Graphical abstract
Clones isolated from a single benznidazole-resistant Trypanosoma cruzi population contain a stop-codon-generating mutation in the nitroreductase gene TcNTR. Clonal variation in resistance suggests that additional mechanisms must also operate.
Keywords: Trypanosoma cruzi, Drug-resistance, Benznidazole, Nitroreductase
Highlights
-
•
Drug-resistance in T. cruzi can arise independently within a single population.
-
•
Distinct mechanisms can contribute to benznidazole-resistance in T. cruzi.
-
•
Stop-codon generating mutations in the TcNTR gene linked to benznidazole-resistance.
Abstract
Benznidazole is the main drug used to treat Trypanosoma cruzi infections. However, frequent instances of treatment failure have been reported. To better understand potential resistance mechanisms, we analysed three clones isolated from a single parasite population that had undergone benznidazole-selection. These clones exhibited differing levels of benznidazole-resistance (varying between 9 and 26-fold), and displayed cross-resistance to nifurtimox (2 to 4-fold). Each clone had acquired a stop-codon-generating mutation in the gene which encodes the nitroreductase (TcNTR) that is responsible for activating nitroheterocyclic pro-drugs. In addition, one clone had lost a copy of the chromosome containing TcNTR. However, these processes alone are insufficient to account for the extent and diversity of benznidazole-resistance. It is implicit from our results that additional mechanisms must also operate and that T. cruzi has an intrinsic ability to develop drug-resistance by independent sequential steps, even within a single population. This has important implications for drug development strategies.
Chagas disease, which is caused by infection with Trypanosoma cruzi, is the most important parasitic infection in South America. As a result of mobility and migration, the disease is also becoming a global public health issue [1,2], with large numbers of infected individuals now resident in the USA and Europe, for example. The nitroheterocyclic agents benznidazole and nifurtimox are the only drugs currently used to treat T. cruzi infections. However, they have several drawbacks. Their efficacy against chronic stage disease remains uncertain, their therapeutic schedules can stretch over several months, and toxic side effects are a common problem [3]. In addition, parasite strains refractory to treatment are frequently observed [4].
Benznidazole and nifurtimox are pro-drugs and both are activated within the parasite by a mitochondrial NADH-dependent type-I nitroreductase (TcNTR), which utilises FMN as a co-factor [5]. The reduction of benznidazole results in the generation of the cytotoxic metabolite glyoxal [6], while nifurtimox reduction leads to the production of an unsaturated open-chain nitrile which has trypanocidal properties [7]. Cross-resistance to nitroheterocyclic drugs can be readily selected in the laboratory [5,8,9], and can be associated with the loss of one copy of the chromosome containing the TcNTR gene [5,8]. Mutations within the FMN-binding region of TcNTR, which result in loss of enzyme activity, have also been identified in drug-resistant parasites [8]. However, other resistance mechanisms, not involving TcNTR, might operate. For example, instances where resistance to nifurtimox occurs independently of benznidazole-resistance have been reported [10]. Likewise, wide natural variations in benznidazole-sensitivity have been identified in Colombian isolates (EC50 4–30 μM), which are not associated with changes in the TcNTR sequence [8].
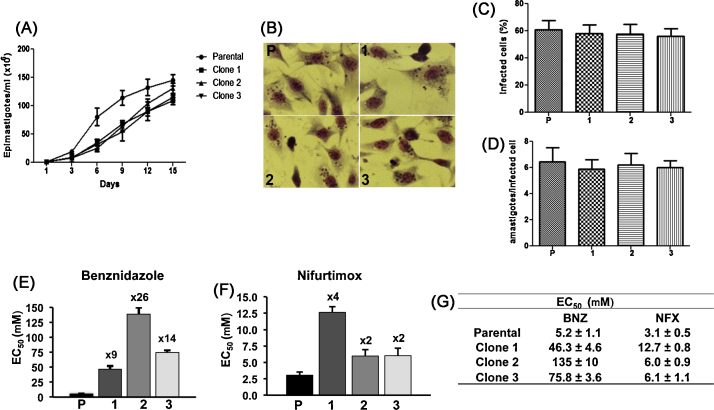
To gain new insights into mechanisms that give rise to resistance against nitroheterocylic drugs, we examined three T. cruzi clones (Y strain) derived from a single population which had been selected for resistance by exposure to increasing concentrations of benznidazole [11]. When these parasites were grown in culture in the absence of benznidazole, the drug-resistant clones grew at about half the rate of the parental non-resistant cells, which had a doubling time of 33 h (Fig. 1A). However, there were no significant differences in the rate of amastigote replication or the extent of infectivity between the parental parasites and any of the drug-resistant clones (Fig. 1B–D). Surprisingly, assessment of the level of benznidazole-resistance revealed significant differences between clones, ranging from 9 to 26-fold (Fig. 1E and G). These results suggest that different and/or additional mechanisms must be contributing to benznidazole-resistance in each case. We also observed that each clone was cross-resistant to nifurtimox, at levels ranging from 2 to 4-fold (Fig. 1F and G). It can be inferred that the mechanism responsible for cross-resistance is insufficient of itself, to account for the complete extent and variability of benznidazole-resistance. In previous cases of cross-resistance, a clear linkage between the levels observed for benznidazole and nifurtimox has been reported [5,8].
Fig. 1.
Characterisation of benznidazole-resistant T. cruzi clones. (A) Growth curves of parental and drug-resistant clones. Epimastigotes were cultured as described previously [15] in 25 cm2 cell culture flasks (in triplicate) at 28 °C and parasite proliferation monitored by counting using a Neubauer chamber. (B–D) Rat skeletal myoblast L6 cells were cultured on glass cover slips in 24-well plates (3 × 105 cells per well) for 24 h in RPMI-1640/10% FBS at 37 °C in 5% CO2. These cells were then infected overnight with metacyclic trypomastigotes obtained from stationary phase epimastigote cultures (5:1 parasite/host cell). Non-internalised parasites were removed by at least three washes. After 5 days, cells were washed twice with PBS, fixed with methanol and stained with Giemsa. Average values for the percentage (%) infected cells and the number of amastigotes were established by examination of at least 200 L6 cells for each parasite clone. Experiments were performed in triplicate, and the values shown are the mean ± standard deviation of the three experiments. No statistical difference was observed among parental parasites and clones following one-way analysis of variance (ANOVA). (E–G) To determine the level of drug-resistance, epimastigotes were seeded into 48-well microtitre plates at 2.5 × 106 parasites ml−1 at a range of benznidazole (BNZ) (Rochagan) or nifurtimox (NFX) (Lampit®, Bayer) concentrations for 6 days at 28 °C. Cell counting was carried out to determine the EC50 values (half maximal effective concentration). Experiments were performed in triplicate (with 3 independent counts in each case) and the values shown are the mean ± standard deviation. The fold resistance of each clone to benznidazole and nifurtimox are indicated above the bars. The benznidazole-resistance levels of the three clones were significantly different from each other (P < 0.0001). In terms of nifurtimox-resistance, each of the clones was significantly more resistant than the parental parasites (P < 0.0001). The levels of resistance displayed by clones 2 and 3 were not statistically different from each other, but both were significantly less than clone 1 (P < 0.0001).
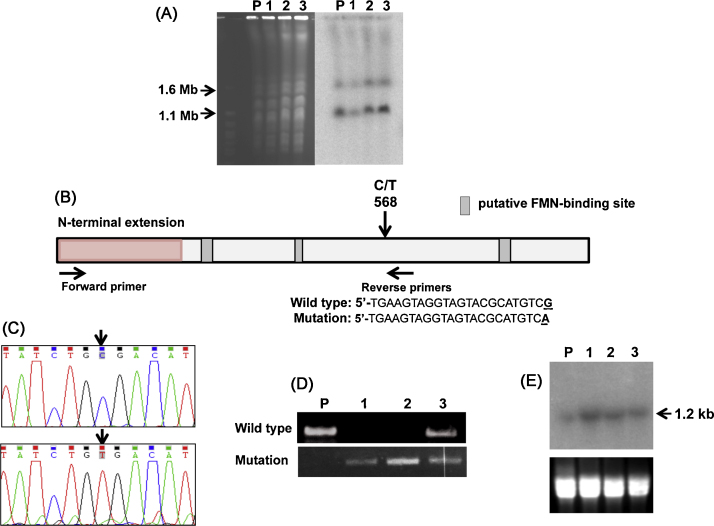
As TcNTR is the main activator of both benznidazole and nifurtimox, we examined the structure and expression of the corresponding gene in each of the resistant clones. Southern blotting revealed an absence of gross rearrangements in the structure of TcNTR. Similarly, karyotypic analysis failed to reveal any major changes in chromosome organisation in the resistant clones, although there was variation in the ploidy of the 1.1 and 1.6 Mb chromosome homologues that contain the TcNTR gene (Fig. 2A). In the parental Y strain and drug-resistant clones 2 and 3, the smaller homologue is present in at least 2 copies, whereas in clone 1, the ratio equates to 1:1. Genome plasticity has been commonly observed in T. cruzi [12] and chromosome loss has been associated with benznidazole-resistance [5,8]. In Y strain, the existence of triploidy in the case of some chromosomes has also been reported [13].
Fig. 2.
Analysis of the structure and expression of the TcNTR gene from benznidazole-resistant parasites. (A) Chromosomal DNA from the parental T. cruzi Y strain (P) and three drug resistant clones (1–3) was immobilised in agar blocks and fractionated by a Bio-Rad CHEF Mapper system using an auto-algorithm set to the designated molecular mass range [12] (left hand image). Chromosomes from Saccharomyces cerevisiae (Bio-Rad, Hercules, CA, USA) were used as molecular mass standards. After Southern hybridisation using a radiolabelled TcNTR probe, the membrane was autoradiographed (right hand image). (B) Schematic of the TcNTR gene showing the region corresponding to the N-terminal extension and putative FMN-binding regions [8,16]. The position of the C/T transition that generates a stop codon and the locations of primers used to differentiate between wild type and mutated genes are shown, together with the sequence of the reverse primers. (C) Electropherograms identifying the C/T transition in wild type (upper) and mutated (lower) genes (GenBank accession number KF731779). (D) Products generated following amplification of the TcNTR gene fragment using the primers shown in (B). PCRs were carried out using 30 amplification cycles; 96 °C for 30 s, 70 °C (wild type reverse primer) or 65 °C (mutation-containing reverse primer) for 30 s, 72 °C for 90 s, and a final extension step at 72 °C for 10 min. Upper inset, wild type reverse primer; lower inset, mutant reverse primer. (E) Northern blot analysis of RNA from T. cruzi Y strain (P) and three drug resistant clones (1–3). Upper inset, autoradiograph following hybridisation with radiolabelled TcNTR probe; lower inset, ethidium bromide stained gel as loading control.
When we examined the sequence of the TcNTR genes in the drug-resistant clones, we found a C/T transition at position 568 in each case (Fig. 2B and C). This generates a stop codon (TGA) in the middle of the gene, resulting in a truncated protein deficient in the putative carboxyl terminal FMN-binding site. To determine whether the parasites were homozygous for this mutation, we used a diagnostic PCR with primers designed to specifically amplify the parental and mutated copies of the gene (Fig. 2D). The results showed that clones 1 and 2 were homozygous, whereas clone 3 had retained a copy of the non-mutated parental allele. These results were confirmed by sequencing multiple amplicon fragments derived from 3 independent PCRs in the case of each clone. Interestingly, when we examined the level of TcNTR RNA expression by Northern blotting, we observed an increased abundance in each of the resistant clones (Fig. 2E). One possibility is that translation of an inactive protein results in a metabolic feedback loop that acts to stabilise the TcTNR transcript, or to otherwise increase the level of transcript expression. The biological role of TcNTR has yet to be unequivocally demonstrated, but evidence suggests that it may function as a NADH-ubiquinone oxidoreductase [6,14].
Our data indicate that several sequential events had a role in producing the range of resistance phenotypes. The most parsimonious explanation is that the first step involved the generation of a stop codon in a TcNTR allele (Fig. 2B) resulting in a progenitor cell with reduced capacity to activate nitroheterocycles. It can be inferred that the remaining intact copies of TcNTR were then removed by gene conversion (clones 1 and 2) and/or loss of one copy of the 1.1 Mb chromosome homologue (clone 1). Furthermore, additional mechanisms must also have acted independently in the cases of clones 1 and 2, and contributed to the enhanced level of benznidazole (but not nifurtimox) resistance. Previous studies [5,8] have shown that TcNTR null mutants display 6- to 10-fold benznidazole-resistance, whereas with clone 2 and the TcNTR heterozygote clone 3, we observed 26 and 14-fold resistance, respectively. These additional mechanisms are unknown, but it is clear that they do not result in cross-resistance to nifurtimox. In the absence of evidence for active uptake in T. cruzi, such resistance mechanisms could involve enzymes or pathways that operate to protect the parasite from toxic products of benznidazole reduction [6], or from enhanced efflux mechanisms [11].
In this paper, we show that at least three distinct mechanisms act in concert to generate high levels of benznidazole-resistance within a single population. This intrinsic capacity of T. cruzi to rapidly acquire drug-resistance by different routes may explain the widely reported treatment failures with this drug. Furthermore, it may have serious implications for the prospects of developing anti-parasitic drugs to treat Chagas disease.
Acknowledgements
This work was supported by funds from the Wellcome Trust (Grant 084175), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). We thank Shane Wilkinson and Laura Muller for comments on the manuscript.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
Contributor Information
Mônica C.O. Campos, Email: monica.caroline@lshtm.ac.uk.
Leonor L. Leon, Email: lleon@ioc.fiocruz.br.
Martin C. Taylor, Email: martin.taylor@lshtm.ac.uk.
John M. Kelly, Email: john.kelly@lshtm.ac.uk.
References
- 1.Bern C., Montgomery S.P. An estimate of the burden of Chagas disease in the United States. Clin Infect Dis. 2009;49:e52–e54. doi: 10.1086/605091. [DOI] [PubMed] [Google Scholar]
- 2.Lee B.Y., Bacon K.M., Bottazzi M.E., Hotez P.J. Global economic burden of Chagas disease: a computational simulation model. Lancet Infect Dis. 2013;13:342–348. doi: 10.1016/S1473-3099(13)70002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilkinson S.R., Kelly J.M. Trypanocidal drugs: mechanisms, resistance and new targets. Expert Rev Mol Med. 2009;11(31):1–24. doi: 10.1017/S1462399409001252. [DOI] [PubMed] [Google Scholar]
- 4.Castro J.A., de Mecca M.M., Bartel L.C. Toxic side effects of drugs used to treat Chagas’ disease (American trypanosomiasis) Hum Exp Toxicol. 2006;25:471–479. doi: 10.1191/0960327106het653oa. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson S.R., Taylor M.C., Horn D., Kelly J.M., Cheeseman I. A mechanism for cross-resistance to nifurtimox and benznidazole in trypanosomes. Proc Natl Acad Sci U S A. 2008;105:5022–5027. doi: 10.1073/pnas.0711014105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall B.S., Wilkinson S.R. Activation of benznidazole by trypanosomal type I nitroreductases results in glyoxal formation. Antimicrob Agents Chemother. 2012;56:115–123. doi: 10.1128/AAC.05135-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hall B.S., Bot C., Wilkinson S.R. Nifurtimox activation by trypanosomal type I nitroreductases generates cytotoxic nitrile metabolites. J Biol Chem. 2011;286:13088–13095. doi: 10.1074/jbc.M111.230847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mejia A.M., Hall B.S., Taylor M.C., Gómez-Palacio A., Wilkinson S.R., Triana-Chávez O. Benznidazole-resistance in Trypanosoma cruzi is a readily acquired trait that can arise independently in a single population. J Inf Dis. 2012;206:220–228. doi: 10.1093/infdis/jis331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murta S.M., Gazzinelli R.T., Brener Z., Romanha A.J. Molecular characterization of susceptible and naturally resistant strains of Trypanosoma cruzi to benznidazole and nifurtimox. Mol Biochem Parasitol. 1998;93:203–214. doi: 10.1016/s0166-6851(98)00037-1. [DOI] [PubMed] [Google Scholar]
- 10.Filardi L.S., Brener Z. Susceptibility and natural resistance of Trypanosoma cruzi strains to drugs used clinically in Chagas disease. Trans R Soc Trop Med Hyg. 1987;81:75575–75579. doi: 10.1016/0035-9203(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 11.Campos M.C., Castro-Pinto D.B., Ribeiro G.A., Berredo-Pinho M.M., Gomes L.H., da Silva Bellieny M.S. P-glycoprotein efflux pump plays an important role in Trypanosoma cruzi drug resistance. Parasitol Res. 2013;112:2341–2351. doi: 10.1007/s00436-013-3398-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Obado S.O., Taylor M.C., Wilkinson S.R., Bromley E.V., Kelly J.M. Functional mapping of a trypanosome centromere by chromosome fragmentation identifies a 16 kb GC-rich transcriptional strand-switch domain as a major feature. Genome Res. 2005;15:36–43. doi: 10.1101/gr.2895105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Souza R.T., Lima F.M., Barros R.M., Cortez D.R., Santos M.F., Cordero E.M. Genome size, karyotype polymorphism and chromosomal evolution in Trypanosoma cruzi. PLoS ONE. 2011;6:e23042. doi: 10.1371/journal.pone.0023042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alsford S., Eckert S., Baker N., Glover L., Sanchez-Flores A., Leung K.F. High-throughput decoding of antitrypanosomal drug efficacy and resistance. Nature. 2012;482:232–236. doi: 10.1038/nature10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kendall G., Wilderspin A.F., Ashall F., Miles M.A., Kelly J.M. Trypanosoma cruzi glycosomal glyceraldehyde-3-phosphate dehydrogenase does not conform to the hotspot topogenic signal model. EMBO J. 1990;9:2751–2758. doi: 10.1002/j.1460-2075.1990.tb07462.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parkinson G.N., Skelly J.V., Neidle S. Crystal structure of FMN-dependent nitroreductase from Escherichia coli B: a prodrug-activating enzyme. J Med Chem. 2000;43:3624–3631. doi: 10.1021/jm000159m. [DOI] [PubMed] [Google Scholar]