “pseudo-atomic” model for which the quality assessment is not trivial. Approaches that begin to address this issue include the use of confidence intervals and quantifying the best-fitting model relative to a distribution of different fits (Henderson et al., 2012; Tung et al., 2010; Volkmann, 2009; Roseman, 2000; Rossmann et al., 2005; Vasishtan and Topf, 2011). Additionally, if the models are calculated by different methods a question arises regarding their consensus. A recent paper pioneered the issue of consensus among different flexible fitting approaches and proposed to use this information to improve the quality of the fitted models (Ahmed et al., 2012).
Here, we developed a protocol to aid flexible fitting and assessment of virus capsids into cryoEM maps at sub-nanometer resolution. The protocol is designed to use multiple flexible fitting programs, compare and assess the quality of the fit locally, at the level of individual secondary structure elements (SSEs). It also highlights the possibility of producing an improved fit guided by the comparison of multiple independent programs. The protocol is generic and could also be used for systems other than virus capsids.
First, in order to demonstrate the effects of modelling errors on flexible fitting we fitted a homology model of an actin subunit into a density map simulated from a known actin crystal structure in different conformation. Second, to address the challenge of fitting a structure in one conformation into a corresponding EM map in a different conformation, we fitted the crystal structure of EV71 mature (full) capsid into the procapsid map of EV71 strain 1095 (Cifuente et al., 2013). Finally, we applied the protocol to characterise the conformational states of the mature (full) and empty capsid of Coxsackievirus A7 (CAV7). We had previously calculated homology models of the same virus and fitted them into the sub-nanometer resolution cryoEM maps representing the empty (6.09 Å) and full (8.23 Å) CAV7 capsids (Seitsonen et al., 2012).
CAV7 belongs to the Human enterovirus A species within the Picornaviridae family (Oberste et al., 2004). It is an important pathogen with different strains varying in their pathogenicity and tropism (Seitsonen et al., 2012). The CAV7-USSR strain is associated with flaccid paralysis (Voroshilova and Chumakov, 1959) whereas CAV7-275/58 causes aseptic meningitis (Richter et al., 1971). Our original models were based on remote homologs to the virus (Seitsonen et al., 2012) and were refined within the corresponding cryoEM maps using a single flexible fitting method (Flex-EM) (Topf et al., 2008). Here, to improve our original models, we used as templates, recently published crystal structures of the empty and full capsids of the much closer homolog, EV71 (Plevka et al., 2012; Wang et al., 2012) with capsid protein sequence identity of 60% for VP1, 84% for VP2, and 76% for VP3. This time we refined the homology models using two flexible fitting programs, Flex-EM (Topf et al., 2008) and iMODfit (Lopez-Blanco and Chacon, 2013). The different fits were assessed and compared, and new hybrid pseudo-atomic models were generated using the results from both programs. Finally, the conformational changes between the empty and full capsids were characterised based on the new models.
2. Methods
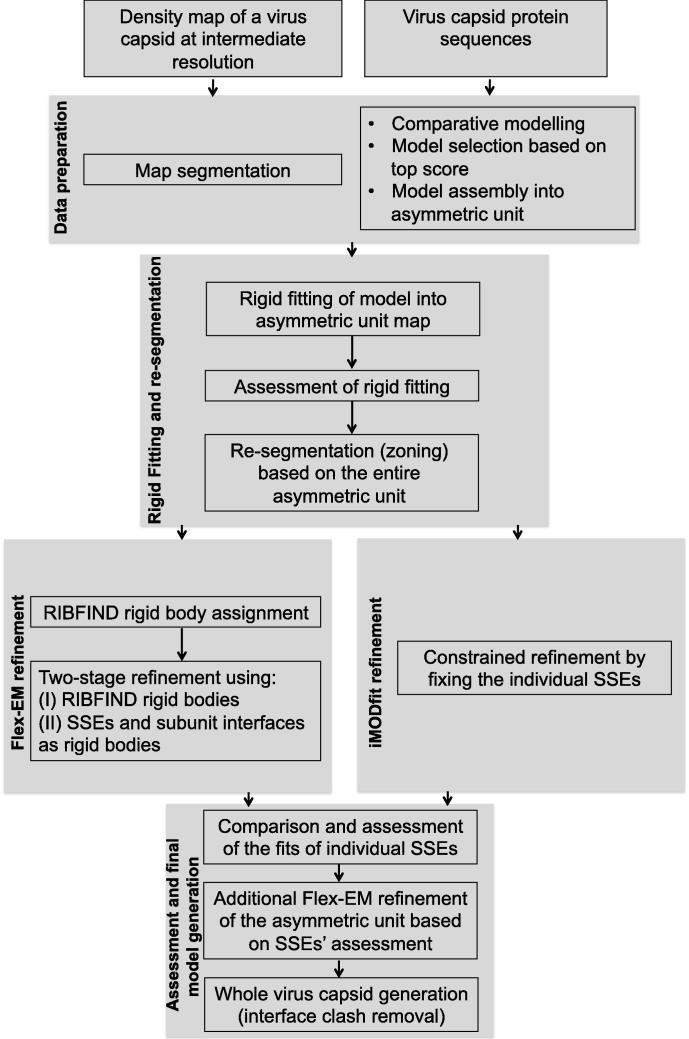
We describe a protocol for modelling and fitting of virus capsids into the cryoEM maps at intermediate resolution using two different flexible fitting programs (Fig. 1). The main feature of the protocol is its ability to compare and assess the quality of the fits produced by independent programs. This approach allows the identification of reliable local fits as well as those that could be further improved by additional stages of refinement. The assessment/refinement protocol can also be applied to systems other than virus capsids. Below we describe the various steps involved in the protocol.
Fig.1.
Protocol describing various stages involved in the modelling of viral capsids in the context of cryoEM data. The protocol starts with the data preparation step, which involves segmenting the asymmetric unit density from the virus capsid map and obtaining the atomic model derived using comparative modelling (in case there is no model available from an experimental technique). The rigid fitting and re-segmentation step provides a good starting fit for flexible fitting, which is performed in the next step by two independent methods (here Flex-EM (Topf et al., 2008) and iMODfit (Lopez-Blanco and Chacon, 2013). The final step involves the local assessment of fits produced by the two different methods, further refinement of identified regions needing improvement, and generation of the whole capsid model (including the identification and removal of clashes). In general, for a given input map and a rigid fit, except for the capsid assembly generation, the steps in the protocol can also be applied to non-viral capsid systems.
2.1. Data preparation
2.1.1. Density map segmentation
The capsid of a mature CAV7 and EV71 virion (full) is made of icosahedrally-arranged viral proteins VP1, VP2, VP3 and VP4 with encapsidated RNA. The empty capsid is also icosahedral but lacks VP4 and RNA. The five-fold vertex is composed of VP1 whereas the three- and the two-fold symmetry axes are made of alternating VP2 and VP3. VP4, a small protein characterised by an extended chain (possibly with a small helix in the middle), is present below the shell of VP1, VP2 and VP3. To help the initial rigid fitting of the asymmetric unit of CAV7 we used the manually segmented maps of the individual viral proteins VP1–VP3 from the density of both empty and full capsids, as described in our previous study (Seitsonen et al., 2012). In the CAV7 full map, VP4 could not be segmented unambiguously and therefore we decided that there were not enough density features to accurately model it. For fitting the EV71 full capsid, the procapsid map was segmented around the asymmetric unit using the fit deposited in PDB (PDB ID: 3VBU; EMD-5557) (Cifuente et al., 2013; Wang et al., 2012).
2.1.2. Homology modelling
CAV7 modelling: From the three target sequences of CAV7-USSR, homology models of the capsid proteins (VP1–VP3) were built using the I-TASSER server (Roy et al., 2010). For a given sequence, I-TASSER builds fragments of template proteins using threading and/or ab initio techniques. The fragments are assembled and refined into a complete model using replica-exchange Monte Carlo simulation (Roy et al., 2010). The template structures used for the modelling were the respective viral proteins in the enterovirus 71 (EV71) crystal structures of empty (PDB ID: 3VBO) and full (PDB ID: 3VBF) capsid forms (Wang et al., 2012). The server generated five different models for each of the two conformations of the three capsid proteins (30 in total) and we selected the model with the top I-TASSER score (out of the five) for further analysis (six models in total). Additionally, the qualitative model energy analysis (QMEAN) scores (Benkert et al., 2008) were used to evaluate both the global and local quality of the selected models and were compared with the previously published models (Seitsonen et al., 2012). Briefly, the QMEAN score for a given protein model is calculated using a combination of the geometrical structural descriptors that include the torsion angle, pairwise residue and solvation potentials. The best I-TASSER models for the three capsid proteins (VP1, VP2 and VP3) obtained using the template structure of the empty capsid (PDB ID: 3VBO) were assembled into an empty capsid asymmetric unit (“empty asymmetric unit”) by superposing the individual VP proteins onto their respective VP proteins in the template structures. Similarly, a full capsid asymmetric unit (“full asymmetric unit”) was assembled using the I-TASSER model (VP1, VP2 and VP3) obtained using the full capsid (mature virus) as the template (PDB ID: 3VBF). The superposition was done using the superpose command in Chimera (Pettersen et al., 2004).
Actin modelling: a homology model of actin was generated from the actin sequence (UniProt: P68135) with MODELLER (Sali and Blundell, 1993) based on the crystal structure of actin-related protein 3 from the Arp2/3 complex, (PDB ID: 1K8K: A) (Robinson et al., 2001). The two proteins share sequence identity of ∼38%.
Below, we describe the general procedure we used for fitting the models into the density maps.
2.2. Rigid fitting and re-segmentation
The actin model was rigidly fitted into the simulated map of the native structure with the Chimera fit_in_map tool (Goddard et al., 2007). For EV71 test case, the initial rigid fit was obtained by superposing the asymmetric unit onto the asymmetric fit deposited in PDB (PDB ID: 3VBU; EMD-5557).
In real-case scenarios of virus capsids, however, a rigid fit can be obtained by fitting individual subunits or the whole asymmetric unit into the density (either of the whole virus or segmented around the asymmetric unit). The former approach is followed when the arrangements of the subunits within the asymmetric unit is unknown. The latter approach is more appropriate when the knowledge of the intra-subunit interactions within the asymmetric unit is already available, for example, from the crystal structure of a homologous virus. The latter approach also helps to minimise rigid and flexible fitting issues arising due to segmentation error/bias within the densities of the asymmetric units. In contrast to our previous study where we used the first approach, here, we adopted the second approach to perform the initial fit in the CAV7 capsids using a recent homologous crystal structure (EV71) describing the entire asymmetric unit (Wang et al., 2012). The empty and full asymmetric units were manually placed on the respective segmented densities.
All initial rigid fits were assessed using an independent global cross-correlation coefficient (CCC) score as described previously and implemented in our in-house code, TEMPy (Vasishtan and Topf, 2011; Vasishtan, Farabella, Pandurangan, and Topf, in preparation). TEMPy code is based on Python and standard Numpy (http://www.numpy.org/) and Scipy (http://www.scipy.org/) python libraries.
For both CAV7 and EV71, following rigid fitting, segmentation of the density map was performed using the zone tool in Chimera (Pettersen et al., 2004), by using the whole map (empty and full) and zoning 9 Å around the asymmetric unit.
2.3. Flexible fitting
2.3.1. Flex-EM/RIBFIND
Next, we employed Flex-EM to optimise the conformation of the atomic structure in a cryoEM map using real-space refinement (Topf et al., 2008). The method is flexible, allowing the optimisation procedures (a conjugate-gradients minimisation and simulated annealing molecular dynamics) to be applied to any groups of rigid bodies, including user-defined rigid bodies (for example, based on prior knowledge of the structure or visual inspection in the context of the density). However, in a recent paper we showed that Flex-EM refinement could be considerably improved using a careful selection of clustered sets of rigid bodies obtained by RIBFIND (Pandurangan and Topf, 2012a). For a given atomic model, RIBFIND clusters the α-helices and β-sheets (set of β-strands) denoting the individual SSEs into a set of rigid bodies. The SSE definitions are obtained using DSSP (Kabsch and Sander, 1983) and the clustering is done based on parameters defining the spatial proximity between SSEs (Pandurangan and Topf, 2012a).
In the current study, the asymmetric unit proteins (VP1–VP3) of each of the CAV7 comparative models as well as the asymmetric unit proteins of the full EV71 crystal structure were submitted separately to the RIBFIND server (Pandurangan and Topf, 2012b) to calculate sets of rigid bodies. Next, a two-stage refinement protocol was employed (Pandurangan and Topf, 2012a) during flexible fitting of each of the asymmetric units in the respective re-segmented (zoned) maps (empty, full and procapsid at 6.09, 8.23, and 8.78 Å resolution, respectively). In the first stage of refinement, the RIBFIND rigid body set with maximal number of rigid bodies was given as an input to Flex-EM. In the second stage, the fits were further “relaxed” by keeping only the SSEs (and some interface loops) as rigid bodies. In both stages Flex-EM refinement cycles were carried out until the CCC values converged. Similarly for actin, the RIBFIND rigid bodies were obtained for the homology model and flexibly fitted into a simulated map at 9 Å resolution using the same two-stage refinement protocol. This protocol had been shown to significantly improve flexible fitting (Pandurangan and Topf, 2012a,b).
2.3.2. iMODfit
To increase the confidence in our results, we also employed a different flexible fitting method – iMODfit (Lopez-Blanco and Chacon, 2013). The method works on the principle of normal mode analysis using internal coordinates (Lopez-Blanco et al., 2011). In general, internal coordinates are used to describe the molecular geometry using the properties including the bond length, bond angle and dihedral angles. In iMODfit, the ϕ and ψ dihedral angles are used to explore the internal coordinate space. Fixing some of the dihedral angles (for instance in α-helices and β-sheets) can reduce the search space. The main advantage of iMODfit is the computational speed. The speed depends on the number of normal modes taken into account during fitting and the percentage of fixed dihedral angles.
Here, fitting was performed on the asymmetric units (VP1–VP3) of CAV7 models (empty and full), EV71 full and the actin homology model into their respective density maps using the default setting given on the program’s web page (http://chaconlab.org/methods/fitting/imodfit). The dihedral coordinates of α-helices and β-sheets were fixed during fitting. The following input density cutoff values (threshold) were used: 2.7 for CAV7 empty map, 2.5 for CAV7 full map, 3.0 for EV71 procapsid map and 0.005 for simulated actin map. All density levels below the threshold were not considered in the calculations. The thresholds were selected by visual inspection of the atomic models in the EM maps using Chimera to best describe the EM density.
2.4. Model assessment and final model generation
2.4.1. Segment-based CCC
To quantify and compare the local quality of fits, a segment-based cross correlation score (SCCC) was calculated between the simulated map of each selected local segment of the fit and its corresponding target map using TEMPy. The simulated map of each selected local segment was obtained by convoluting its atomic coordinates into a grid using a Gaussian function. The resolution, box size and the voxel size of the simulated map were kept similar to the target map. Only grid points in the simulated map with values above its lowest threshold value were included in the SCCC calculation. The lowest threshold values represent the lowest positive density value among all the map grid points. A score based on a similar principle has been used previously to dock domains of GroEL into the cryoEM map (Roseman, 2000). SCCC was calculated for two different kind of local segments, one representing individual SSEs and the other representing the individual proteins in the asymmetric unit for the case of virus capsid. In addition, we calculated the global CCC score for the entire asymmetric unit (including all SSEs and loops).
2.4.2. Generation of an improved fit
The fits from the two different programs and the corresponding SSEs’ SCCC scores were used to generate an improved fit. To obtain the improved fit, the likely best fit between the two programs was selected based on the analysis of the global CCC and SCCC score. Starting from the selected fit from one program, an additional round of refinement was performed in Flex-EM by relaxing every SSE that was shown to have a poor fit (in terms of SCCC score) relative to the fit produced by the other program while the rest of the structure was kept rigid.
2.4.3. Final model generation for a whole virus capsid
For the case of virus capsids (CAV7 and EV71), the asymmetric unit of the improved fit was used to generate a 60-mer containing the whole capsid with the oligomer generator utility in VIPERdb (Carrillo-Tripp et al., 2009). From these capsid models, three adjacent asymmetric units were selected with one unit sharing two unique interfaces. Using backbone atoms only, the selected asymmetric units were inspected for clashes on the interface loops using the Find Clashes/Contacts tool in Chimera. The identified clashes were resolved using the Flex-EM CG refinement protocol. The complete refined asymmetric unit was used to construct a final whole virus capsid with VIPERdb.
3. Results
We have outlined a general modelling and fitting protocol for inserting atomic models into intermediate resolution EM maps. The modelling and fitting protocol was tested on two different case studies for which the target fits were known. The first test case was fitting of actin homology model into the simulated map from a known actin crystal structure (PDB ID: 2A40) (Chereau et al., 2005). The idea was to try and separate modelling errors from errors resulting from conformational differences. The second case was the fitting of the crystal structure of EV71 full capsid (PDB ID: 3VBF) into the EV71 strain 1095 procapsid map (EMD-5557). Here the effects of conformational difference in flexible fitting were addressed. The tested protocol was then applied to generate new and improved models of the full and empty capsid of Coxsackievirus A7 (CAV7) and the two conformational states were characterised. The results of the studies are discussed below.
3.1. Homology modelling and fitting of actin
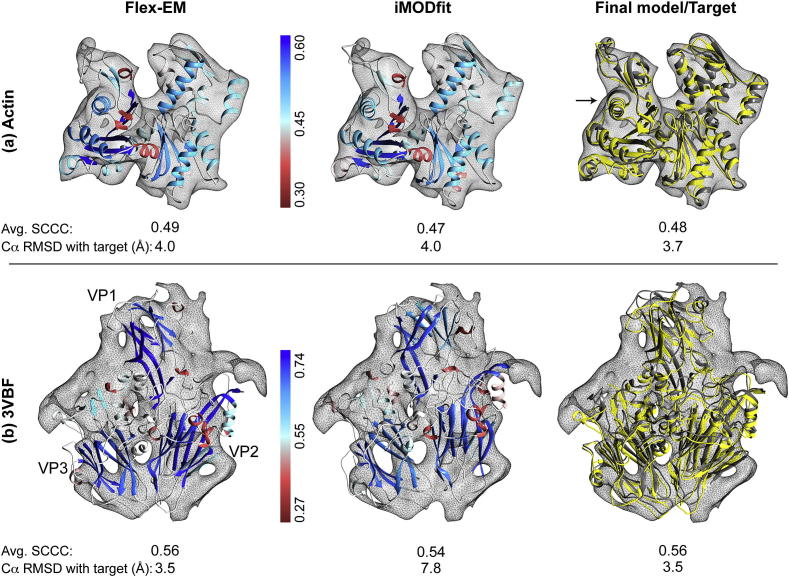
The actin homology model was flexibly fitted using Flex-EM and iMODfit into a 9 Å simulated map from actin crystal structure (Target fit PDB ID: 2A40, chain A). The map was generated using the Chimera molmap command (Pettersen et al., 2004). The Cα RMSD and the global CCC between the rigidly fitted model and the target fit (2A40) are 5.0 Å and 0.89 respectively (Table 1). After flexible fitting, the global CCC values for Flex-EM and iMODfit improved to 0.94 and 0.93 respectively (Table 1). The SCCC scores of individual SSEs were calculated for the Flex-EM and iMODfit and represented on the respective fits (Fig. 2a). In the case of Flex-EM fit, for 84% (16/19) of SSEs, the SCCC values are higher or equal to iMODfit (Table S1). The average SCCC values of SSEs are 0.49 and 0.47 for Flex-EM and iMODfit, respectively.
Table 1.
Comparison of cross correlation scores for actin and EV71.
| Test case | Cross correlation score | Rigid fit | Flex-EM | iMODfit | Final a |
|---|---|---|---|---|---|
| Actin | CCCb | 0.89 | 0.94 | 0.93 | 0.94 |
| EV71 | |||||
| VP1 | SCCCc | 0.75 | 0.81 | 0.80 | 0.82 |
| VP2 | 0.83 | 0.85 | 0.83 | 0.85 | |
| VP3 | 0.84 | 0.84 | 0.83 | 0.84 | |
| VP1, VP2, VP3 | CCCb | 0.67 | 0.73 | 0.73 | 0.73 |
“Final” refers to the model resulting from a final refinement step of Flex-EM using information from the assessment of fits by Flex-EM and iMODfit.
CCC is the global CCC calculated for the asymmetric unit composing VP1, VP2 and VP3.
SCCC is the segment-based CCC calculated separately for VP1, VP2 and VP3.
Fig.2.
Comparison of fits obtained using Flex-EM, iMODfit and the final refined model. (a) Fits of the homology model of actin into the simulated map obtained using Flex-EM (left) and iMODfit (middle), and the final refined model (shown in yellow) in comparison with the target fit (PDB ID: 2A40, shown in grey) (right). (b) Fits of the asymmetric unit of EV71 mature capsid into the procapsid map of EV71 obtained using Flex-EM (left), iMODfit (middle) and the final refined model (shown in yellow) in comparison with the target fit (PDB ID: 4GMP, shown in grey) (right). In (a) and (b) the Flex-EM and iMODfits models are coloured based on their respective segment-based cross correlation score of individual SSEs (SCCC, see Methods). The colour gradient for each SSE was selected based on its respective SCCC score using the Render by Attribute function in Chimera (Pettersen et al., 2004). The averaged SCCC score over all SSEs is indicated below each fit. The colour gradient scales in panel (a) and (b) are shown as vertical bars. In (a), the arrow points to the fit (helix residues 76–88) that improved during the refinement of the final model.
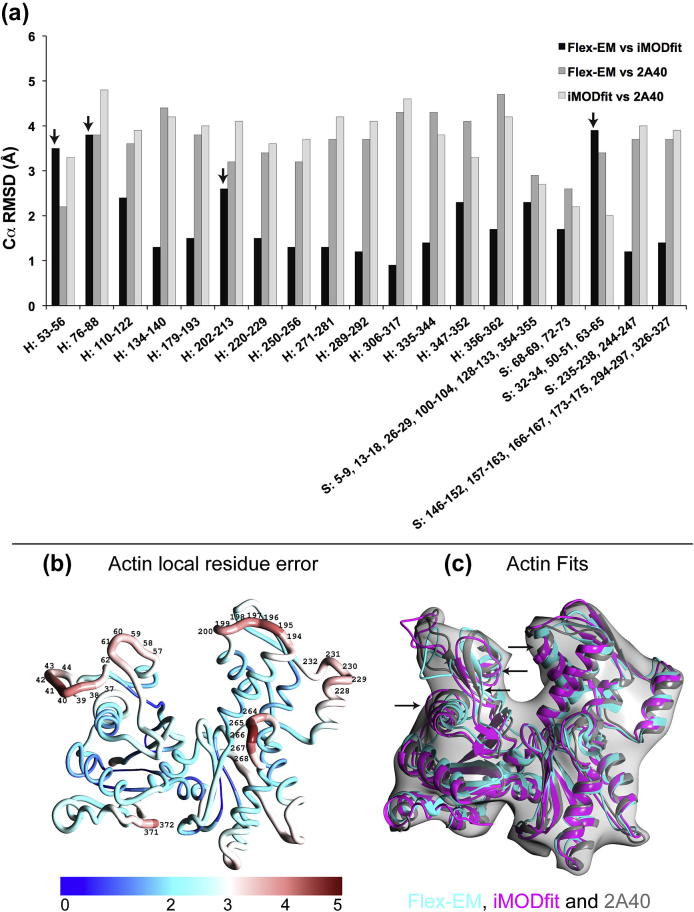
Fig. 3a shows the Cα RMSD between Flex-EM and iMODfit for individual SSEs. The figure also shows the Cα RMSDs of Flex-EM and iMODfit with respect to the target fit (2A40). The average Cα RMSD of all SSEs of Flex-EM and iMODfit with respect to the target fit is 3.6 and 3.7 Å, respectively (Table 2).
Fig.3.
Analysis of Cα RMSDs for individual SSEs and modelling errors for the case of flexible fitting of actin subunit homology model into the simulated map. In (a) three different RMSD comparisons are shown (Flex-EM vs. iMODfit, Flex-EM vs. target fit and iMODfit vs. target fit). The target fit corresponds to the PDB ID 2A40. (b) The actin homology model coloured using the QMEAN local residue error values (in Å) from the lowest (blue) to the highest (red). The range of local residue error values and its corresponding colour gradient is shown below (b). Error values above 3.5 Å that are considered unreliable are labelled. (c) Comparison of flexible fits obtained using Flex-EM (cyan), iMODfit (magenta) and the target fit (PDB ID 2A40) (grey). The arrows in (c) shows SSEs (helices 53–56, 76–88 and 202-213, and the sheet 32–34, 50–51, 63–65) with low consensus fit (Cα RMSD between Flex-EM and iMODfit >2.50 Å). The four SSEs are directly linked to the unreliable loops shown in (b).
Table 2.
Comparison of Cα RMSDs of rigid, Flex-EM, iMODfit and final fits with the target fit for actin and EV71.
| Test case | Cα RMSD (Å) |
|||||||
|---|---|---|---|---|---|---|---|---|
| All-atom |
Average over all SSEs |
|||||||
| Rigid fit | Flex-EM | iMODfit | Finala | Rigid fit | Flex-EM | iMODfit | Finala | |
| Actin | 5.0 | 4.0 | 4.0 | 3.7 | 3.8 | 3.6 | 3.7 | 3.6 |
| EV71 | 5.6 | 3.5 | 7.8 | 3.5 | 5.0 | 2.9 | 6.3 | 2.8 |
“Final” refers to the model resulting from a final refinement step of Flex-EM using information from the assessment of fits by Flex-EM and iMODfit.
Overall the Cα RMSD from the target fit decreased from 5.0 to 4.0 Å with both Flex-EM and iMODfit (Table 2). The difficulty in convergence may be due to the inherent modelling errors in the starting homology model. To understand the effects of modelling errors on the quality of the fit, the QMEAN server was used to calculate the local residue error for the initial homology model. Residues with an estimated error above 3.5 Å were considered to be unreliable (Benkert et al., 2008). Accordingly, six loop segments in the homology model were identified to be unreliable (residues 37–44, 57–62, 194–200, 228–232, 264–268 and 371–372) (Fig. 3b). All the four SSEs (Helices 53–56, 76–88 and 202–213; sheet 32–34, 50–51, 63–65) with low consensus fits (Cα RMSD between Flex-EM and iMODfit >2.50 Å) were linked to the loop segments identified to be unreliable (Fig. 3a and c) demonstrating how errors in the model can impose limitations on the fitting programs to converge to the target fit.
In order to emphasise the usefulness of the step involved in generating the hybrid final model, we took the fit obtained from iMODfit and refined it using Flex-EM by relaxing all the SSEs that had lower SCCC values compared to Flex-EM fit (Table S1). In the hybrid final model obtained, the SCCC values remained either the same or improved for 84% of the cases with an average SCCC value of 0.48 (Fig. 2a and Table S1). The all-atom Cα RMSD and the Cα RMSD averaged over all SSEs between the final model and the target fit decreased to 3.7 and 3.6 Å respectively (Table 2). Particularly, the SCCC values of SSE 76–88 improved from 0.51 to 0.54 (Table S1). This improvement in the SCCC score corresponds to conformational changes leading to an improved fit in the final model (after refinement, the Cα RMSD of the SSE to the target structure decreased by 0.7 Å) (Fig. 2a).
3.2. Fitting the crystal structure of mature EV71 into procapsid map
The full asymmetric unit of EV71 was rigidly fitted into the zoned density map and flexible fitting was performed using Flex-EM and iMODfit. The global CCC values for the initial rigid fit and the two flexible fits were 0.67 (Chimera), 0.73 (Flex-EM) and 0.73 (iMODfit) (Table 1). For both Flex-EM and iMODfit, the SCCC scores for VP1 improved when compared to the corresponding rigid fit (Table 1). The average Cα RMSD over all SSEs between initial rigid fit, Flex-EM and iMODfit with respect to the target fit was 5.0, 2.9 and 6.3 Å, respectively (Table 2). iMODfit refinement resulted in the fit that has the largest deviation from the target fit. The average SCCC score of all SSEs is similar between Flex-EM and iMODfit, with values of 0.56 and 0.55, respectively (Table S2). However, further analysis shows that the Cα RMSD of the individual SSEs between Flex-EM and iMODfit showed considerable differences (with average and standard deviation over all SSEs is 6.7 and 4.1 Å respectively) (Fig. S1) making the choice of the better fit based on cross correlation scores only particularly challenging. In the case of Flex-EM fit, for 69% (18/26) of SSEs, the SCCC values are higher or equal to iMODfit (Table S2). A direct one to one comparison of the SSEs’ SCCC scores between two different fits may act as an indicator to access the quality of the local fit relative to one another. For example, overall the β-sheets forming part of the core β-sandwich in VP1 (S6 and S4), VP2 (S9 and S10) and VP3 (S7 and S8) have higher SCCC values for Flex-EM fit than iMODfit (Table S2). It is interesting to note that for VP1 and VP2, the Cα RMSD between the core β-sheets (VP1 residues 106–110, 150–156, 178–182, 232–237 and VP2 residues 101–102, 133–140, 168–180, 259–264, 301–317) and their corresponding β-sheets in the target fit (4GMP) are considerable lower for Flex-EM than iMODfit (Fig. S1) suggesting that the core of the VP proteins are fitted better by Flex-EM (which is also correlated with the higher SCCC values).
Comparing fits from two or more different programs may result in the identification of the regions of similar fits (consensus) as well as those of different fits (non-consensus) and hence their reliability (Ahmed and Tama, 2013). The lower the Cα RMSD values between the corresponding SSEs refined by Flex-EM and iMODfit the better the consensus between two fits. In Fig. S1 for most of the SSEs showing good consensus fit (<5 Å) between Flex-EM and iMODfit, the corresponding Cα RMSD from the target fit is significantly lower (in both Flex-EM and iMODfit) compared to SSEs with non-consensus fit. The SSEs with non-consensus fits indicate possible spurious fits. For example, for the SSEs in VP1 (H:169–172) and VP2 (H:126–128 and H:159–167), Flex-EM and iMODfit produced non-consensus fits (Cα RMSD between Flex-EM and iMODfit >10 Å) (Fig. S1). After constructing the whole capsid with VIPERdb oligomer generator tool using the fit produced by iMODfit, the above mentioned SSEs were found to be involved in the interface clashes between the asymmetric unit. Additionally, for the β-hairpin found in VP2 (S:83–87, 90–94), both Flex-EM and iMODfit did not produce a consensus fit. The SCCC score for the β-hairpin using Flex-EM (0.56) was slightly lower than iMODfit (0.58). However, Cα RMSD of the β-hairpin with the target fit by Flex-EM (4.1 Å) was considerably lower than iMODfit (11.0 Å), which demonstrates a situation of over-fitting. Thus, the knowledge about the variations of individual local fits with the model (consensus as well as non consensus) produced by multiple programs can be used as a tool for validating fits (Ahmed and Tama, 2013). In conjunction with the comparison of SCCC values of individual SSEs, the fits could possibly be improved using a further hybrid refinement.
Starting from the Flex-EM fit, we tried to generate a hybrid final model (using Flex-EM) by further refining only the SSEs that have lower SCCC values compared to the corresponding SSEs in the iMODfit fit. Following this step, SCCC values either marginally increased or remained unchanged for 22 out of 24 cases. However, the average SCCC value remained the same (0.56) before and after the final refinement (Table S2 and Fig. 2b) and the marginal improvements in the individual SCCC values after refinement suggested possible convergence.
From the final model of the asymmetric unit, the whole capsid was constructed using VIPERdb. It is worth noting that the proposed hybrid refinement step will be more advantageous when the individual SSEs undergoing refinement have significantly worse local fits.
3.3. Modelling and characterising the conformational states of CAV7
We have previously modelled three of the proteins of CAV7 capsid (VP1–VP3) using I-TASSER in two conformations with (full) and without RNA (empty) icosahedral reconstructions at sub-nanometer resolution (Seitsonen et al., 2012). One comparative model for each of the three proteins was generated based on remotely-related templates (Seitsonen et al., 2012). All the templates used were mature (full) capsid forms except for one empty capsid template (PDB: 1POV). The best sequence identity of those templates is 42% (VP1), 58% (VP2) and 52% (VP3) to the respective CAV7 sequences. Since then, crystal structures for empty (PDB: 3VBO) and full states (PDB: 3VBF) of a Human enterovirus A species, EV71, became available (Wang et al., 2012) with significantly higher sequence identity to CAV7 of 60% (VP1), 84% (VP2) and 76% (VP3). Using the latter structures as templates in the current work gave more reliable CAV7 comparative models for refinement in the maps. The I-TASSER score (C-score) for all the six comparative models were considered good except for the empty model of VP3 (Table 3). Additionally, we calculated the QMEAN scores to access the quality of the models. For all six comparative models the QMEAN scores were higher than the corresponding previous models (Table 3). The QMEAN error values for individual residues in the new models were compared with the previous models. Overall, the local residue error is similar between the old and new models. However, the average of residue error of the residues in the core β-sandwich of VP1, VP2 and VP3 show lower residue error in the new models than the old ones. In addition, there are more errors in the C-terminal regions of the old homology models compared to new ones. Among the three proteins, the most improved models were of VP1. We still considered the N-terminal (1–73) and C-terminal residues (278–296) in VP1 as well as the N-terminal residues (1–40) in VP3 as unreliable and therefore removed them, but the models contained 47 more amino acids in VP1 and 12 more in VP3 than previously (Seitsonen et al., 2012).
Table 3.
Assessment scores for previous and current comparative models of CAV7 proteins.
| Protein name | Previous modela |
Current model |
||||
|---|---|---|---|---|---|---|
| Full |
Empty |
|||||
| C-score | QMEAN | C-score | QMEAN | C-score | QMEAN | |
| VP1 | 0.60 | 0.33 | 1.93 | 0.42 | 1.87 | 0.47 |
| VP2 | 1.08 | 0.42 | 0.91 | 0.53 | 1.49 | 0.52 |
| VP3 | 1.33 | 0.50 | 1.36 | 0.54 | 1.31 | 0.51 |
Descriptions for the items are: C-score: a confidence score to estimate the quality of the predicted I-TASSER models (C-score of higher value signifies a model with a high confidence and vice-versa); QMEAN: a model quality estimation score based on a single model (It ranges from 0 to 1 with higher values indicating reliable models); “Full” and “Empty” refer to models based on the EV71 template proteins from the full (PDB ID: 3vbf) and empty capsids (PDB ID: 3vbo), respectively.
The previous models for VP1, VP2 and VP3 were obtained from I-TASSER using multiple template structure before the availability of the crystal structure of EV71 empty and full capsid.
3.3.1. Assessment of fits
The starting rigid fits of the comparative models of both empty and full maps optimised in Chimera were assessed using the global CCC (Vasishtan and Topf, 2011). The global CCC values for the asymmetric unit of the empty and full maps were 0.59 and 0.60, respectively. These values are higher than the corresponding asymmetric unit rigid fits of the previously published models using the current segmented maps (0.56 and 0.54, respectively).
The results of the flexible fitting of the individual CAV7 capsid proteins (VP1-VP3) starting from the asymmetric unit rigid fits were compared between Flex-EM and iMODfit. The SCCC scores of the fits of the individual VP proteins are comparable between Flex-EM and iMODfit for both empty and full maps (Table 4). The global CCC score for the asymmetric unit is 0.72 in the empty map using both Flex-EM and iMODfit, whereas in the full map the corresponding scores are 0.72 and 0.73, respectively (Table 4).
Table 4.
Comparison of cross correlation scores calculated for CAV7.
| Protein name | Cross correlation score | Empty |
Full |
||||
|---|---|---|---|---|---|---|---|
| Flex-EM | iMODfit | Final a | Flex-EM | iMODfit | Final a | ||
| VP1 | SCCCb | 0.74 | 0.72 | 0.75 | 0.77 | 0.78 | 0.78 |
| VP2 | 0.75 | 0.76 | 0.75 | 0.77 | 0.79 | 0.79 | |
| VP3 | 0.78 | 0.77 | 0.79 | 0.81 | 0.80 | 0.82 | |
| VP1,VP2,VP3 | CCCc | 0.72 | 0.72 | 0.72 | 0.73 | 0.74 | 0.73 |
“Final” refers to the model resulting from a final refinement step of Flex-EM using information from the assessment of fits by Flex-EM and iMODfit.
SCCC is the segment-based CCC calculated separately for VP1, VP2 and VP3.
CCC is the global CCC calculated for the asymmetric unit composing VP1, VP2 and VP3.
3.3.2. Comparison of pairs of corresponding SSEs in multiple fits
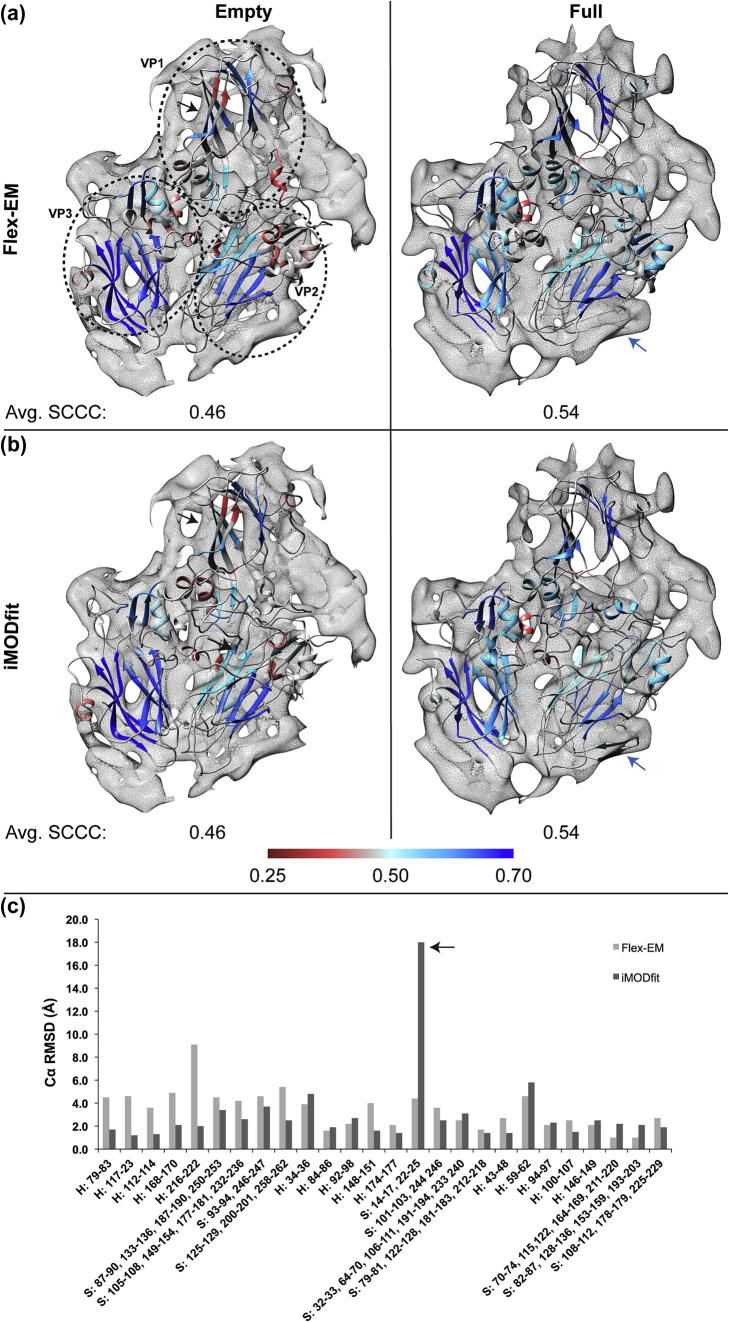
The results of the flexible fitting of the CAV7 capsid proteins (VP1–VP3) were also assessed using the SCCC score of the individual SSEs (see Methods). Fig. 4 shows the comparison of iMODfit and Flex-EM fits in the empty and full asymmetric maps. In all three proteins (VP1–VP3), the largest fraction of the SSEs in each protein corresponds to the core β-sandwich fold composed of eight strands (which is the fold common to the Picornaviridae-like VP family, SCOP entry: 88634).
Fig.4.
Comparison of Flex-EM and iMODfit based model fitting in asymmetric maps of CAV7 empty and full capsid. (a) Fitting of VP1, 2 and 3 models into the asymmetric unit of empty and full maps using Flex-EM. Each protein is shown within a circle (left). (b) Fitting of VP1, 2 and 3 models into the asymmetric unit of empty and full map using iMODfit. The individual SSEs within the fitted models are coloured based on their segment-based cross correlation score (SCCC, see Methods). The averaged SCCC score of all SSEs is indicated below each fit. The colour gradient for each SSE was selected based on its respective SCCC score using the Render by Attribute function in Chimera and its scale is described below the figure. Black arrows indicate a β-sheet (strands 87–90, 133–136, 187–190, 250–253), which is fitted better using Flex-EM. Blue arrows indicate the β-hairpin (residues 14–17, 22–25) that is likely to be overfitted by iMODfit. (c) Comparison of Cα RMSDs for individual SSEs between the CAV7 empty and full fits of Flex-EM and iMODfit. X-axis indicates the SSE residue range with prefix indicating the type of SSE (H: for helix and S: for β-sheet). The arrow highlights the large conformational change observed by iMODfit for β-hairpin (residues 14–17, 22–25), which is likely to be a result of overfitting (see also in (a) and (b)).
3.3.2.1. Empty map
Comparison of the fits in the empty map indicated that on average the β-sandwich fitted equally well using both methods in the cases of VP2 and VP3 (similar gradient colouring based on SCCC, Fig. 4a and b). However in the case of VP1, the Flex-EM fit of one of the two sheets of the β-sandwich was better than the iMODfit fit (strands 87–90, 133–136, 187–190, 250–253; Fig. 4a (left) in blue and Fig. 4b (left) in light blue) with respective SCCC values of 0.62 and 0.58. The Cα RMSD between the Flex-EM and iMODfit fits for VP1 in the empty map is the highest among the three VP proteins (4.6 Å). There are some additional helices and sheets in all three proteins (VP1, 2 and 3) that were not well fitted using either Flex-EM or iMODfit. For instance, in Flex-EM, helices 216–222, 92–98, 43–48 and sheet 14–17/22–25 have a low SCCC relative to their respective iMODfit results (Table S3). Their corresponding SCCC values with Flex-EM are 0.32, 0.42, 0.39 and 0.42 and with iMODfit are 0.44, 0.45, 0.44 and 0.49. Similarly, with iMODfit, helices 79–83, 117–123, 146–149 and sheet 108–112/178–179/225–229 have a low SCCC relatively to their respective Flex-EM results. The corresponding SCCC values with iMODfit are 0.40, 0.39, 0.38 and 0.61 and with Flex-EM are 0.43, 0.44, 0.41 and 0.64.
3.3.2.2. Full map
In the full map, the fitting results of Flex-EM and iMODfit are more consistent in general, except for VP2. Although the fit of the β-sandwich of VP2 is very similar in both methods, the fit of a β-hairpin present in the C-terminus at the interface between the asymmetric units (residues 14–25) is very different resulting in a relatively higher Cα RMSD between the two fits (4.9 Å) (Fig. 4a and b). The SCCC values of the hairpin for Flex-EM and iMODfit are 0.43 and 0.48, respectively. However, it is worth noting that even though the β-hairpin fit using iMODfit appears to be better with a higher SCCC, overfitting of the hairpin may be inferred from the low consensus between the fits (higher Cα RMSD between them) (see Discussion). Similar overfitting by iMODfit for the hairpin was observed while fitting the EV71 full asymmetric unit into the procapsid map.
Interestingly, the average SCCC for overall SSEs is very similar between Flex-EM and iMODfit (0.46 ± 0.13 for the empty map and 0.54 ± 0.09 for the full map) suggesting that the quality of fits from the two programs is similar. The average Cα RMSD of all SSEs between the fits obtained by the two methods is 2.83 Å for the empty and 2.37 Å for the full map.
3.3.3. Conformational changes observed between empty and full fits
Examining the refined models within both empty and full maps allowed us to observe conformational changes between the two states at the level of individual SSEs (Fig. 4c). In 16 out of 26 SSEs, Flex-EM showed more conformational variability between the empty and full fits relative to iMODfit. Out of these 16 SSEs, 14 showed higher SCCC in Flex-EM for both empty and full fits (Table S3). With iMODfit, out of the 10 SSEs that showed more variability, only 7 had higher SCCC for both empty and full fits (Table S3).
In the case of the β-hairpin mentioned above (strands 14–17, 22–25), the Cα RMSD between the empty and full fits obtained using Flex-EM and iMODfit was 4.4 and 18.0 Å, respectively (Fig. 4c). Interestingly, the homologous β-hairpin found in the crystal structure of EV71 virus showed a deviation of 4.4 Å RMSD between the two forms.
3.3.4. Generation of final models of the whole virus capsid
To generate an improved final fit, we used the two final fits (empty and full) of Flex-EM and refined every SSE that was shown to have a worse fit than the corresponding iMODfit fit, while keeping all the loops connecting all the SSEs flexible. The resulting fits were used to generate the whole capsid models with VIPERdb (Carrillo-Tripp et al., 2009). Clashes between the asymmetric units were identified using Chimera (see Methods).
For the empty capsid model, interface loop residues of VP1 (141–148 and 236–245), VP2 (37–63 and 219–231) and VP3 (170–192 and 204–210) were found to have clashes. For the full capsid model, clashes were only observed in the interface loop residues of VP1 (141–148 and 236–245). For each of the empty and full capsids, all the interface clashes were resolved (see Methods) and the final capsid model was generated using VIPERdb (Fig. 5a and b). The new updated coordinates have been deposited in the PDB with the accession codes 4BIP and 4BIQ for full and empty capsid, respectively.
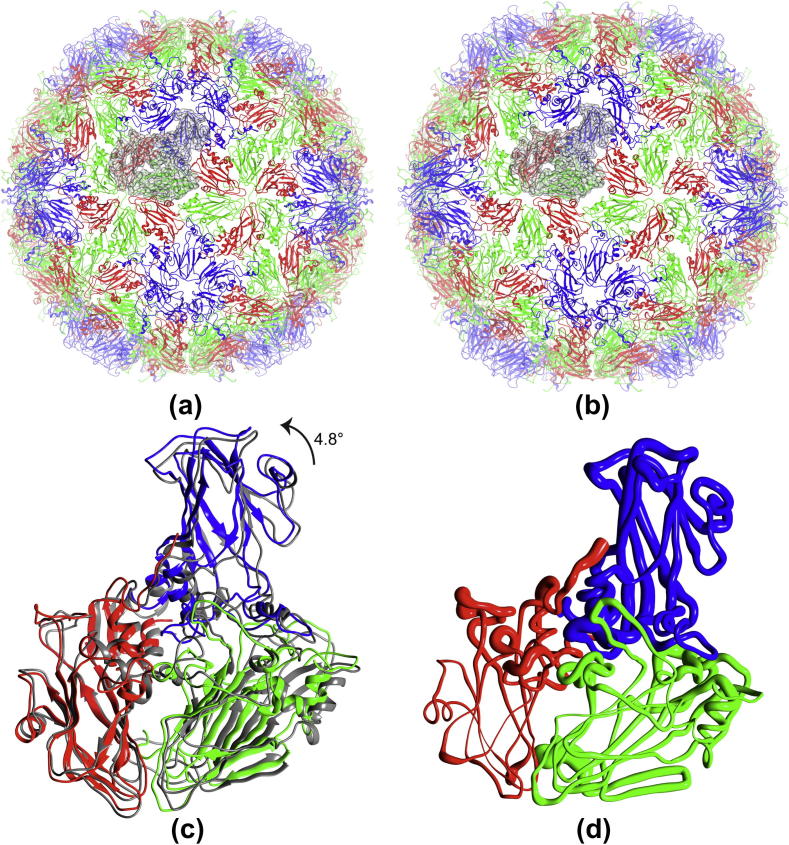
Fig.5.
Pseudo atomic models of CAV7 empty and full capsids. (a and b) Fitted model for the complete full and the empty capsids, respectively. EM density for one asymmetric unit is shown as transparent surface in the background in both. (c) Conformational changes at an asymmetric unit level between full (coloured grey) and empty (non-grey) capsids shown by superposing the final models of the full and empty asymmetric unit. (d) Structural differences mapped onto the empty asymmetric unit using the worm representation. The thickness of the worm from smallest to largest reflects the local deviation (per-residue backbone RMSD) from smallest to largest between the empty and full asymmetric units. The backbone RMSD ranges between 0.46 and 12.45 Å. In (a–d) VP1, 2 and 3 are coloured as blue, green and red, respectively.
The global CCCs of the final fits of both empty and full asymmetric units were similar to the original respective Flex-EM and iMODfit fits (Table 4 and Fig. S2). The SCCC scores of the individual proteins VP1-3, in both empty and full for the final fit is shown in Table 4. Among 26 SSE elements of the empty asymmetric unit, 19 had equal or higher SCCC in the final fit compared to the iMODfit fit and 14 compared to the Flex-EM fit. In the full capsid, 18 SSEs had equal or higher SCCC compared to the iMODfit fit and 14 compared to the Flex-EM fit (Table S3). The average SCCCs of all SSE fits for the empty and full asymmetric units remained unchanged in comparison to the Flex-EM fit (0.46 and 0.54, respectively). However, further analysis of the SCCC scores in the improved final fit shows that while most of the individual SSE fits remained approximately the same (either improved by 40% or worsened by 20%, relative to the models refined by each method individually) there was one fit in VP1, of helix 216–222, which was improved more significantly, especially for the empty case (40% for empty and 9% for full).
Based on the improved final models of empty and full capsids, the capsid expansion seems to be accompanied by a ∼4.8° counter clockwise rotation of the asymmetric unit (viewed perpendicular to the plane of Fig. 5c). This change is in close agreement with the 5.4° rotation observed in EV71 crystal structures (Wang et al., 2012). The component placement scores (CPS) for the individual viral proteins VP1, 2 and 3 (Seitsonen et al., 2012; Zhang et al., 2010) and the Cα RMSDs between the full and empty capsids indicate that the largest conformational change during capsid expansion corresponds to VP1 (4.4 Å RMSD, CPS: 3.8 Å, 1.8°) (Table 5 and Fig. 5d). This observation is in agreement with the analysis of the EV71 crystal structures, where the capsid protein VP1 was found to be predominantly associated with capsid expansion (Wang et al., 2012). Additionally, the area-based component placement score (ACPS, (Pandurangan and Topf, 2012a)) indicates that the conformational changes observed between empty and full capsids for VP1 are almost two-fold larger than those observed for VP2 and VP3 (Table 5).
Table 5.
Comparison of Cα RMSDs, component placement score (CPS) and area based CPS (ACPS) for the individual CAV7 proteins.
| Protein name | Flex-EM vs. iMODfit (Cα RMSD in Å) |
Final empty model vs. final full model |
|||
|---|---|---|---|---|---|
| Empty | Full | Cα RMSD (Å) | CPSa (Å, °) | ACPSb (Å2) | |
| VP1 | 4.6 | 3.8 | 4.4 | 3.8, 1.8 | 0.23 |
| VP2 | 4.0 | 4.9 | 3.3 | 1.4, 7.6 | 0.13 |
| VP3 | 3.5 | 4.0 | 2.6 | 1.5, 5.0 | 0.10 |
CPS is the component placement score (Seitsonen et al., 2012; Zhang et al., 2010). The pair of values in the CPS score (Å, °) corresponds to the component’s translation in Angstrom and rotation in degrees respectively.
ACPS score combines the values of translation and rotation into a net score by calculating the area of the sector whose radius and angle correspond to the translation and rotation values, respectively (Pandurangan and Topf, 2012a).
4. Discussion
Like in the fitting of many structures of assembly components into the lower-resolution density map of their assembly, fitting into a virus capsid map can be quite challenging, particularly when the crystal structures of the components are not available and an atomic model has to be predicted prior to the fitting. Additionally, due to conformational changes occurring in the virus morphology, the resulting maps often represent multiple conformations. To accurately model these conformations, flexible fitting of the atomic models into EM maps is necessary. Although, there are many different methods available to perform flexible fitting (Beck et al., 2011; Esquivel-Rodriguez and Kihara, 2013), a general approach for assessment of fits produced from such methods is lacking in the field. In this paper, we proposed a protocol for comparative modelling, multi-step hybrid flexible fitting and assessment of pseudo-atomic models within intermediate-resolution EM maps. We tested it on two model cases – one virus capsid (EV71) and one non-viral protein (actin), and applied it to the experimental case of CAV7 virus capsid expansion. The protocol can be extended to other assemblies including, clearly, those cases where comparative modelling is not required. Additionally, it is not restricted to the fitting methods used here (Flex-EM and iMODfit) but could be applied to any combination of flexible fitting methods (Ahmed and Tama, 2013).
In the following sections, we have attempted to point out various technical issues that might arise during model building, fitting and assessment as well as emphasise the advantages of using a hybrid approach such as the one adopted in this paper.
4.1. Model truncation
In virus capsid proteins that form icosahedrally-symmetric capsids, the terminals are highly flexible so that they can easily interact with the encapsidated genome, can perform protein-protein interactions spanning across the capsid, and conformational switching required for quasi-equivalent interactions (Abrescia et al., 2004; Seitsonen et al., 2010; Stehle et al., 1996; Williams et al., 2004; Xing et al., 2004). Such flexible ends are not easily resolved as the chains are often in an extended conformation (Seitsonen et al., 2012), which is likely to lead to low confidence in their fit. A successful attempt to include flexible termini strongly depends on the resolution of the density map. However, even with a sub-nanometer resolution map (5–10 Å), this remains a challenge. Due to the above reasons, we removed the highly flexible terminal loops of the models of CAV7 and EV71 capsid proteins prior to fitting.
4.2. Segmentation issues
There are a number of automated segmentation methods available, which are predominantly based on watershed (Pintilie et al., 2010; Volkmann, 2002) or fast-marching algorithms (Bajaj et al., 2003; Zhang et al., 2012). However, the accuracy of these methods depends on various factors including the resolution, parameters inherent to the segmentations methods as well as manual intervention in specific cases (Pintilie et al., 2010). Unguided manual segmentation is time-consuming but can be more accurate when carried out iteratively. For example, for a cryoEM map of the whole virus capsid, manual segmentation relies on the knowledge of the icosahedrally-arranged protein subunits in the capsid.
Regions that are difficult to segment often lie at the interfaces of the subunits spanning between and within the asymmetric units. This challenge comes from the fact that flexible terminal regions of one subunit often intertwine with other subunits (Jäälinoja et al., 2007; Seitsonen et al., 2012). Here, we were able to manually segment out unambiguously all the capsid proteins of CAV7 except VP4 (which is highly unstructured and lies close to the flexible VP1 N-terminal) and the termini of VP1 and VP3. Rigid fitting guided by this initial segmentation allowed us to re-zone around the asymmetric unit (Seitsonen et al., 2012), thereby reducing some interface errors between subunits, and allowing identification of some of the segmentation errors on the interfaces between adjacent asymmetric units.
4.3. A hybrid approach for flexible fitting
Comparing fits obtained from independent programs can be ideally used as a tool for identifying spurious local fits and to aid the generation of an improved model. Recently, the idea of combining different fitting programs in order to identify a consensus fit and measure its local reliability using root mean square fluctuations (RMSF) has been introduced (Ahmed and Tama, 2013; Ahmed et al., 2012). Here, we applied the principle of consensus between fits based on multiple methods in a different way. First, we calculated a different local reliability measure – the SCCC score – for each pair of corresponding SSE fits generated by two methods (Flex-EM and iMODfit) and identified local variations between them. The scores became even more informative when mapped onto the structure and used as a comparison tool within Chimera (Pettersen et al., 2004). Based on the comparison, we selected one of the fits and improved it by only refining the SSEs that had low SCCC values compared with the other fit. Although we used only two methods, ideally our approach can potentially be expanded to multiple methods and combined with the RMSF measure described in Ahmed et al. (Ahmed and Tama, 2013; Ahmed et al., 2012) to achieve even better results.
4.4. Modelling errors and fitting
Flexible fitting of atomic models into the density map provides insight into the function and the dynamics of the system under study. The interpretation becomes more challenging as the number of errors in the atomic model increases. Identifying those errors and their potential effect on the outcome of the flexible fitting procedure can be helpful in fit assessment. In the actin homology model we identified six loops with modelling errors (identified by QMEAN local residue score) and showed that Flex-EM and iMODfit could not produce consensus fit for most of the SSEs attached to those loops. The study not only suggests the possibility of incorporating the information about modelling errors to improve flexible fitting, but also demonstrates how useful this information can be in combination with the use of multiple flexible fitting programs.
4.5. Over-fitting
In general, overfitting can occur when the fit that is being optimised has neighbourhood densities that are not well resolved (for example, in virus capsids this could occur on the interface between asymmetric units if the proteins are fitted into a map segmented around the asymmetric unit). In this situation, the fit may be optimised into an incorrect position in the density which is termed overfitting. Using Flex-EM in conjunction with multiple sets of rigid bodies (assigned by RIBFIND), we previously showed that a two-stage refinement protocol can reduce over-fitting and thereby improve flexibly fitted models (Pandurangan and Topf, 2012a). This idea has shown to be useful in the current study as well. Although, on average both Flex-EM and iMODfit produced similar fits, the use of a two-stage refinement protocol helped avoiding over-fitting in Flex-EM, for example in fitting the β-hairpin found in VP2 protein of CAV7 and EV71. Additionally, here we show that using a “local” score, such as the SCCC, in combination with structural comparison of fits from different programs can help in identifying regions that might raise ambiguity (such as the β-hairpin). Additionally, by refining the fit of the asymmetric unit as a whole rather than the individual proteins we avoided fitting errors within the interfaces (compared to our previous study) (Seitsonen et al., 2012).
4.6. Capsid asymmetric unit interface
Refining loops at the interface between the asymmetric units is challenging as they can often clash. Here, clashes were identified when we constructed the whole capsid from the asymmetric unit. The loops were refined considering only the symmetrically-related neighbouring asymmetric units. Symmetry-based refinement programs may be a better solution to avoid such problems (Chan et al., 2011) at sub-nanometer resolution. However, for intermediate to low-resolution maps, the refinement of asymmetric unit interface (especially flexible loops) remains a challenge.
4.7. Current vs. previous CAV7 models
It is interesting to observe that the improved final fits obtained using the new models for the empty and full asymmetric units were quite similar to their respective initial rigid fits. As the new models were more complete, there was less ambiguity for movement of subunits within the densities. As a result, the overall changes now seen between the two states are more moderate than previously reported (Seitsonen et al., 2012) and the interaction interfaces are better defined. These new findings are significant if, for example, one tries to inhibit the interaction with neutralising antibodies that would recognise one of the states. Nevertheless, the conclusions about the important regions for the release of RNA are still in agreement with the previous report and with the movement seen in the case of EV71 when it goes from an immature state to a mature, RNA-filled state (Seitsonen et al., 2012; Wang et al., 2012).
5. Conclusion
In this paper we describe a protocol for comparative modelling, fitting and assessment of atomic structures into sub-nanometer resolution cryoEM density maps and highlighted various important issues pertaining to it. We applied the protocol in order to improve the modelling of CAV7 virus capsids in two conformations, which resulted in better agreement between the model and the experimental data of both CAV7 and its homolog EV71. Ideally, the protocol could be applied to any system and is not restricted to capsid modelling (as demonstrated for the actin test case). We showed that the refinement process is worth addressing in multiple progressive steps combined with model and local fit assessment. Such an approach would provide more control and allow the check of model quality at various steps leading to more accurate and complete pseudo-atomic models.
Acknowledgments
We thank Drs. Daven Vasishtan and Irene Farabella for helpful discussions and Dr. David Houldershaw and Richard Westlake for computer support. This work was supported by an MRC Centenary Award (G0600084 to M.T.), the Leverhulme Trust (RPG-2012-519 to M.T.) and BBSRC (BB/K01692X/1 to M.T.), the Academy of Finland (1139178 to S.J.B.), Sigrid Juselius Foundation (S.J.B.) and Helsinki Graduate Program in Biotechnology and Molecular Biology (S.S.).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Appendix A. Supplementary data
Analysis of Cα RMSDs for individual SSEs for the flexible-fitted atomic model of the EV71 mature asymmetric unit into the procapsid map. Three different RMSD comparisons are shown (Flex-EM vs. iMODfit, Flex-EM vs. target fit and iMODfit vs. target fit). The target fit corresponds to the PDB ID 4GMP.
Asymmetric unit fits of Flex-EM, iMODfit and the improved final fit in the empty and full map of CAV7. (a) VP1, 2 and 3 models fitted into the map of an asymmetric unit of the empty (a) and full (b) capsids (shown as mesh) using Flex-EM, iMODfit and the final improved model. The global cross correlation score (CCC) for the asymmetric unit is given below each fit. VP1, 2 and 3 are coloured as blue, green and red, respectively.
Segment-based cross correlation scores (SCCC) for SSEs as calculated for the actin subunit models.
Segment-based cross correlation scores (SCCC) for SSEs as calculated for the asymmetric unit of 3vbf fitted into procapsid map of EV71.
Segment-based cross correlation scores (SCCC) for SSEs as calculated for the models of empty and full capsid proteins refined by Flex-EM and iMODfit.
References
- Abrescia N.G., Cockburn J.J., Grimes J.M., Sutton G.C., Diprose J.M. Insights into assembly from structural analysis of bacteriophage PRD1. Nature. 2004;432:68–74. doi: 10.1038/nature03056. [DOI] [PubMed] [Google Scholar]
- Ahmed A., Tama F. Consensus among multiple approaches as a reliability measure for flexible fitting into cryo-EM data. J. Struct. Biol. 2013;182:66–67. doi: 10.1016/j.jsb.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Ahmed A., Whitford P.C., Sanbonmatsu K.Y., Tama F. Consensus among flexible fitting approaches improves the interpretation of cryo-EM data. J. Struct. Biol. 2012;177:561–570. doi: 10.1016/j.jsb.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj C., Yu Z., Auer M. Volumetric feature extraction and visualization of tomographic molecular imaging. J. Struct. Biol. 2003;144:132–143. doi: 10.1016/j.jsb.2003.09.037. [DOI] [PubMed] [Google Scholar]
- Baker T.S., Olson N.H., Fuller S.D. Adding the third dimension to virus life cycles: three-dimensional reconstruction of icosahedral viruses from cryo-electron micrographs. Microbiol. Mol. Biol. Rev. 1999;63:862–922. doi: 10.1128/mmbr.63.4.862-922.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Topf M., Frazier Z., Tjong H., Xu M. Exploring the spatial and temporal organization of a cell’s proteome. J. Struct. Biol. 2011;173:483–496. doi: 10.1016/j.jsb.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert P., Tosatto S.C., Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Carrillo-Tripp M., Shepherd C.M., Borelli I.A., Venkataraman S., Lander G. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37:D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.Y., Gumbart J., McGreevy R., Watermeyer J.M., Sewell B.T. Symmetry-restrained flexible fitting for symmetric EM maps. Structure. 2011;19:1211–1218. doi: 10.1016/j.str.2011.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K. Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. USA. 2005;102:16644–16649. doi: 10.1073/pnas.0507021102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuente J.O., Lee H., Yoder J.D., Shingler K.L., Carnegie M.S. Structures of the procapsid and mature virion of enterovirus 71 strain 1095. J. Virol. 2013;87:7637–7645. doi: 10.1128/JVI.03519-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquivel-Rodriguez J., Kihara D. Computational methods for constructing protein structure models from 3D electron microscopy maps. J. Struct. Biol. 2013;184:93–102. doi: 10.1016/j.jsb.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard T.D., Huang C.C., Ferrin T.E. Visualising density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Henderson R., Sali A., Baker M.L., Carragher B., Devkota B. Outcome of the first electron microscopy validation task force meeting. Structure. 2012;20:205–214. doi: 10.1016/j.str.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäälinoja H.T., Huiskonen J.T., Butcher S.J. Electron cryomicroscopy comparison of the architectures of the enveloped bacteriophages phi6 and phi8. Structure. 2007;15:157–167. doi: 10.1016/j.str.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Lawson C.L., Baker M.L., Best C., Bi C., Dougherty M. EMDataBank.org: unified data resource for CryoEM. Nucleic Acids Res. 2011;39:D456–464. doi: 10.1093/nar/gkq880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Blanco J.R., Chacon P. IMODFIT: efficient and robust flexible fitting based on vibrational analysis in internal coordinates. J. Struct. Biol. 2013;184:261–270. doi: 10.1016/j.jsb.2013.08.010. [DOI] [PubMed] [Google Scholar]
- Lopez-Blanco J.R., Garzon J.I., Chacon P. IMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics. 2011;27:2843–2850. doi: 10.1093/bioinformatics/btr497. [DOI] [PubMed] [Google Scholar]
- Oberste M.S., Penaranda S., Maher K., Pallansch M.A. Complete genome sequences of all members of the species Human enterovirus A. J. Gen. Virol. 2004;85:1597–1607. doi: 10.1099/vir.0.79789-0. [DOI] [PubMed] [Google Scholar]
- Orlova E.V., Saibil H.R. Structural analysis of macromolecular assemblies by electron microscopy. Chem. Rev. 2011;111:7710–7748. doi: 10.1021/cr100353t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandurangan A.P., Topf M. Finding rigid bodies in protein structures: application to flexible fitting into cryoEM maps. J. Struct. Biol. 2012;177:520–531. doi: 10.1016/j.jsb.2011.10.011. [DOI] [PubMed] [Google Scholar]
- Pandurangan A.P., Topf M. RIBFIND: a web server for identifying rigid bodies in protein structures and to aid flexible fitting into cryo EM maps. Bioinformatics. 2012;28:2391–2393. doi: 10.1093/bioinformatics/bts446. [DOI] [PubMed] [Google Scholar]
- Patwardhan A., Carazo J.M., Carragher B., Henderson R., Heymann J.B. Data management challenges in three-dimensional EM. Nat. Struct. Mol. Biol. 2012;19:1203–1207. doi: 10.1038/nsmb.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pintilie G.D., Zhang J., Goddard T.D., Chiu W., Gossard D.C. Quantitative analysis of cryo-EM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plevka P., Perera R., Cardosa J., Kuhn R.J., Rossmann M.G. Crystal structure of human enterovirus 71. Science. 2012;336:1274. doi: 10.1126/science.1218713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter F.A., Rhodes A.J., Macpherson L.W., Labzoffsky N.A. A possible new enterovirus serotype isolated in Ontario. Arch. Gesamte Virusforsch. 1971;35:218–222. doi: 10.1007/BF01249713. [DOI] [PubMed] [Google Scholar]
- Robinson R.C., Turbedsky K., Kaiser D.A., Marchand J.B., Higgs H.N. Crystal structure of Arp2/3 complex. Science. 2001;294:1679–1684. doi: 10.1126/science.1066333. [DOI] [PubMed] [Google Scholar]
- Roseman A.M. Docking structures of domains into maps from cryo-electron microscopy using local correlation. Acta Crystallogr. D Biol. Crystallogr. 2000;56:1332–1340. doi: 10.1107/s0907444900010908. [DOI] [PubMed] [Google Scholar]
- Rossmann M.G., Morais M.C., Leiman P.G., Zhang W. Combining X-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Kucukural A., Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Sali A., Glaeser R., Earnest T., Baumeister W. From words to literature in structural proteomics. Nature. 2003;422:216–225. doi: 10.1038/nature01513. [DOI] [PubMed] [Google Scholar]
- Seitsonen J., Susi P., Heikkilä O., Sinkovits R.S., Laurinmäki P. Interaction of alphaVbeta3 and alphaVbeta6 integrins with human parechovirus 1. J. Virol. 2010;84:8509–8519. doi: 10.1128/JVI.02176-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitsonen J.J., Shakeel S., Susi P., Pandurangan A.P., Sinkovits R.S. Structural analysis of Coxsackievirus A7 reveals conformational changes associated with uncoating. J. Virol. 2012;86:7207–7215. doi: 10.1128/JVI.06425-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehle T., Gamblin S.J., Yan Y., Harrison S.C. The structure of simian virus 40 refined at 3.1 A resolution. Structure. 1996;4:165–182. doi: 10.1016/s0969-2126(96)00020-2. [DOI] [PubMed] [Google Scholar]
- Topf M., Lasker K., Webb B., Wolfson H., Chiu W. Protein structure fitting and refinement guided by cryo-EM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung C.C., Lobo P.A., Kimlicka L., Van Petegem F. The amino-terminal disease hotspot of ryanodine receptors forms a cytoplasmic vestibule. Nature. 2010;468:585–588. doi: 10.1038/nature09471. [DOI] [PubMed] [Google Scholar]
- Vasishtan D., Topf M. Scoring functions for cryoEM density fitting. J. Struct. Biol. 2011;174:333–343. doi: 10.1016/j.jsb.2011.01.012. [DOI] [PubMed] [Google Scholar]
- Volkmann N. A novel three-dimensional variant of the watershed transform for segmentation of electron density maps. J. Struct. Biol. 2002;138:123–129. doi: 10.1016/s1047-8477(02)00009-6. [DOI] [PubMed] [Google Scholar]
- Volkmann N. Confidence intervals for fitting of atomic models into low-resolution densities. Acta Crystallogr. D Biol. Crystallogr. 2009;65:679–689. doi: 10.1107/S0907444909012876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voroshilova M.K., Chumakov M.P. Poliomyelitis-like properties of AB-IV-coxsackie A7 group of viruses. Prog. Med. Virol. 1959;2:106–170. [Google Scholar]
- Wang X., Peng W., Ren J., Hu Z., Xu J. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 2012;19:424–429. doi: 10.1038/nsmb.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.H., Kajander T., Hyypiä T., Jackson T., Sheppard D. Integrin alpha v beta 6 is an RGD-dependent receptor for Coxsackievirus A9. J. Virol. 2004;78:6967–6973. doi: 10.1128/JVI.78.13.6967-6973.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing L., Huhtala M., Pietiäinen V., Käpylä J., Vuorinen K. Structural and functional analysis of integrin alpha2I domain interaction with echovirus 1. J. Biol. Chem. 2004;279:11632–11638. doi: 10.1074/jbc.M312441200. [DOI] [PubMed] [Google Scholar]
- Zhang S., Vasishtan D., Xu M., Topf M., Alber F. A fast mathematical programming procedure for simultaneous fitting of assembly components into cryoEM density maps. Bioinformatics. 2010;26:i261–268. doi: 10.1093/bioinformatics/btq201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Bettadapura R., Bajaj C. Macromolecular structure modeling from 3D EM using VolRover 2.0. Biopolymers. 2012;97:709–731. doi: 10.1002/bip.22052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of Cα RMSDs for individual SSEs for the flexible-fitted atomic model of the EV71 mature asymmetric unit into the procapsid map. Three different RMSD comparisons are shown (Flex-EM vs. iMODfit, Flex-EM vs. target fit and iMODfit vs. target fit). The target fit corresponds to the PDB ID 4GMP.
Asymmetric unit fits of Flex-EM, iMODfit and the improved final fit in the empty and full map of CAV7. (a) VP1, 2 and 3 models fitted into the map of an asymmetric unit of the empty (a) and full (b) capsids (shown as mesh) using Flex-EM, iMODfit and the final improved model. The global cross correlation score (CCC) for the asymmetric unit is given below each fit. VP1, 2 and 3 are coloured as blue, green and red, respectively.
Segment-based cross correlation scores (SCCC) for SSEs as calculated for the actin subunit models.
Segment-based cross correlation scores (SCCC) for SSEs as calculated for the asymmetric unit of 3vbf fitted into procapsid map of EV71.
Segment-based cross correlation scores (SCCC) for SSEs as calculated for the models of empty and full capsid proteins refined by Flex-EM and iMODfit.