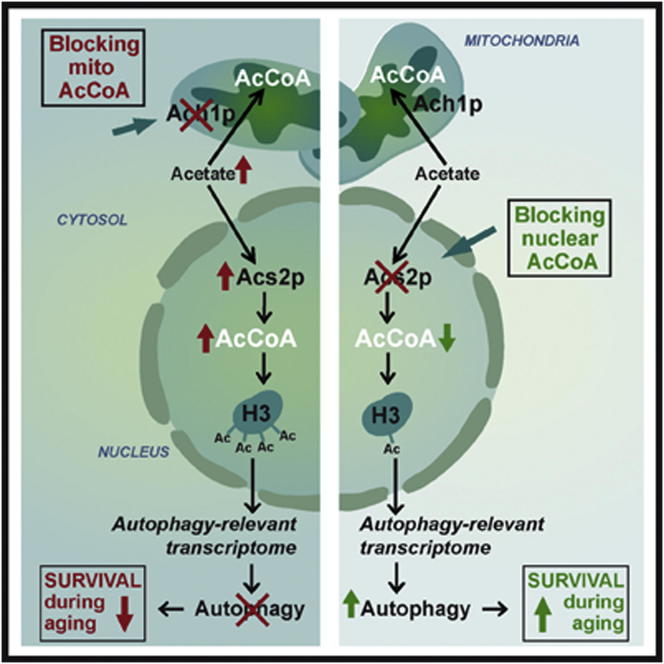
Summary
Healthy aging depends on removal of damaged cellular material that is in part mediated by autophagy. The nutritional status of cells affects both aging and autophagy through as-yet-elusive metabolic circuitries. Here, we show that nucleocytosolic acetyl-coenzyme A (AcCoA) production is a metabolic repressor of autophagy during aging in yeast. Blocking the mitochondrial route to AcCoA by deletion of the CoA-transferase ACH1 caused cytosolic accumulation of the AcCoA precursor acetate. This led to hyperactivation of nucleocytosolic AcCoA-synthetase Acs2p, triggering histone acetylation, repression of autophagy genes, and an age-dependent defect in autophagic flux, culminating in a reduced lifespan. Inhibition of nutrient signaling failed to restore, while simultaneous knockdown of ACS2 reinstated, autophagy and survival of ach1 mutant. Brain-specific knockdown of Drosophila AcCoA synthetase was sufficient to enhance autophagic protein clearance and prolong lifespan. Since AcCoA integrates various nutrition pathways, our findings may explain diet-dependent lifespan and autophagy regulation.
Graphical Abstract
Highlights
-
•
Acetyl-CoA (AcCoA) metabolism regulates autophagy during aging
-
•
Autophagy regulation by AcCoA metabolism acts downstream of nutrient signaling
-
•
Brain-specific knockdown of Drosophila AcCoA synthetase prolongs lifespan
-
•
Histone point mutations permanently activate autophagy during aging
Autophagy plays a crucial role in healthy aging. By blocking mitochondrial AcCoA production, Eisenberg et al. show that accumulation of nucleocytosolic AcCoA inhibits autophagy and reduces lifespan through a conserved epigenetic mechanism involving histone acetylation of specific autophagy genes in yeast and flies.
Introduction
Aging is accompanied by accumulation of cellular damage, changes in the repair and detoxification processes, and a shifting homeostatic balance in conflicting lethal and vital signaling programs (Madeo et al., 2010a; Rubinstein and Kimchi, 2012). Macroautophagy (hereafter referred to as “autophagy”) is a bulk degradation pathway in which parts of the cytosol or cytoplasmic organelles are encapsulated into double-membraned vesicles, so-called autophagosomes, which ultimately fuse with vacuoles/lysosomes where the cytoplasmic material becomes degraded (He and Klionsky, 2009). Autophagy plays a major role in the maintenance of cellular homeostasis, recycling energy reserves in the context of dwindling external resources and contributing to the removal of damaged organelles and potentially harmful protein aggregates (Chen and White, 2011; Kroemer et al., 2010; Mizushima and Komatsu, 2011). Autophagy therefore has emerged as a pivotal cellular process that can delay the pathogenic manifestations of aging and age-associated disease (Gelino and Hansen, 2012; Madeo et al., 2010a; Rubinsztein et al., 2011). While regulation of autophagy during short-term induction conditions (up to several hours) is well-investigated, understanding the long-term regulation of autophagy during the process of aging remains a challenge.
Autophagy is profoundly influenced by nutrient-responsive kinases, including the target of rapamycin (TOR), protein kinase A (PKA), and AMP-activated protein kinase (AMPK), all of which are also well-known regulators of aging and lifespan (Fontana et al., 2010; He and Klionsky, 2009; Kenyon, 2010). However, recent studies suggest that protein acetylation, a process that is lately recognized as a posttranslational modification that rivals phosphorylation in importance (Choudhary et al., 2009; Guan and Xiong, 2011; Sadoul et al., 2011), may also regulate autophagy at targets distinct from that of known kinase regulators (Geeraert et al., 2010; Lee et al., 2008; Morselli et al., 2011; Xie et al., 2010; Yi et al., 2012). While acetylation of ATG3 and of tubulin is required for autophagy (Geeraert et al., 2010; Xie et al., 2010; Yi et al., 2012), ATG7 must be deacetylated by sirtuin 1 for the initiation of starvation-induced autophagy (Lee et al., 2008). Autophagy is in principle a cytoplasmic process that does not require a nuclear-localized transcriptional response for short-term activity, as demonstrated by the ability of cytoplasts (i.e., enucleated cells) to undergo starvation-induced autophagy (Morselli et al., 2011). Nevertheless, regulation of age-associated autophagy may depend on epigenetic processes (including that of histone acetylation) as well as on specific transcription factors such as FoxO3 that transactivate the autophagy-relevant transcriptome necessary for enduring autophagic activity that exceeds beyond a few hours (Eisenberg et al., 2009; Settembre et al., 2011; Zhao et al., 2007). Acetylation at lysine 16 of histone H4 has only recently been demonstrated to determine the outcome of autophagy (i.e., cytotoxic versus protective), possibly by influencing the transcriptional status of ATG genes (Füllgrabe et al., 2013), and the Ume6p transcription factor regulates the size of autophagosomes through control of ATG8 expression levels (Bartholomew et al., 2012).
Acetyl-coenzyme A (AcCoA) serves as an acetyl-group donor for protein and histone acetylation in eukaryotic cells and at the same time represents a central metabolite of cellular energy metabolism. It has been proposed that metabolism may connect to various cellular functions by modulating intracellular metabolites (including that of AcCoA), which in turn act as cofactors for posttranslational modifications affecting enzyme function or epigenetic status of the chromatin (Kaelin and McKnight, 2013; Lu and Thompson, 2012). Given the increasingly recognized role of protein acetylation as well as histone acetylation in the regulation of autophagy, we therefore aimed at evaluating the hypothesis that AcCoA biosynthetic availability would affect autophagy in a long-term context, in chronological aging.
For more than a decade now, budding yeast has significantly contributed to our understanding of aging, unraveling mechanisms of cellular senescence and age-induced death of postmitotic cells in a model of chronological aging (Kaeberlein, 2010; Longo et al., 2012). In budding yeast, de novo synthesis of AcCoA is facilitated by two distinguishable metabolic routes, namely the mitochondrial (ACS1-, ACH1-, or MPC1-dependent) versus the nucleocytosolic (ACS2-dependent) pathways (Bricker et al., 2012; Fleck and Brock, 2009; Takahashi et al., 2006). Based on these premises, we determined the autophagy-modulatory effects of manipulating the de novo biosynthetic pathways of AcCoA generation in a model of chronological aging.
Results
Knockdown of ACS2 Is Sufficient to Promote Autophagy during Aging
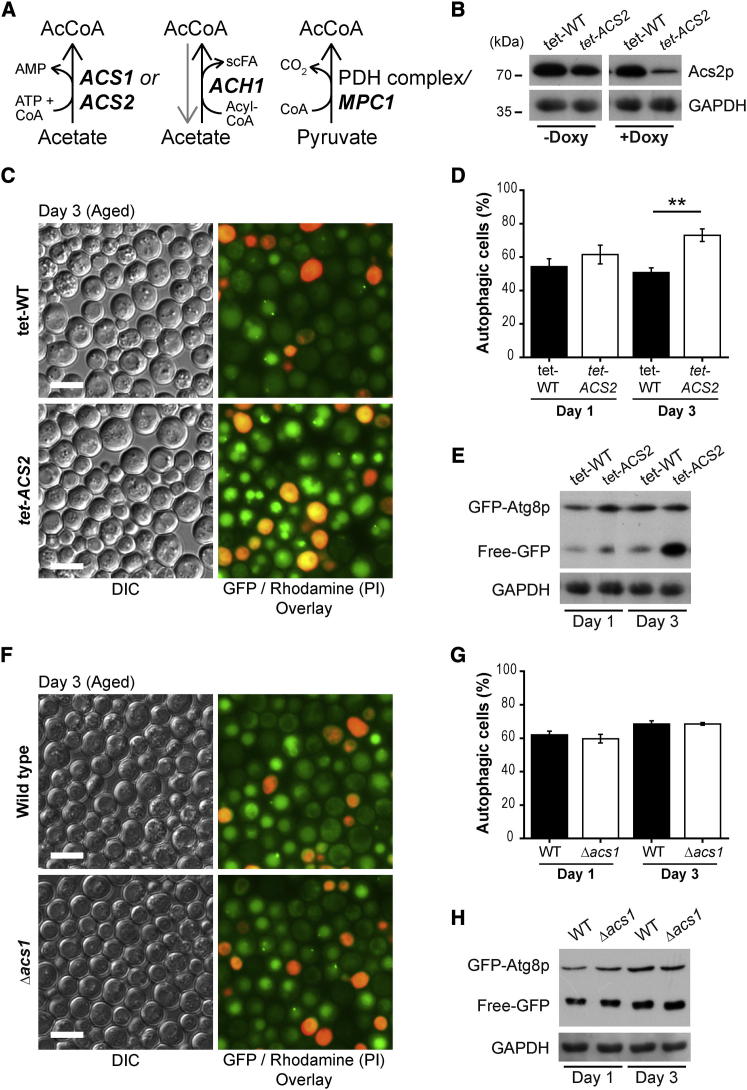
To test if de novo AcCoA biosynthesis from acetate or pyruvate modulated the autophagic response during cellular aging, we screened deletion or knockdown mutants of genes encoding enzymes involved in AcCoA formation (see scheme in Figure 1A). To this end, we first assessed the potential influence of the AcCoA synthetases Acs1p and Acs2p that generate AcCoA through the ATP-dependent condensation of acetate and coenzyme A (CoA). We optimized the experimental conditions for knockdown of the growth essential ACS2 gene using a doxycycline-repressible tetO7-promoter (tet-ACS2) so that cells would manifest a significant depletion of the Acs2 protein (Figure 1B) yet only partially reduce their growth during the logarithmic phase, yielding a rather modest reduction of cell densities upon entry into the stationary phase as compared to wild-type controls (tet-WT) (1 ng/ml doxycycline, Figure S1, available online). Knockdown of ACS2 led to a strong induction of autophagy upon chronological aging, as monitored by assessing the autophagy-dependent translocation of a cytosolic green fluorescent protein (GFP)-Atg8p fusion to the vacuole (Figures 1C and 1D). Both the number of autophagic cells, defined as cells that displayed clearly vacuolar localization of GFP (Figure 1C; quantified in Figure 1D), and vacuolar GFP intensities increased upon knockdown of ACS2. To strengthen these findings, we used a complementary assay based on the immunoblot detection of liberated GFP that is generated upon the autophagy-associated delivery of GFP-Atg8p to the vacuole and subsequent proteolytic removal of Atg8p (Klionsky et al., 2007). This GFP liberation assay demonstrated enhanced autophagic flux with active vacuolar proteolysis in aged tet-ACS2 cells compared to its wild-type (tet-WT) counterpart (Figure 1E). In contrast to knockdown of ACS2, age-associated autophagy was not affected by deletion of ACS1 (Figures 1F–1H).
Figure 1.
ACS2 Depletion Ameliorates Age-Associated Autophagy in Yeast
(A) Scheme of known major acetyl-CoA (AcCoA)-producing enzymes (bold characters) modulated in this study.
(B) Representative immunoblot of GFP-Atg8p expressing yeast. Wild-type cells (tet-WT) were compared to strains carrying a doxycycline-repressible tet-O7 promoter controlling ACS2 transcription (tet-ACS2). Cells were grown in SC 2% glucose medium for 24 hr (day 1 of aging) in the presence (+Doxy) or absence (−Doxy) of 1 ng/ml doxycycline to induce knockdown of ACS2 (see also Figure S1).
(C) Fluorescence microscopy of GFP-Atg8p expressing (under control of its natural pATG8 promoter) wild-type (tet-WT) and ACS2 knockdown (tet-ACS) cells grown in the presence of 1 ng/ml doxycycline (as shown in A) and chronologically aged for 3 days. Propidium iodide (PI) counterstaining served to visualize dead cells. Scale bars represent 5 μm.
(D) Quantification of cells depicted in (C) (day 3) and of young (day 1) cells with 150–300 counts (blinded) for each replicate. Autophagic cells were defined as cells with clear vacuolar GFP fluorescence. Data represent means ± SEM (n = 4). ∗∗p < 0.01.
(E) Representative immunoblot analysis of day 1 (young controls) and day 3 (aged) cells shown in (C) and (D) using anti-GFP and anti-GAPDH (loading control) antibodies to detect “free-GFP” indicative of autophagic flux.
(F–H) Representative micrographs (F), respective quantification (G), and immunoblot analysis (H) of wild-type (WT) and ACS1-deleted (Δacs1) yeast aged to day 3 compared to young (day 1) cells expressing GFP-Atg8p chimera as in (C)–(E). Data represent means ± SEM (n = 4).
Thus, while deletion of ACS1, the gene which is known to be suppressed by glucose (van den Berg et al., 1996), was dispensable for age-dependent autophagic activity, knockdown of ACS2—the crucial gene for AcCoA production on glucose conditions (Takahashi et al., 2006)—was sufficient to enhance the autophagic response to aging.
Mitochondrial AcCoA Production Pathways Are Required for Age-Associated Autophagy
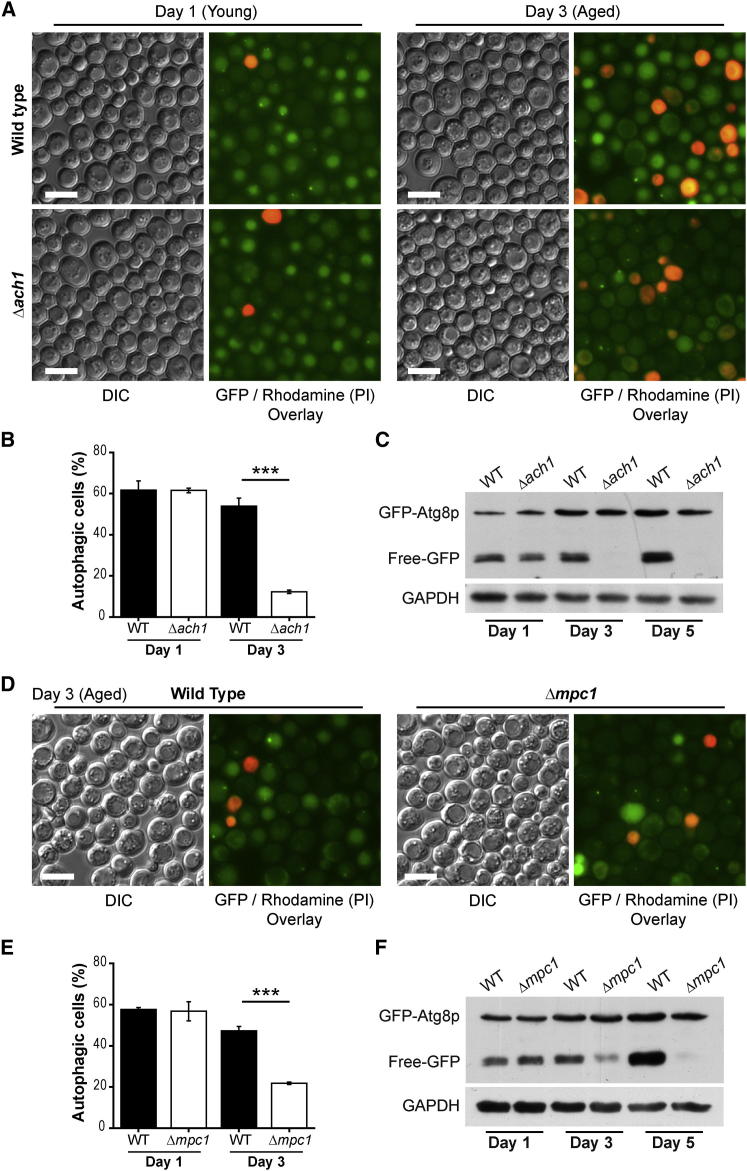
We next asked if blocking AcCoA synthesis through Ach1p, which transfers CoA from short-chain Acyl-CoAs to acetate generating AcCoA (Fleck and Brock, 2009), would also affect the autophagic response of aging cells. Remarkably, deletion of ACH1 completely blocked the autophagic activity after 3 days of aging, while young (day 1) cultures presented comparable levels of autophagy-competent cells (manifesting vacuolar localization of GFP-Atg8p as well as GFP liberation activity) in both wild-type and ach1 mutant (Figures 2A–2C). In addition, we assessed the autophagic flux by measuring vacuolar alkaline phosphatase (vac. ALP) activity using yeast strains carrying an engineered form of PHO8 (pho8ΔN60), encoding a cytosolically localized phosphatase, which becomes activated through autophagic delivery to the vacuole (Noda and Klionsky, 2008). While showing comparable levels of vac. ALP activity early during aging, ach1 mutant cells developed a strong defect in vac. ALP activity after 3 days (>72 hr) of aging (Figure S2A). We also measured cell stress by assessing the fraction of cells staining positive upon incubation with dihydroethidine (DHE), which is converted into fluorescent ethidium in ROS-overproducing cells but also gives rise to strong fluorescence in cells that have lost their plasma membrane integrity. This assay revealed that Δach1 cells lost their autophagic capacity before actual cell stress or death occurred (>88 hr; Figure S2A). These observations rule out that the autophagy defect was simply a result of the previously reported increase in cell death of ach1 mutant cells (Orlandi et al., 2012). Since Ach1p has been suggested to function primarily in mitochondria (Fleck and Brock, 2009), we next assessed whether block of AcCoA production through mitochondrial pyruvate dehydrogenase (PDH) pathway would also impair autophagy of aging yeast. Deletion of MPC1, a recently identified transporter of pyruvate into mitochondria crucial for PDH-derived AcCoA formation (Bricker et al., 2012), indeed caused a comparable defect of age-associated autophagy as observed for Δach1 (Figures 2D–2F). The combined deletion of ACH1 and MPC1 led to similar kinetics of the age-dependent loss of autophagy (Figure S2B). Together, this strongly suggests that neither the specific CoA-transferase function of Ach1p nor mitochondrial pyruvate supply by Mpc1p was specifically responsible for the autophagic deficiency of ach1 or mpc1 mutant cells. Our data demonstrate a strict requirement of mitochondrial AcCoA production pathways for maintaining autophagic flux during aging.
Figure 2.
Mitochondrial AcCoA Production by Ach1p- or Mpc1p-Associated Pathways Is Required for Autophagy during Aging
(A) Fluorescence microscopy of GFP-Atg8p expressing wild-type (WT) and ACH1-deleted (Δach1) yeast grown to day 1 (young) and aged for 3 days in SC 2% glucose medium; propidium iodide (PI) counterstaining served to visualize dead cells. Scale bars represent 5 μm.
(B) Quantification of cells depicted in (A) with 150–300 counts (blinded) for each replicate. Autophagic cells were defined as cells with clear vacuolar GFP fluorescence. Data represent means ± SEM (n = 4). ∗∗∗p < 0.001.
(C) Representative immunoblot analysis of cells shown in (A) and further aged to day 5 using anti-GFP and anti-GAPDH (loading control) antibodies to detect “free-GFP” indicative of autophagic flux (see also Figure S2).
(D–F) Representative micrographs (D), respective quantification (E), and immunoblot analysis (F) of wild-type (WT) and MPC1 deleted (Δmpc1) yeast expressing GFP-Atg8p chimera as in (A)–(C) and aged to indicated time points. Data represent means ± SEM (n = 4). ∗∗∗p < 0.001.
Mitochondrial AcCoA Biosynthesis Pathways Are Required for Healthy Aging
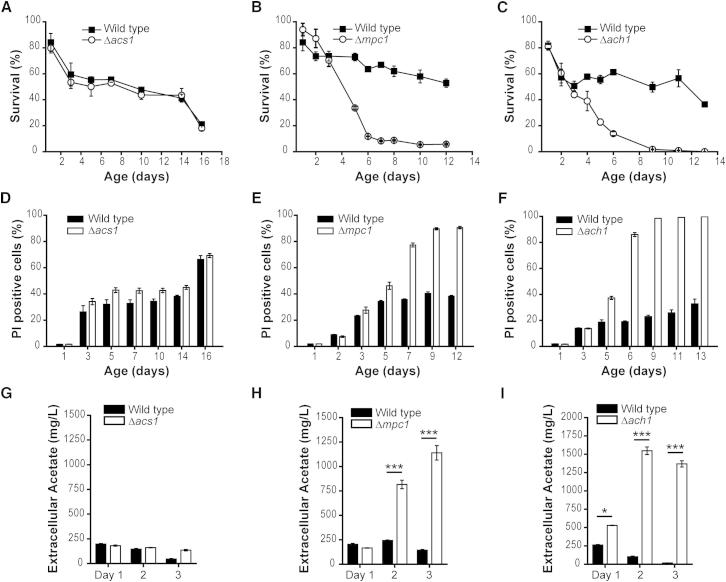
We next asked if the repression of autophagy induced by blockade of the mitochondrial route to AcCoA biosynthesis would culminate in increased cell death and a reduced lifespan. While deletion or knockdown of ACS1 or ACS2, respectively, barely affected yeast survival during chronological aging as measured by clonogenic assays (Figures 3A and S3A–S3D), deletion of either ACH1 or MPC1 dramatically reduced lifespan (Figures 3B and 3C). The frequency of cell death (as determined by propidium iodide-positive cells and further characterized to be of apoptotic and secondary necrotic morphology, see Figure S3I and respective legend for details) consistently increased in short-lived mutant conditions (Δach1 and Δmpc1) compared to wild-type cells and remained largely unchanged upon deletion or knockdown of ACS1 or ACS2, respectively (Figures 3D–3F, S3E, and S3F). Similar to the comparable kinetics of age-associated autophagy impairment, the combined deletion of ACH1 and MPC1 showed an epistatic effect on lifespan and age-induced cell death (Figures S3G and S3H). Thus, the consequences of these mutations on lifespan and cell death correlate with the (in)ability to induce a permanently activated autophagy response during aging.
Figure 3.
Deletion of ACH1 or MPC1 Shortens Chronological Lifespan and Leads to Excess Release of Cellular Acetate
(A–C) Chronological lifespan (CLS) analyses in SC 2% glucose medium of wild-type (WT) cells compared to Δacs1 (A), Δmpc1 (B), or Δach1 (C) cells (see also Figure S3). Survival was determined by colony-forming capacity (clonogenicity). Data represent means ± SEM (n = 4) of a representative aging experiment.
(D–F) Propidium iodide (PI)-positive cells analyzed by flow cytometry to quantify age-induced cell death of experiments shown in (A)–(C). Data represent means ± SEM (n = 4) (see also Figure S4).
(G–I) Extracellular acetate assessed from crude culture supernatants obtained at indicated time points of experiments shown in (A)–(C). Data represent means ± SEM (n = 4).
∗p < 0.05 and ∗∗∗p < 0.001.
Accumulating extracellular acetate upon deletion of ACH1 (Fleck and Brock, 2009; Orlandi et al., 2012) has been suggested to trigger the death of ach1 mutant cells (Orlandi et al., 2012). Accordingly, both ach1 and mpc1 mutant cells exhibited an age-dependent and comparable increase in extracellular acetate (Figures 3H and 3I). Deletion of ACS1 instead entailed rather minor changes in extracellular acetate concentration (Figure 3G), consistent with its lack of an effect on lifespan or autophagic activity.
Deletion of MPC1 or ACH1 Causes Upregulation of the Nucleocytosolic, Acs2p-Mediated Pathway of AcCoA Production
Excess acetate released from cells most likely reflects an overflow of acetate within the cytosol, which may fuel and trigger a cytosolic acetate-metabolizing pathway. Since Acs2p, the nucleocytosolic acetate-converting AcCoA synthetase, appeared limiting for autophagy (remember knockdown provoked autophagy during aging; compare to Figure 1), we hypothesized that upregulation of the Acs2p pathway would mechanistically explain the autophagy defect of ach1 and mpc1 mutant cells.
To test this hypothesis, we first analyzed the protein level of Acs2p in these mutants. While the level of Acs2p decreased during aging in wild-type cells, deletion of ACH1 or MPC1 clearly increased the relative amount of Acs2p compared to wild-type cells, suggesting an upregulation of the nucleocytosolic AcCoA pathway under both conditions (Figures 4A, 4B, S4A, and S4B). Interestingly, the sole addition of acetate to aging cultures was similarly efficient in increasing Acs2p protein levels as well as in the inhibition of autophagy, suggesting that acetate may indeed represent the trigger of its downstream metabolic pathway (Figure S4C). To test if the increased protein level of Acs2p also resulted in higher enzyme activity, we determined the AcCoA synthetase (ACS) activity from crude protein extracts using an established biochemical enzyme assay (van den Berg et al., 1996), using Δacs1 background conditions to specifically address Acs2p activity (for details, see Supplemental Experimental Procedures and Figures S3J–S3L). In agreement with the idea that deletion of ACH1 hyperactivates the nucleocytosolic AcCoA pathway, ach1 acs1 double mutant cells developed an age-dependent increase in ACS activity compared to acs1 control cells (Figure S4D).
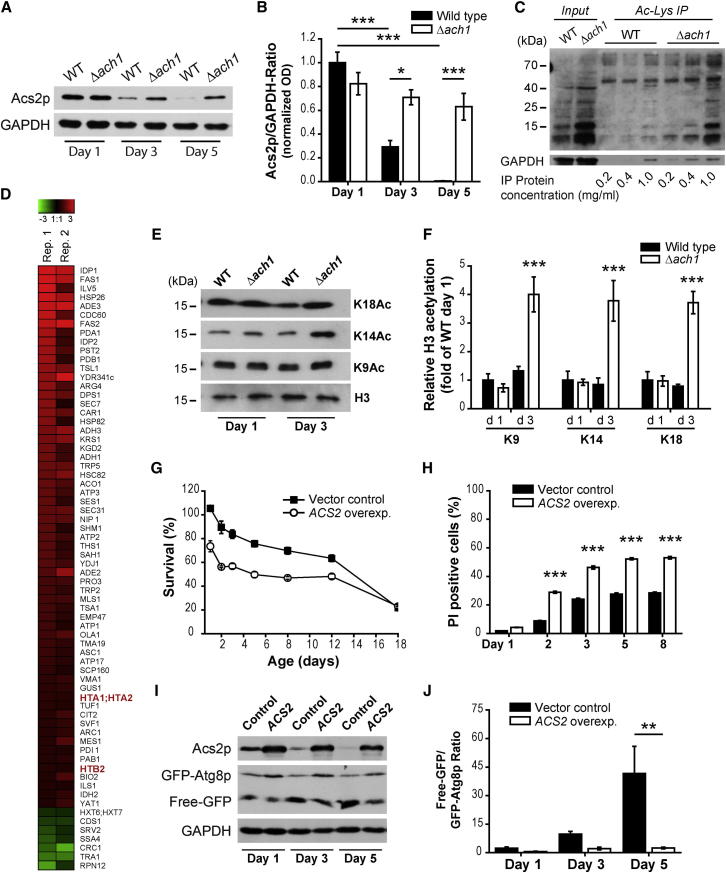
Figure 4.
Deletion of ACH1 or MPC1 Results in Upregulation of the Nucleocytosolic Acs2p Pathway, Causing Histone Hyperacetylation
(A and B) Representative immunoblot (A) and densitometric quantification expressed as normalized Acs2p/GAPDH ratios (B) of protein extracts from wild-type (WT) and Δach1 yeast chronologically aged to indicated time points in SC 2% glucose medium (see also Figure S4).
(C) Representative immunoblot analysis of 3-day-old WT and Δach1 cells similar to (A) using pan-acetyl-lysine antibodies. Crude protein extracts (Input) obtained in the presence of histone deacetylase and sirtuin inhibitors (see Supplemental Experimental Procedures) were subjected to immunoprecipitation (IP) at indicated protein concentrations using pan-acetyl-lysine antibodies to enrich acetylated proteins (Ac-Lys IP). GAPDH served as loading control.
(D) Heatmap of protein ratios (Δach1 versus WT, log2 scale) corresponding to indicated genes. Data were obtained from mass spectrometric analyses of acetylated proteins enriched by IP similar to (C, 1 mg/ml protein). Extracts from stable-isotope (SILAC)-labeled cells were mixed prior to IP (prelysate), and mean SILAC-protein ratios after IP were normalized to ratios of prelysates to correct for changes in general protein abundance. Two biological replicates were performed (Rep. 1 and Rep. 2) with a regression coefficient of 0.76 of observed protein ratios. Heatmap represents identified proteins with reproducible 1.5-fold upregulation (red color) or downregulation (green color).
(E and F) Representative immunoblots of whole cell acid extracts of wild-type (WT) and Δach1 (E) cells chronologically aged to designated time points. Blots were probed with antibodies against total histone H3 (loading control) or H3 acetylated lysines (K9Ac, K14Ac, K18Ac). Densitometric quantification (F) of relative acetylation was calculated as Ac-K/total H3 ratios normalized to ratios of WT at day 1. Data represent means ± SEM (n = 7–8) (see also Figure S5).
(G and H) Survival analyses in SC 1.25% galactose/0.75% glucose medium of wild-type cells ectopically overexpressing ACS2 (ACS2 overexp.) compared to vector control cells (Vector control). Survival (I) was determined by colony-forming capacity (clonogenicity) and cell death (J) assessed by propidium iodide (PI)-positive cells analyzed by flow cytometry. Data represent means ± SEM (n = 4) of a representative aging experiment.
(I and J) Autophagic flux determination by vacuolar protease-dependent GFP liberation after ACS2 overexpression compared to empty vector controls (Control) similar to (G) and (H). Representative immunoblots at indicated time points (G) and quantification by densitometry of free-GFP/GFP-Atg8p signal ratios (H) are shown. Data represent means ± SEM (n = 4).
∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Simple measurement of AcCoA levels is not applicable to our experimental question, as crude extracts reflect the total cellular content of AcCoA, not allowing for discrimination of the mitochondrial versus nuclear-cytosolic pools, which constitute two separate subcellular sites of AcCoA generation in yeast (Takahashi et al., 2006). Therefore, we decided to investigate the changes in AcCoA metabolism by alternative technologies that were based on the consideration that AcCoA represents an important cofactor of protein acetylation in eukaryotic cells and may directly affect the level of protein acetylation, enzymatically or nonenzymatically (Guan and Xiong, 2011). Consistent with the increased Acs2p activity, ach1 mutants exhibited increased overall acetylation based on immunoblot analysis using pan-acetyl-lysine antibodies (Figure 4C, Input). In a more sophisticated approach using SILAC-based proteomics of crude protein extracts that were enriched for acetylated proteins by means of immunoprecipitation (IP) (Figure 4C, Ac-Lys IP), we identified potential targets affected by deletion of ACH1. Under the selected cutoff conditions (reproducible 1.5-fold regulation; for details, see Experimental Procedures), 61 proteins were found to be hyperacetylated upon ACH1 deletion, while only 7 proteins were hypoacetylated (Figure 4D). Among these hyperacetylated target proteins, we identified several histones (Figure 4D, red highlighted text), in line with the prior observation that histone acetylation is associated with cellular AcCoA fluctuations in yeast and mammals (Cai et al., 2011; Wellen et al., 2009) as well as to the activity of Acs2p (Takahashi et al., 2006). The most abundant immunoblot signals detected after IP using pan-acetyl-lysine antibodies indeed corresponded to the size of histones at about 10–15 kDa (Figure 4C).
Next, we directly assessed the levels of histone H3 acetylation by means of acetylation-site-specific antibodies (i.e., histone marks that we have previously connected to autophagy during aging of yeast; Eisenberg et al., 2009). In line with the mass spectrometry results, Δach1 but also Δmpc1 cells exhibited consistent hyperacetylation of all tested N-terminally located lysine residues in histone H3 compared to wild-type cells (Figures 4E, 4F, S4E, and S4F). As identified by our mass spectrometry analysis, the histones H2A and H2B appeared similarly hyperacetylated (Figure 4D, HTA1/2 and HTB2), as observed for H3. Since acetylation of histone H4, in particular that of lysine 16, was only recently found to regulate the outcome of autophagy in yeast and mammals (Füllgrabe et al., 2013) and may determine yeast replicative lifespan (Dang et al., 2009), we asked whether this histone was also affected by deletion of ACH1. Interestingly, histone H4 acetylation was unaffected by deletion of ACH1 after 3 days of aging (Figure S4G), pointing toward a role of histone H2A/B and H3 acetylation under conditions of chronological aging that is distinct from that of the control of replicative lifespan by acetylation targets at histone H4.
In conclusion, impaired mitochondrial AcCoA production resulting from deletion of ACH1 or MPC1 causes upregulation of the nucleocytosolic pathway of AcCoA biosynthesis, as indicated by a concomitant increase in the Acs2p substrate acetate, the expression of Acs2p protein, the enzymatic activity of Acs2p, and hyperacetylation of specific histones.
Episomal Overexpression of ACS2 Mimics the Effects of ACH1 Deletion
To exclude that the effects of ACH1 or MPC1 deletion were simply due to a general defect in mitochondrial AcCoA supply, we tested whether genetic overexpression of ACS2 mimicked the effects of blocking mitochondrial AcCoA pathways. Indeed, Gal10p-driven overexpression of ACS2 led to early onset cell death (Figures 4G and 4H) and limited the autophagic response during aging (Figures 4I and 4J). Importantly, ectopic expression of a mutant acs2-Ts1 allele, which is stably expressed but shows reduced catalytic activity upon increased temperature (Takahashi et al., 2006), failed to induce cell death and caused reduced histone acetylation compared to expression of its wild-type counterpart that showed toxicity compared to BY4741 controls (Figures S4I–S4K). Of note, under our aging conditions (using only moderately increased permissive temperature to facilitate normal growth of the otherwise lethal acs2-Ts mutation), both the ACS2 wild-type (Wt-ACS2) and the Acs2-Ts1 mutant displayed a comparable increase in Acs2p levels compared to its BY4741 background strain (Figure S4H).
Histone Hyperacetylation Is Associated with Impaired Transcription of Autophagy-Essential Genes
We previously reported that histone hypoacetylation induced by supplementation of cultures with spermidine (an inhibitor of histone acetyl transferases) or simultaneous genetic inactivation of two histone acetyl transferase complexes correlates with enhanced expression of ATG genes and increased autophagy during aging (Eisenberg et al., 2009). Based on this premise, we hypothesized that histone hyperacetylation induced by upregulation of the Acs2p pathway could repress the transcription of autophagy-essential ATG genes. We therefore determined the relative mRNA abundance of five ATG transcripts (ATG5, ATG7, ATG8, ATG11, and ATG14) by quantitative reverse-transcriptase PCR. ACH1 deletion significantly reduced the abundance of several ATG transcripts, including that of ATG7, whose mRNA level decreased to ∼40% compared to the wild-type level (Figures 5A and S5A). In contrast, the ATG8 mRNA level increased in Δach1 cells (Figure S5A). The protein levels of Atg7p (assessed by immunoblotting of pATG7-ATG7-6HA fusion strains) consistently decreased upon deletion of ACH1 during aging (Figure 5B), while Atg8p levels remained constant (compare immunoblots of pATG8-EGFP-ATG8 in Figure 2C). The level of total Atg7p also decreased significantly in aged wild-type cells (Figure 5B), again in clear contrast to Atg8p (Figure 2C). Thus, as exemplified by Atg7p, transcriptional control of the autophagy-relevant proteome may be crucial for enduring autophagic activity during chronological aging.
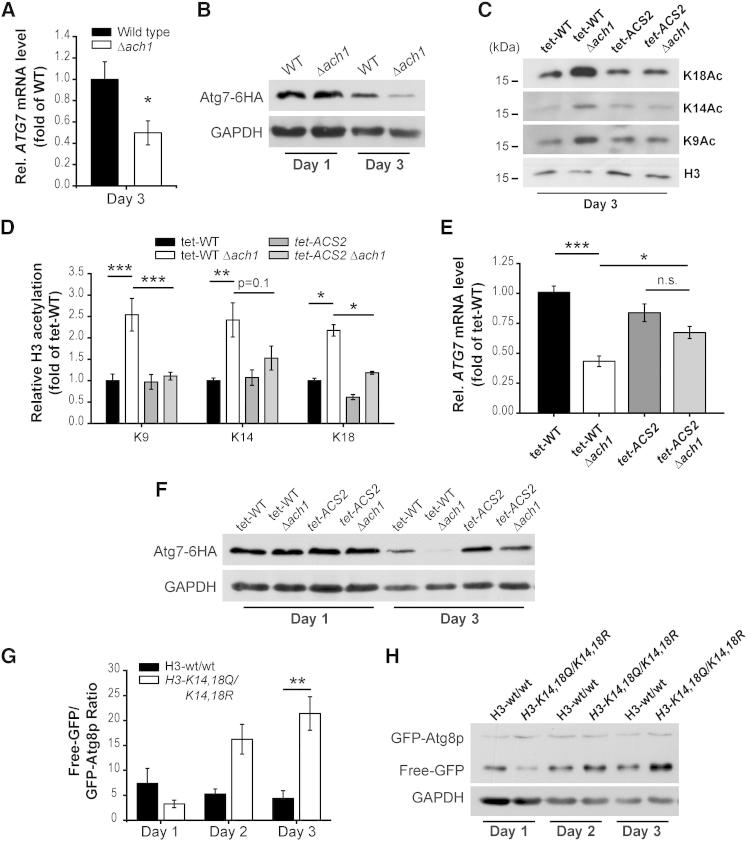
Figure 5.
Acs2p Controls ATG7 Transcription through Epigenetic Histone Acetylation
(A) ATG7 mRNA levels by quantitative reverse-transcriptase PCR (RT-qPCR) of wild-type and Δach1 cells aged to day 3 (see also Figure S5). Rel. mRNA levels are expressed as ratios of 18S rRNA normalized to wild-type cells by ΔΔCt-method. Data represent means ± SEM (n = 8).
(B) Representative immunoblot analysis from wild-type (WT) and ACH1-deleted (Δach1) yeast expressing chromosomally tagged ATG7 by C-terminal 6HA fusion aged to indicated time points. Blots were probed with anti-HA and anti-GAPDH (loading control) antibodies.
(C and D) Representative immunoblots (C) and densitometric quantification (D) of whole-cell acid extracts of wild-type and Δach1 cells combined with or without knockdown of ACS2 (tet-ACS2). Cells were chronologically aged to day 3 in the presence of 1 ng/ml doxycycline. Blots were probed with antibodies against total histone H3 (loading control) or H3 acetylated lysines (K9Ac, K14Ac, K18Ac). Data represent means ± SEM (n = 8).
(E and F) ATG7 mRNA levels (E) by RT-qPCR and representative immunoblot analysis (F) of wild-type and Δach1 cells with or without knockdown of ACS2 (tet-ACS2) as in (C) and (E) aged to day 3. Rel. mRNA levels (E) are expressed as ratios to 18S rRNA normalized to wild-type cells by ΔΔCt method. Data represent means ± SEM (n = 7–8).
(G and H) Representative immunoblot (H) and densitometric quantification (G) of histone H3 wild-type (H3-wt/wt) and H3 mutated (H3-K14,18Q/-K14,18R) strains carrying the GFP-Atg8p fusion to calculate “free-GFP/GFP-Atg8p Ratio” indicative of autophagic flux.
Data represent means ± SEM (n = 4) (for strain details and supplemental data, see Figures S5D–S5F).
∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; n.s., not significant.
ACS2 Depletion Reinstalls Histone Deacetylation and ATG7 Transcription in ach1 Mutants
In order to test if Acs2p-induced histone acetylation was indeed causally linked to the transcriptional repression of ATG7, we tested if knockdown of ACS2 in the background of ACH1 deletion would (1) reverse the observed histone hyperacetylation phenotype and (2) reinstate normal ATG7 transcription. Using the conditions depicted in Figure 1, knockdown of ACS2 almost completely restored physiological (wild-type-like) levels of histone acetylation in ach1 mutant cells (tet-ACS2 Δach1), while deletion of ACH1 alone (tet-WT Δach1) again increased overall acetylation at lysines 9, 14, and 18 of histone H3 compared to tet-WT controls (Figures 5C and 5D). Consistently, knockdown of ACS2 also partly recovered ATG7 mRNA and protein levels (Figures 5E and 5F, respectively) and almost completely annihilated the transcriptional defect induced by ACH1 deletion (tet-ACS2 Δach1 compared to tet-ACS2; Figure 5E).
Altogether our data are consistent with a causal inverse relationship of histone H3 acetylation with the transcriptional control of autophagy-essential genes, exemplified by ATG7 that is profoundly influenced by the Acs2p-mediated AcCoA production pathway. In support of a deficiency in the Atg7p-dependent Atg8p lipidating machinery, ach1 mutants failed to accumulate lipidated Atg8p normally evident from enlarged GFP-Atg8p punctuate structures that were observed in autophagy-deficient atg1 mutants carrying functional ATG7 (Figure S5C; see also Autophagy Measurements in the Supplemental Experimental Procedures for more details).
In order to demonstrate the principal requirement of histone acetylation in age-associated autophagy control, we created a panel of (nonacetylable) histone H3 lysyl point mutations, mimicking different states of acetylation and deacetylation. In addition to pure deacetylation-mimicking lysine to arginine (KR) mutations (that likely result in quite unphysiological situations), we also rendered mixed KQ/KR mutations introduced to either one of two histone copies present within the genome as a promising strategy to test for more physiological alterations that retain a certain opening of the chromatin. The triple K9,14,18R mutation indeed resulted in generally impaired survival during aging (for summary of results and histone mutants tested, see Figure S5D), but strikingly we identified a mutant (H3-K14,18Q/K14,18R) that continuously enhanced autophagy during aging (Figures 5G, 5H, and S5D–S5F). Although such mutants are per se far away from the highly refined, time- and location-dependent chromatin modifications that occur in vivo, this finding demonstrates that epigenetic modifications by histone acetylation are in principle capable of modulating the cellular autophagic response during aging, and goes in line with our hypothesis that histone acetylation may represent a determining downstream event upon modulation of AcCoA availability.
Depletion of ACS2, but Not Inhibition of Tor or Sch9 Signaling, Restores Autophagy in ach1 Mutants
As knockdown of ACS2 in the background of Δach1 almost completely abolished the transcriptional impairment of ATG7, we next asked whether ACS2 knockdown would also be able to reinstate the autophagic activity of ach1 mutant cells. Strikingly, knockdown of ACS2 almost completely restored age-associated autophagy of ach1 mutants (Figures 6A–6C and S6A), as determined by the frequency of cells with GFP-Atg8p-positive vacuoles (Figures 6A and 6B) and the levels of liberated GFP reflecting autophagic flux (Figures 6C and S6A). Autophagic activity of tet-ACS2 Δach1 cells approached that of the enhanced levels of the corresponding control strain without deletion of ACH1 (tet-ACS2). Importantly, the knockdown of ACS2 did not prevent extracellular accumulation of acetate induced by deletion of ACH1. The single deletion mutant (tet-WT Δach1), as well as ACH1 deletion combined with the knockdown of ACS2 (tet-ACS2 Δach1), displayed almost comparable levels of excess acetate released from cells (Figure S6D). In addition, the autophagy-inhibitory effect of acetate supplementation to aging yeast cultures (compare Figure S4C) was annihilated and autophagic activity almost retained at wild-type levels in the ACS2 knockdown condition (Figure S6E). This excludes a possible toxic effect of extracellular acetate on general cellular functions including autophagy and strongly argues for a specific role of the nucleocytosolic (Acs2p-mediated) pathway of acetate utilization that explains the inhibition of autophagy by ACH1 deletion.
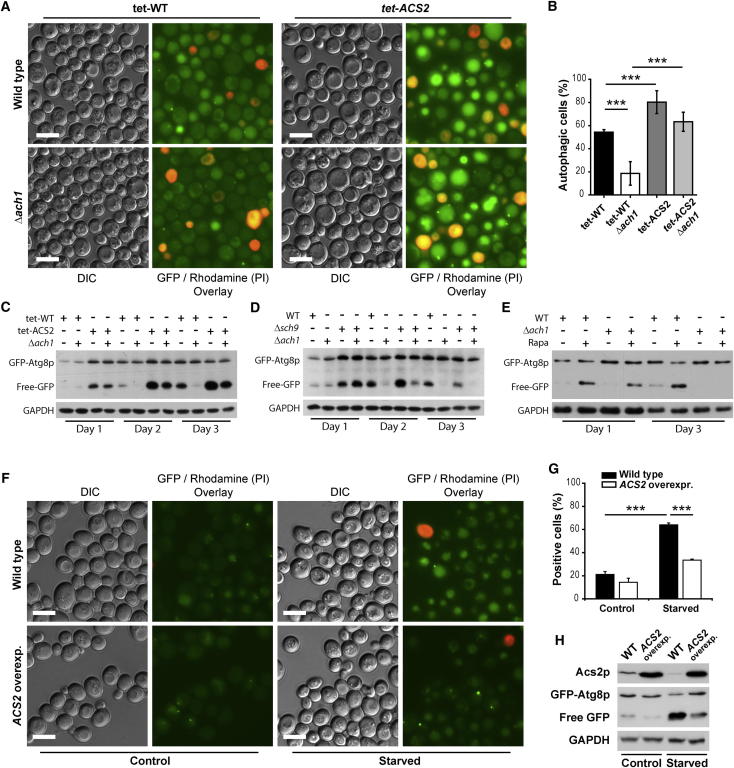
Figure 6.
Knockdown of Acs2p Cures the Autophagy Defect in ach1 Mutants
(A and B) Representative micrographs (A) and respective quantification (B) of wild-type and Δach1 cells expressing GFP-Atg8p chimera combined with or without knockdown of ACS2 (tet-ACS2). Cells were chronologically aged to day 3 in the presence of 1 ng/ml doxycycline and PI counterstained prior to epifluorescence microscopy (see also Figure S6).
(C) Representative immunoblot analysis of cells shown in (A) aged until days 1, 2, and 3 to detect “free-GFP” indicative of autophagic flux (see Figure S6A for quantification).
(D and E) Representative immunoblot analyses of GFP-Atg8p expressing wild-type (WT) and Δach1 cells either combined with deletion of SCH9 (D) or supplemented with or without 20 nM rapamycin (Rapa) (E) and aged until indicated time points. Blots were probed with anti-GFP and anti-GAPDH (loading control) antibodies to detect “free-GFP” indicative of autophagic flux (see Figures S6B and S6C for quantification).
(F and G) Representative micrographs (F) and respective quantification (G) of GFP-Atg8p expressing yeast cells ectopically overexpressing ACS2 (ACS2 overexp.) or carrying the empty vector (wild-type) during nutrient depletion (starved) compared to SC 2% galactose control conditions (control). Counterstaining of cells with propidium iodide was used to visualize dead cells.
(H) Representative immunoblot analyses of the experiment shown in (F) to detect “free-GFP” indicative of autophagic flux.
∗∗∗p < 0.001.
We next investigated whether the autophagy defect of cells lacking ACH1 would be bypassed by inhibition of autophagy-regulatory kinases. Suppression of Tor- or Sch9-function by either genetic or pharmacological means induces autophagy and delays aging in yeast and other model organisms (He and Klionsky, 2009; Madeo et al., 2010a). Using previously reported conditions (Alvers et al., 2009), rapamycin, a potent inhibitor of TORC1, was able to induce autophagy during aging of BY4742 wild-type cells but completely failed to restore the age-induced autophagy defect of Δach1 cells (Figures 6E and S6C). Rapamycin was well capable of inducing autophagy in young (day 1) Δach1 cells (Figures 6E and S6C), excluding that rapamycin would simply fail to inhibit Tor in ach1 mutant cells at the concentrations used for wild-type cells. Similarly, deletion of SCH9, which has been suggested to act in parallel to TORC1 as an autophagy suppressor (Yorimitsu et al., 2007) as well as in the same pathway with TORC1 during lifespan regulation (Wei et al., 2009), transiently amplified the autophagic response of aging wild-type but again failed to reinduce autophagy of aging ach1 cells (Figures 6D and S6B). Thus, knockdown of ACS2—but not inhibition of Tor or Sch9—specifically reinstated autophagy of ach1 mutants, strengthening our conclusion that upregulation of the Acs2p pathway is causally linked to the autophagy defect of Δach1 cells.
Finally, we tested if ectopic overexpression of ACS2 would also impair autophagy induced by nutrient depletion, a known autophagy trigger via nutrient-responsive signaling, including TORC1- or AMPK-dependent pathways (He and Klionsky, 2009). While the level of Acs2p decreased upon starvation (Figure 6H, compare empty vector controls, “WT”), ACS2 overexpression profoundly inhibited autophagic flux after 12 hr of starvation, as evident from reduced vacuolar translocation of GFP-Atg8p (Figures 6F and 6G) as well as reduced amount of liberated GFP (Figure 6H, “Free-GFP”). Similar results were observed after autophagy induction by rapamycin treatment (data not shown).
In sum, enduring autophagic activity during aging (but also that of acute nutrient depletion conditions) is profoundly repressed by nucleocytosolic Acs2p, which appears dominant over autophagy regulation by known kinase regulators such as Sch9 or TORC1.
Reinstating Autophagy of Δach1 Partly Restores Lifespan in an Autophagy-Dependent Manner
Given the crucial role of autophagy for healthy aging, reinstating autophagy in ach1 mutants may recover survival of aging cells, which would hence argue in favor of a causal role of the autophagy deficit in the reduction of lifespan. Knockdown of ACS2, using the same conditions that reactivated autophagy of ach1 cells, indeed significantly enhanced survival (Figure 7A) and reduced markers of cell death (Figure 7B) in aging ach1 cells. Compared to that of wild-type cells, the strong lifespan-shortening effect of ACH1 deletion (tet-WT versus tet-WT Δach1) was significantly inhibited by knockdown of ACS2 (tet-ACS2 versus tet-ACS2 Δach1). Additional deletion of the autophagy-essential gene ATG7 (known to completely abrogate autophagy in aging yeast) again revealed the full lifespan-compromising consequences of ACH1 deletion, even when ACS2 expression was suppressed (Figures 7C and 7D). Knockdown of ACS2 even accelerated death of ach1 mutants when autophagy was absent (compare tet-ACS2 Δach1 Δatg7 to tet-WT Δach1 Δatg7, Figures 7C and 7D). This strongly suggests that loss of autophagy is—at least in part—responsible for the detrimental effects of ACH1 deletion.
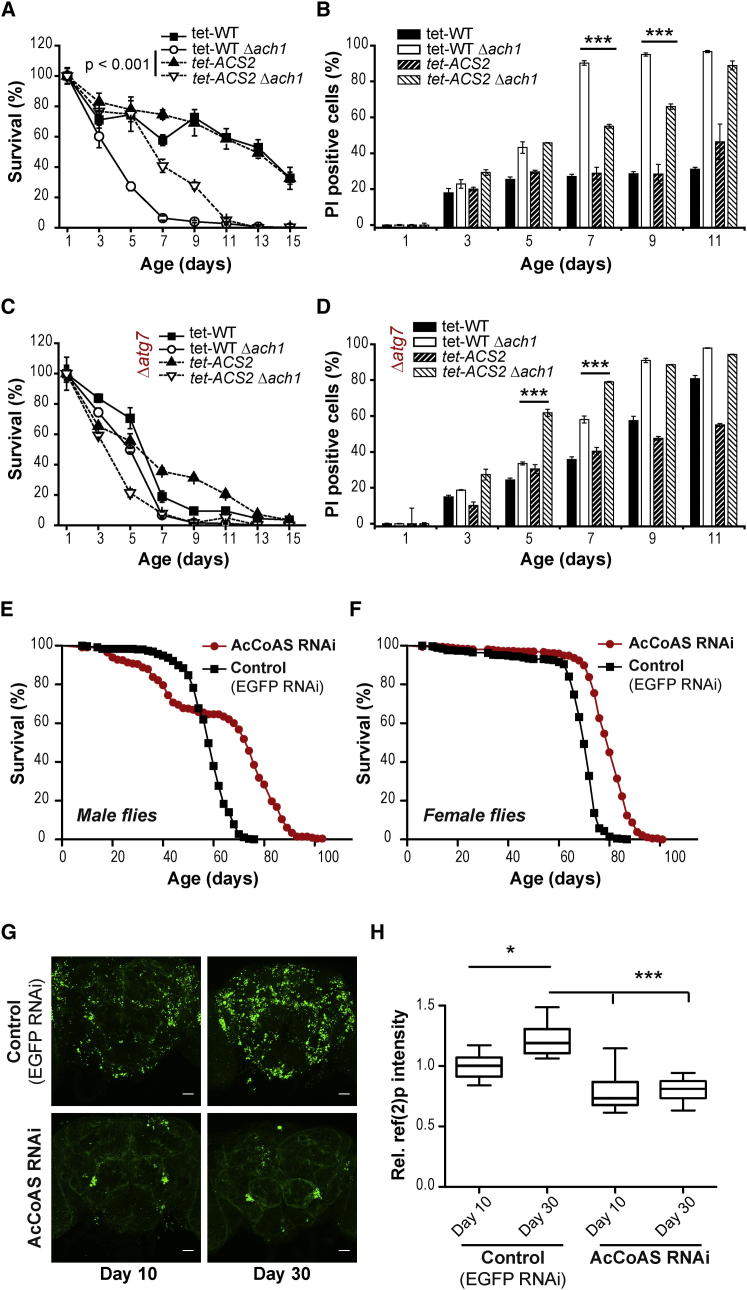
Figure 7.
Knockdown of Acs2p Partly Restores Survival of ach1 in an ATG7-Dependent Manner
(A and B) Chronological aging in SC 2% glucose medium supplemented with 1 ng/ml doxycycline of wild-type and Δach1 cells combined with or without knockdown of ACS2 (tet-ACS2). Survival (A) was determined by colony-forming capacity (clonogenicity). Age-associated cell death (B) was assessed through propidium iodide (PI) staining analyzed by flow cytometry. Data represent day 1 normalized means ± SEM (n = 4).
(C and D) Survival (C) and cell death (D) of chronological aging experiment similar to (A) and (B) but in the background of ATG7-deleted autophagy-incompetent cells (Δatg7). Data represent day 1 normalized means ± SEM (n = 4).
(E and F) Drosophila lifespan analyses of male (E) and female (F) flies depleted for acetyl-CoA synthetase (AcCoAS) using RNAi-mediated knockdown (AcCoAS RNAi) compared to isogenized controls (Control (EGFP RNAi)). Log rank tests revealed p < 0.0001 for both sexes.
(G) Immunofluorescence specific to p62-homolog ref(2)p of adult brain sections from 10-day-old flies compared to 30-day-old flies as depicted in (E). Representative confocal micrographs are shown with scale bars representing 25 μm.
(H) Quantification of total ref(2)P intensity in the central brain region normalized to 10-day-old control flies (Control (EGFP RNAi)). Standard box plot represents data from seven to eight independent brains (whiskers indicate minimum and maximum values).
∗p < 0.05, ∗∗∗p < 0.001.
Knockdown of Acetyl-CoA Synthetase Extends Mean and Maximum Lifespan of Flies
Finally, we tested if inhibition of the nucleocytosolic acetyl-CoA production pathway would affect autophagy and aging in a metazoan organism. In Drosophila, a renowned model of aging in a higher eukaryote, autophagic efficacy was shown to decline with brain aging, while activating the autophagic pathway by brain-specific overexpression of Atg8a using brain-specific appl-gal4 was previously shown to improve lifespan (Simonsen et al., 2008). Therefore, we addressed the effects of brain-specific RNAi-mediated knockdown of the fly acetyl-CoA synthetase gene (AcCoAS).
Remarkably, although initially with adverse effects in males, flies expressing RNAi specific for AcCoAS exhibited mean and maximum lifespan extension in both sexes compared to isogenic control flies that expressed an unrelated RNAi specific for GFP not present in this organism (Figures 7E and 7F, p < 0.0001 for both sexes). We then assessed the levels of the p62-homolog ref(2)p, a protein known to increase with age and to decrease again when autophagy is activated (Gupta et al., 2013). In fact, while both the number of dot-like (aggregated) ref(2)p structures as determined by immunofluorescence (Figure 7G) and overall levels of ref(2)p as quantified by total pixel intensities (Figure 7H) increased in brains of aged (30 days) compared to young (10 days) control animals, this increase was largely abolished in the animals expressing the AcCoAS RNAi by appl-gal4. Thus, our data suggest that, indeed, knockdown of AcCoAS induced the autophagic pathway associated with its longevity-promoting effects.
Discussion
In the present study we identified the nucleocytosolic (Acs2p-mediated) AcCoA production pathway as a suppressor of age-associated autophagy, conserved from yeast to flies. While upregulation of Acs2p activity as a consequence of impaired mitochondrial acetate or pyruvate utilization led to autophagy deficiency, reducing the protein level of Acs2p was sufficient to ameliorate the autophagic response to aging (see Figure S7 for an overview of the mechanistic model). In agreement with this idea, hyperactivation of the Acs2p pathway observed in ach1 or mpc1 mutant cells hampered genetic or pharmacological means of autophagy induction, including deletion of SCH9 or application of rapamycin, respectively. Ectopic overexpression of ACS2 not only limited the autophagic response to aging but also strongly impaired autophagy induced by nutrient depletion, a scenario known to inhibit nutrient signaling. Thus, autophagy observed under distinct conditions is counteracted by a hyperactive Acs2p pathway, which may imply a general requirement of Acs2p inhibition for efficient induction of autophagy.
The activity of Acs2p culminates in its downstream target of histone acetylation. The mechanism by which Acs2p activity affects age-associated autophagy may thus include epigenetic control of ATG gene transcription, as exemplified in this study by regulation of ATG7 mRNA and protein levels. ATG7 mRNA and protein levels inversely correlated with the acetylation of N-terminally located lysyl residues of histone H3. However, regulation of autophagy by changes in the relevant transcriptome most likely involves more than just one ATG transcript. The precise mechanism of how global hyperacetylation modulates the autophagy-relevant transcriptome remains to be clarified but is in agreement with our previous finding that global hypoacetylation can induce ATG7 transcription and autophagy upon treatment with the natural polyamine spermidine (Eisenberg et al., 2009; Madeo et al., 2010b). Our findings add evidence to a previously underestimated pathway of autophagy regulation that may particularly apply to enduring autophagic responses and that involves epigenetic and/or transcriptional alterations (Füllgrabe et al., 2014). Acetylation of histone H4 at lysine 16 has only recently been shown to direct the decision of whether or not autophagy can become toxic (Füllgrabe et al., 2013). The transcription factor FoxO3 controls the expression of autophagy-related genes, including that of LC3—the mammalian ortholog of yeast ATG8—and is required for autophagy induction of mammalian muscle cells with implications to muscle atrophy (Zhao et al., 2007). In yeast, ATG8 transcription is regulated by the Ume3-Sin3-Rpd3-complex, which suppresses induction of ATG8 under nonstarved conditions and modulates the size of autophagosomes during starvation-induced autophagy (Bartholomew et al., 2012).
In this line and as a proof of principle we introduced a histone point mutation that displayed enhanced autophagy during aging. However, we cannot exclude that in combination with transcriptomic alterations driven by epigenetic histone modifications also acetylation of nonhistone proteins may explain the observed phenotypes. This could, for instance, include the acetylation of Atg proteins or autophagy-relevant transcription factors that affect ATG gene transcription more directly. Apart from protein acetylation and/or histone acetylation, more indirect effects on metabolism downstream of AcCoA generation may also explain some of the consequences on autophagy and lifespan and need to be investigated in the future.
The connection of metabolism to posttranslational modifications of histones has been recognized and linked to disease-relevant situations (Kaelin and McKnight, 2013; Lu and Thompson, 2012). However, the connection of AcCoA metabolism to age-relevant autophagy has remained elusive. Here, we describe how energy metabolism controls autophagic activity during cellular aging through modulation of a central metabolite, AcCoA, presumably via its connection to epigenetic changes in chromatin.
The role of acetate in promoting yeast aging has been critically discussed recently (Burhans and Weinberger, 2009; Burtner et al., 2009; Longo et al., 2012). Our data strongly suggest that acetate exerts its proaging effects on yeast not only through rather unspecific events resulting from the primary toxic activity of extracellular acetate, but also through the Acs2p-mediated conversion of acetate to AcCoA. Importantly, acetate metabolism was proposed to contribute to aging in mammals, again via its impact on protein acetylation by generation of AcCoA and subsequent regulation of metabolism (Shimazu et al., 2010).
Interestingly, many lifespan-prolonging regimens (e.g., caloric restriction, rapamycin treatment) or age-associated disease-preventing supplements (e.g., resveratrol) that are known to induce autophagy were also reported to modulate mitochondrial function and respiration (Bonawitz et al., 2007; Houtkooper et al., 2010; Ocampo et al., 2012; Pan et al., 2011). Hence, it is tempting to speculate that efficient utilization of AcCoA by mitochondrial TCA cycle activity and oxidative phosphorylation prevents overflow of AcCoA into protein acetylation and/or histone acetylation pathways that would negatively influence autophagy and other prosurvival processes during aging and age-associated disease.
In summary, we propose that the nucleocytosolic acetyl-CoA pool functions as a crucial, evolutionary conserved, and dominant inhibitor of autophagy and healthy aging. Inhibition of major nutrient signaling pathways (i.e., TORC1 or Sch9) fails to induce autophagy when the Acs2p pathway is activated, arguing for a fundamental function of this pathway downstream of nutrient signaling. Since AcCoA integrates various nutrition pathways, our findings have implications for the understanding of lifespan and autophagy regulation in the context of excessive, restricted, or unbalanced feeding behaviors.
Experimental Procedures
Yeast Strains, Molecular Biology, and Chronological Aging Experiments
Experiments were carried out in S. cerevisiae BY4741 or BY4742 wild-type yeast and respective mutant strains depicted in Table S1. Chronological aging experiments were performed in SC 2% glucose medium, survival determined by plating cells on YPD agar (clonogenicity), and cell death quantified by propidium iodide staining. For further details on yeast strains and genetics (Table S1), pharmacological treatments, cell death assays, and culture conditions, please refer to Supplemental Information.
Immunoblotting and Quantification of Histone Acetylation
Immunoblotting was performed using standard protocols. Details on antibodies and on quantification of histone acetylation by blotting serial dilutions of acid extracts are depicted in Supplemental Information.
Yeast Autophagy Measurements
Autophagy was measured either by monitoring the cytosol to vacuole translocation of Atg8p using fluorescence microscopy or immunoblotting (GFP liberation assay) of cells/cell extracts from strains carrying a GFP-Atg8p fusion protein (Kirisako et al., 1999; Klionsky et al., 2007) expressed from its endogenous promoter and natural chromosomal locus (pATG8-EGFP-ATG8 strains) or by alkaline phosphatase (ALP) activity (Noda and Klionsky, 2008). Propidium iodide (PI) staining served to exclude analyses of dead, potentially autofluorescent cells visualized by standard rhodamine filters. For more details also on quantification of results, refer to Supplemental Information.
Determination of Acetyl-Proteome from SILAC Yeast Cultures
To assess changes in the acetyl-proteome, yeast protein extracts were first enriched for acetylated proteins by IP. Stable isotope-labeled amino acid cell cultures (SILAC) from yeast (Lys0, Lys4, or Lys8 labeled) were subjected to cell disruption by glass beads in the presence of histone deacetylase (trichostatin A, 30 mM) and sirtuin (nicotinamid, 2 μM) inhibitors. Crude extracts from wild-type and Δach1 cultures aged to day 3 were mixed in equal amounts of total protein (determined by Bio-Rad protein assay, Bio-Rad) and subjected to IP using pan-acetyl-lysine antibodies (1:100, Cell Signaling; an equal mix of Ac-K [#9441] and Ac-K2 [#9814] antibodies was used) and subsequently analyzed by mass spectrometry. For details on IP procedure and on quantification and identification of SILAC ratios by mass spectrometry, see Supplemental Information.
Acetyl-CoA-Synthetase Activity
Acetyl-CoA synthetase (ACS) activity was determined using an established biochemical enzyme assay (van den Berg et al., 1996). For details, see Supplemental Information.
Acetic Acid Detection
Enzymatic measurement of acetic acid from crude culture supernatants (appropriately diluted with water) was conducted using the Acetic Acid (Acetate Kinase Manual Format) kit (Megazyme) following the manufacturer’s protocol adapted to a volume of 100 μl for readout in a TECAN plate reader.
Quantitative Reverse-Transcriptase PCR
Target mRNA quantification by quantitative reverse-transcriptase PCR using ΔΔCt-method with 18S rRNA as an internal standard was performed on an ABI StepOnePlus using SYBR Select Master Mix (Life Tech, Invitrogen) ΔΔCt-method. Primers (Table S3) and details can be found in Supplemental Experimental Procedures.
Drosophila Lifespan Analyses and Brain Immunofluorescence
UAS-RNAi lines for acetyl-CoA Synthetase (P{TRiP.HMS02314}attP2) and EGFP (P{VALIUM20-EGFP.shRNA.1}attP2) serving as a background-matched control were obtained from Bloomington Drosophila Stock Center. Isogenized panneural driver line APPL-Gal4 was used to drive RNAi expression in fly brains. Details on housing, survival assessment, and ref(2)p-specific immunofluorescence can be found in Supplemental Information.
Statistical Analyses
One-factor analysis of variance (ANOVA) corrected by the post-hoc Bonferroni test was used for all experiments (multiple comparisons as appropriate) except for mass spectrometric and for chronological lifespan (CLS) analyses. If not otherwise stated, representative CLS experiments are shown with three to four biologically independent samples (as indicated) aged at the same time. CLS experiments have been performed at least three times with similar outcome. For comparison of CLS, a two-factor ANOVA with time and strain as independent factors was used. Log rank tests were performed for Drosophila lifespan analyses. Mass spectrometry was performed twice from two independent chronological aging experiments. Error bars represent SEM of biological replicates as indicated. Whiskers in box plots (Figure 7H) indicate minimum and maximum values.
Acknowledgments
We thank Drs. Jef D. Boeke, Daniel Klionsky, and Günther Daum for providing antibodies. We are also grateful for strains provided by Dr. Boeke. We thank Harald Hofbauer for support with quantitative PCR; and Lydia Opriessnig, Christian Pendl, and Birgit Michelitsch for assistance. This work was supported by the Austrian Science Fund FWF (grants T414-B09 and V235-B09 to S.B.; grants LIPOTOX, I1000, P23490-B12, and P24381-B20 to F.M.). T.E. is recipient of an APART fellowship of the Austrian Academy of Sciences at the Institute of Molecular Biosciences, University of Graz. This work was also supported by grants to G.K. from the Ligue Contre le Cancer (Équipe Labelisée), Agence National de la Recherche (ANR), Association Pour la Recherche sur le Cancer (ARC), Cancéropôle Ile-de-France; AXA Chair for Longevity Research, Institut National du Cancer (INCa), Fondation Bettencourt-Schueller, Fondation de France, Fondation pour la Recherche Médicale (FRM), the European Commission (ArtForce), the European Research Council (ERC), the LabEx Immuno-Oncology, the SIRIC Stratified Oncology Cell DNA Repair and Tumor Immune Elimination (SOCRATE), the SIRIC Cancer Research and Personalized Medicine (CARPEM), and the Paris Alliance of Cancer Research Institutes (PACRI).
Footnotes
This is an open access article under the CC BY license (http://creativecommons.org/licenses/by/3.0/).
Contributor Information
Guido Kroemer, Email: kroemer@orange.fr.
Frank Madeo, Email: frank.madeo@uni-graz.at.
Supplemental Information
References
- Alvers A.L., Wood M.S., Hu D., Kaywell A.C., Dunn W.A., Jr., Aris J.P. Autophagy is required for extension of yeast chronological life span by rapamycin. Autophagy. 2009;5:847–849. doi: 10.4161/auto.8824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew C.R., Suzuki T., Du Z., Backues S.K., Jin M., Lynch-Day M.A., Umekawa M., Kamath A., Zhao M., Xie Z. Ume6 transcription factor is part of a signaling cascade that regulates autophagy. Proc. Natl. Acad. Sci. USA. 2012;109:11206–11210. doi: 10.1073/pnas.1200313109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonawitz N.D., Chatenay-Lapointe M., Pan Y., Shadel G.S. Reduced TOR signaling extends chronological life span via increased respiration and upregulation of mitochondrial gene expression. Cell Metab. 2007;5:265–277. doi: 10.1016/j.cmet.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricker D.K., Taylor E.B., Schell J.C., Orsak T., Boutron A., Chen Y.-C., Cox J.E., Cardon C.M., Van Vranken J.G., Dephoure N. A mitochondrial pyruvate carrier required for pyruvate uptake in yeast, Drosophila, and humans. Science. 2012;337:96–100. doi: 10.1126/science.1218099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burhans W.C., Weinberger M. Acetic acid effects on aging in budding yeast: are they relevant to aging in higher eukaryotes? Cell Cycle. 2009;8:2300–2302. doi: 10.4161/cc.8.14.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtner C.R., Murakami C.J., Kennedy B.K., Kaeberlein M. A molecular mechanism of chronological aging in yeast. Cell Cycle. 2009;8:1256–1270. doi: 10.4161/cc.8.8.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai L., Sutter B.M., Li B., Tu B.P. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol. Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.-Y., White E. Role of autophagy in cancer prevention. Cancer Prev. Res. 2011;4:973–983. doi: 10.1158/1940-6207.CAPR-10-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M.L., Rehman M., Walther T.C., Olsen J.V., Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Dang W., Steffen K.K., Perry R., Dorsey J.A., Johnson F.B., Shilatifard A., Kaeberlein M., Kennedy B.K., Berger S.L. Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature. 2009;459:802–807. doi: 10.1038/nature08085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T., Knauer H., Schauer A., Büttner S., Ruckenstuhl C., Carmona-Gutierrez D., Ring J., Schroeder S., Magnes C., Antonacci L. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- Fleck C.B., Brock M. Re-characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA-transferases are involved in acetic acid detoxification. Fungal Genet. Biol. 2009;46:473–485. doi: 10.1016/j.fgb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Fontana L., Partridge L., Longo V.D. Extending healthy life span—from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J., Lynch-Day M.A., Heldring N., Li W., Struijk R.B., Ma Q., Hermanson O., Rosenfeld M.G., Klionsky D.J., Joseph B. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013;500:468–471. doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Füllgrabe J., Klionsky D.J., Joseph B. The return of the nucleus: transcriptional and epigenetic control of autophagy. Nat. Rev. Mol. Cell Biol. 2014;15:65–74. doi: 10.1038/nrm3716. [DOI] [PubMed] [Google Scholar]
- Geeraert C., Ratier A., Pfisterer S.G., Perdiz D., Cantaloube I., Rouault A., Pattingre S., Proikas-Cezanne T., Codogno P., Poüs C. Starvation-induced hyperacetylation of tubulin is required for the stimulation of autophagy by nutrient deprivation. J. Biol. Chem. 2010;285:24184–24194. doi: 10.1074/jbc.M109.091553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelino S., Hansen M. Autophagy—an emerging anti-aging mechanism. J. Clin. Exp. Pathol. Suppl. 2012;4:006. doi: 10.4172/2161-0681.s4-006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K.-L., Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem. Sci. 2011;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V.K., Scheunemann L., Eisenberg T., Mertel S., Bhukel A., Koemans T.S., Kramer J.M., Liu K.S.Y., Schroeder S., Stunnenberg H.G. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat. Neurosci. 2013;16:1453–1460. doi: 10.1038/nn.3512. [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D.J. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 2009;43:67–93. doi: 10.1146/annurev-genet-102808-114910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper R.H., Williams R.W., Auwerx J. Metabolic networks of longevity. Cell. 2010;142:9–14. doi: 10.1016/j.cell.2010.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–519. doi: 10.1038/nature08981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaelin W.G., Jr., McKnight S.L. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C.J. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999;147:435–446. doi: 10.1083/jcb.147.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Cuervo A.M., Seglen P.O. Methods for monitoring autophagy from yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., Levine B. Autophagy and the integrated stress response. Mol. Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I.H., Cao L., Mostoslavsky R., Lombard D.B., Liu J., Bruns N.E., Tsokos M., Alt F.W., Finkel T. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc. Natl. Acad. Sci. USA. 2008;105:3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo V.D., Shadel G.S., Kaeberlein M., Kennedy B. Replicative and chronological aging in Saccharomyces cerevisiae. Cell Metab. 2012;16:18–31. doi: 10.1016/j.cmet.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C., Thompson C.B. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeo F., Tavernarakis N., Kroemer G. Can autophagy promote longevity? Nat. Cell Biol. 2010;12:842–846. doi: 10.1038/ncb0910-842. [DOI] [PubMed] [Google Scholar]
- Madeo F., Eisenberg T., Büttner S., Ruckenstuhl C., Kroemer G. Spermidine: a novel autophagy inducer and longevity elixir. Autophagy. 2010;6:160–162. doi: 10.4161/auto.6.1.10600. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- Morselli E., Mariño G., Bennetzen M.V., Eisenberg T., Megalou E., Schroeder S., Cabrera S., Bénit P., Rustin P., Criollo A. Spermidine and resveratrol induce autophagy by distinct pathways converging on the acetylproteome. J. Cell Biol. 2011;192:615–629. doi: 10.1083/jcb.201008167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T., Klionsky D.J. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol. 2008;451:33–42. doi: 10.1016/S0076-6879(08)03203-5. [DOI] [PubMed] [Google Scholar]
- Ocampo A., Liu J., Schroeder E.A., Shadel G.S., Barrientos A. Mitochondrial respiratory thresholds regulate yeast chronological life span and its extension by caloric restriction. Cell Metab. 2012;16:55–67. doi: 10.1016/j.cmet.2012.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi I., Casatta N., Vai M. Lack of Ach1 CoA-transferase triggers apoptosis and decreases chronological lifespan in yeast. Front. Oncol. 2012;2:67. doi: 10.3389/fonc.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Schroeder E.A., Ocampo A., Barrientos A., Shadel G.S. Regulation of yeast chronological life span by TORC1 via adaptive mitochondrial ROS signaling. Cell Metab. 2011;13:668–678. doi: 10.1016/j.cmet.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein A.D., Kimchi A. Life in the balance—a mechanistic view of the crosstalk between autophagy and apoptosis. J. Cell Sci. 2012;125:5259–5268. doi: 10.1242/jcs.115865. [DOI] [PubMed] [Google Scholar]
- Rubinsztein D.C., Mariño G., Kroemer G. Autophagy and aging. Cell. 2011;146:682–695. doi: 10.1016/j.cell.2011.07.030. [DOI] [PubMed] [Google Scholar]
- Sadoul K., Wang J., Diagouraga B., Khochbin S. The tale of protein lysine acetylation in the cytoplasm. J. Biomed. Biotechnol. 2011;2011:970382. doi: 10.1155/2011/970382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Di Malta C., Polito V.A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S.U., Huynh T., Medina D., Colella P. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T., Hirschey M.D., Huang J.Y., Ho L.T., Verdin E. Acetate metabolism and aging: an emerging connection. Mech. Ageing Dev. 2010;131:511–516. doi: 10.1016/j.mad.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Simonsen A., Cumming R.C., Brech A., Isakson P., Schubert D.R., Finley K.D. Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy. 2008;4:176–184. doi: 10.4161/auto.5269. [DOI] [PubMed] [Google Scholar]
- Takahashi H., McCaffery J.M., Irizarry R.A., Boeke J.D. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol. Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- van den Berg M.A., de Jong-Gubbels P., Kortland C.J., van Dijken J.P., Pronk J.T., Steensma H.Y. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- Wei M., Fabrizio P., Madia F., Hu J., Ge H., Li L.M., Longo V.D. Tor1/Sch9-regulated carbon source substitution is as effective as calorie restriction in life span extension. PLoS Genet. 2009;5:e1000467. doi: 10.1371/journal.pgen.1000467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen K.E., Hatzivassiliou G., Sachdeva U.M., Bui T.V., Cross J.R., Thompson C.B. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie R., Nguyen S., McKeehan W.L., Liu L. Acetylated microtubules are required for fusion of autophagosomes with lysosomes. BMC Cell Biol. 2010;11:89. doi: 10.1186/1471-2121-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi C., Ma M., Ran L., Zheng J., Tong J., Zhu J., Ma C., Sun Y., Zhang S., Feng W. Function and molecular mechanism of acetylation in autophagy regulation. Science. 2012;336:474–477. doi: 10.1126/science.1216990. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T., Zaman S., Broach J.R., Klionsky D.J. Protein kinase A and Sch9 cooperatively regulate induction of autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 2007;18:4180–4189. doi: 10.1091/mbc.E07-05-0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Brault J.J., Schild A., Cao P., Sandri M., Schiaffino S., Lecker S.H., Goldberg A.L. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–483. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.