Summary
Objective
To identify osteoarthritis (OA) relevant genes and pathways in damaged and undamaged cartilage isolated from the knees of patients with anteromedial gonarthrosis (AMG) – a specific form of knee OA.
Design
Cartilage was obtained from nine patients undergoing unicompartmental knee replacement (UKR) for AMG. AMG provides a spatial representation of OA progression; showing a reproducible and histologically validated pattern of cartilage destruction such that damaged and undamaged cartilage from within the same knee can be consistently isolated and examined. Gene expression was analysed by microarray and validated using real-time PCR.
Results
Damaged and undamaged cartilage showed distinct gene expression profiles. 754 genes showed significant up- or down-regulation (non-False discovery rate (FDR) P < 0.05) with enrichment for genes involved in cell signalling, Extracellular Matrix (ECM) and inflammatory response. A number of genes previously unreported in OA showed strongly altered expression including RARRES3, ADAMTSL2 and DUSP10. Confirmation of genes previously identified as modulated in OA was also obtained e.g., SFRP3, MMP3 and IGF1.
Conclusions
This is the first study to examine a common and consistent phenotype of OA to allow direct comparison of damaged and undamaged cartilage from within the same joint compartment. We have identified specific gene expression profiles in damaged and undamaged cartilage and have determined relevant genes and pathways in OA progression. Importantly this work also highlights the necessity for phenotypic and microanatomical characterization of cartilage in future studies of OA pathogenesis and therapeutic development.
Keywords: Microarray, Signalling, Gene expression, Cartilage, Knee
Introduction
Knee osteoarthritis (OA) is common with more than 37% of those aged over 60 showing radiographic disease1. Cartilage damage in OA is likely to result from the aggregate effect of multiple genetic, environmental, mechanical and cell biological factors driving changes in gene expression2. These gene expression changes alter chondrocyte activity and phenotype, further driving OA progression. Studies of gene expression in OA have highlighted a number of differentially regulated genes and pathways (e.g., Matrix Metalloproteinases (MMPs) and tissue remodelling) providing valuable clues to the gross changes that differentiate OA from normal cartilage3, 4, 5, 6, 7, 8, 9.
Gene expression analysis of human OA is limited by inter-patient variability due to confounding factors including genetics and drug therapy; cartilage is generally removed from an end-stage joint and compared to normal cartilage from an individual without OA. The site of cartilage excised is not always consistent across patients or controls; and end-stage disease is heterogeneous in both its phenotype and the mechanisms underlying its onset and progression, thus key pathways and gene expression changes may be masked or misrepresented3, 5, 6, 7, 9. In order to overcome the heterogeneity of human OA a number of animal models have been utilized10. These rely on a known mechanism to induce disease (e.g., destabilization of the medial meniscus) thus providing a relatively consistent OA phenotype in terms of cause and time of onset. These models provide important clues to the genes expression changes at different stages of OA pathogenesis4, 11. However comparisons to human disease may be limited due to the lack of equivalent disease phenotype in humans (i.e., acute injury and fast progressing).
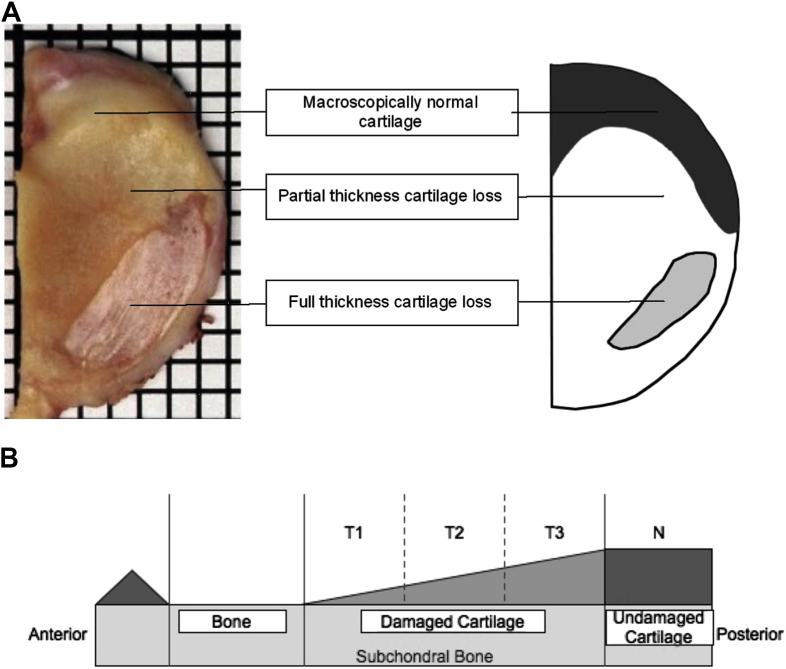
Studying gene expression in damaged and undamaged cartilage from within the same joint compartment would be advantageous. The most common phenotype of knee OA, Anteromedial Gonarthrosis (AMG) provides a spatially reproducible pattern of disease. AMG affects up to 60% of patients presenting with knee OA and is characterized by: full thickness cartilage loss in the anterior third of the medial tibial plateau; partial thickness loss of the middle third; and a macroscopically and histologically normal region in the posterior third12. Thus AMG provides a spatial representation of cartilage degradation in OA, allowing comparison of undamaged and damaged cartilage from microanatomically defined regions [Fig. 1].
Fig. 1.
AMG. A. A medial tibial plateau resection specimen from a patient having Unicompartmental knee arthroplasty (UKA) for AMG. The schematic on the right helps better demonstrate the areas of full thickness cartilage loss, partial thickness cartilage loss and macroscopically normal cartilage. B. Schematic representation of AMG. The damaged area can be divided into equal thirds (T1, T2, T3). The undamaged cartilage is region N. T1-T3 and N were collected for microarray analysis.
This study aims to characterize gene expression in AMG, a defined and common phenotype of OA13. Studying AMG has the advantage of reproducible identification and isolation of damaged and undamaged cartilage from within the same joint. Additionally AMG provides a consistent human phenotype of disease. Through study of these defined regions of damaged and undamaged cartilage we aim to identify genes and pathways that can be therapeutically targeted. We also aim to assess whether regional variation in gene expression occurs across cartilage from the same joint. This study will provide much needed insight into both AMG pathogenesis and OA progression.
Materials and methods
Tissue collection
Patients were identified if attending our specialist centre for unicompartmental knee replacement (UKR) for AMG. Ethical approval was obtained to approach these patients and obtain consent for use of their resected tissue in research (Ethics Reference C01.071). Evidence of these patients having tricompartmental OA made them unsuitable for both UKR and this study.
Ten medial tibial plateaus were collected (median age 64 years, six right knees and four left knees, six females and four males). Cartilage from damaged and undamaged regions was removed and snap frozen for microarray analysis. A uniform and anatomically aligned section was taken through all regions, wax embedded and the blocks stored for confirmation of cartilage phenotype using Safranin-O. A further six patients were recruited and cartilage collected for qPCR confirmation studies.
RNA extraction and preparation
Damaged and undamaged cartilage was identified and isolated using our previously defined spatial model of AMG [Fig. 1]. Cartilage was ground in liquid nitrogen and RNA extracted using the RNeasy lipid kit (Qiagen, Crawley, UK) according to the manufacturers instructions. All samples underwent on-column DNAse treatment. RNA yield and quality was determined using an Agilent Bioanalyzer and Nanodrop Spectrophotometer. The median RNA integrity number (RIN) value was 7.3 (range 6.5–8.8), one patient was excluded due to a low RIN value. Paired (damaged and undamaged) samples were used for microarray at equal concentrations. Prior to labelling, the samples were concentrated to 8.3 μl in a rotary evaporator.
Microarray
Samples were prepared as per Agilent's Two-Colour Microarray-Based Gene Expression Analysis (Quick Amp Labelling v5.7) protocol. RNA was fragmented at 60°C for 30 min and the reaction was stopped by the addition of 2× Agilent Hi-RPM hybridisation buffer. Samples were loaded on to arrays (Aglient G2159F, Design ID14850, 44,000 probes) and hybridised at 65°C for 17 h at 10 RPM. Slides were washed and scanned as per Agilent's protocol.
Complementary DNA synthesis and real-time PCR
First strand cDNA synthesis was performed on 50 ng RNA using Superscript III (Invitogen, Paisley, UK) according to the manufacturers protocol. qPCR for TNFRSF11B (OPG), SOX11, IGF1 and FGF18 was performed using predesigned primers (Quantitect, Qiagen, Crawley, UK) and SYBR green (Qiagen, Crawley, UK) in a Rotor-Gene RG-3,000 qPCR cycler (Qiagen, Crawley, UK). MMP1, MMP3 and MMP13 expression was measured using Taqman primer-probes and Taqman Master Mix (Applied Biosystems, Paisley, UK) on an ABI7900HT.
Statistical analysis
Microarray signal normalisation was carried out using Agilent Feature Extraction 9.5.3, which applied a global Lowess normalisation to remove systematic errors resulting from the fluorescence profiles of the two dyes. The normalised fluorescence data was analysed using GeneSpring GX10 (Agilent Technologies) to identify differentially expressed genes. Transcripts identified as differentially expressed had to show >2 fold differences in damaged compared to undamaged cartilage in all of the nine individuals tested. The fold change value represents average signal intensity of damaged compared to undamaged cartilage across all nine patients. As all samples were paired, the Wilcoxon signed rank test (a non-parametric test) was utilized to identify genes showing significant (P < 0.05) changes in expression using GeneSpring GX10. P-values were not False discovery rate (FDR) corrected as the microarray was utilized as a discovery tool with a more liberal gene selection used to identify whether regulated genes belonged to specific functional clusters.
Differentially expressed genes (fold >2, P < 0.05 not FDR corrected) were taken forward for further analysis. Analysis of enriched pathways and GO groups analysis was undertaken using Ingenuity (Ingenuity Systems, California, USA), DAVID and Webgestalt (Vanderbilt, USA). miRNA target analysis was undertaken using Webgestalt. Fisher Exact test (Ingenuity, DAVID) and hypergometric test (Webgestalt) were used to calculate associated pathways and GO groups. Benjamini-Hochberg was used to correct for multiple testing.
For confirmation qPCR analysis gene expression was assessed using the standard curve method and normalised to 18S (measured using the relevant qPCR chemistry). Paired samples were compared and significance (P < 0.05) assessed by a Wilcoxon test using PASW Statistics 18.0 (IBM, Illonois, USA).
Results
Microarray
754 genes (4% of the 19,365 measured) were differentially expressed (>2 fold, P < 0.05 not FDR corrected) between damaged and undamaged cartilage. In damaged cartilage 390 genes showed decreased expression whilst 364 genes were expressed at a higher level. Table I, Table II show the top 50 up- and down-regulated genes. A full list of regulated genes is available in Supplementary Table I. 11 genes showed >5-fold up-regulation in damaged cartilage and 61 >3-fold. The top upregulated genes included IL11, SOX11, DNER, GFRA2 and FGF18. 38 genes showed >5-fold downregulation in damaged cartilage and 128 >3-fold. Down-regulated genes included LAMB1, SPARCL1, SFRP4 and MMP13 Table III.
Table I.
The top 50 upregulated genes in damaged cartilage compared to undamaged cartilage
| Gene name | Gene symbol | Fold | P-value |
|---|---|---|---|
| Interleukin 11 | IL11 | 15.9 | 0.0109 |
| SRY (sex determining region Y)-box 11 | SOX11 | 8.66 | 0.0077 |
| Delta/notch-like EGF repeat containing | DNER | 8.15 | 0.0077 |
| Chromosome 15 open reading frame 48 | C15orf48 | 7.88 | 0.0077 |
| Chromosome 5 open reading frame 23 | C5orf23 | 6.98 | 0.0209 |
| GDNF family receptor alpha 2 | GFRA2 | 6.19 | 0.0077 |
| Chemokine (C-X-C motif) ligand 14 | CXCL14 | 6.17 | 0.0077 |
| UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 1 | GALNTL1 | 5.98 | 0.0077 |
| Brain and acute leukaemia, cytoplasmic | BAALC | 5.60 | 0.0077 |
| Prostaglandin E receptor 1 (subtype EP1) | PTGER1 | 5.04 | 0.0209 |
| GDNF family receptor alpha 2 | GFRA2 | 4.96 | 0.0077 |
| Fibroblast growth factor 18 | FGF18 | 4.68 | 0.0077 |
| S100 calcium binding protein A2 | S100A2 | 4.41 | 0.0077 |
| Coagulation factor C homologue, cochlin | COCH | 4.33 | 0.0284 |
| Chloride intracellular channel 3 | CLIC3 | 4.23 | 0.0077 |
| Transglutaminase 2 | TGM2 | 4.10 | 0.0077 |
| Chloride channel Ka | CLCNKA | 3.98 | 0.0109 |
| KIAA1622 transcript variant 2 | KIAA1622 | 3.93 | 0.0077 |
| Glutamate receptor, ionotropic, AMPA 2 | GRIA2 | 3.88 | 0.0077 |
| Cartilage oligomeric matrix protein | COMP | 3.86 | 0.0077 |
| Urocanase domain containing 1 | UROC1 | 3.86 | 0.0152 |
| Growth differentiation factor 6 | GDF6 | 3.85 | 0.0152 |
| Angiopoietin-like 4 | ANGPTL4 | 3.84 | 0.0077 |
| Annexin A8 | ANXA8 | 3.81 | 0.0152 |
| Ring finger protein 152 | RNF152 | 3.77 | 0.0077 |
| Cytoplasmic FMR1 interacting protein 2 | CYFIP2 | 3.77 | 0.0077 |
| Olfactory-like receptor PJCG2 | OR7E13P | 3.77 | 0.0077 |
| Tumour necrosis factor receptor superfamily, member 11b | TNFRSF11B | 3.69 | 0.0077 |
| Rho guanine nucleotide exchange factor (GEF) 15 | ARHGEF15 | 3.63 | 0.0077 |
| Serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 | SERPINE2 | 3.62 | 0.0077 |
| GDNF family receptor alpha 2 | GFRA2 | 3.59 | 0.0284 |
| Heparan sulphate (glucosamine) 3-O-sulfotransferase 3A1 | HS3ST3A1 | 3.52 | 0.0077 |
| HtrA serine peptidase 3 | HTRA3 | 3.48 | 0.0077 |
| Progestin and adipoQ receptor family member IX | PAQR9 | 3.45 | 0.0077 |
| Laminin, beta 3 | LAMB3 | 3.44 | 0.0077 |
| LIM domain only 6 | LMO6 | 3.43 | 0.0077 |
| ADAMTS-like 2 | ADAMTSL2 | 3.40 | 0.0077 |
| BTB (POZ) domain containing 16 | BTBD16 | 3.34 | 0.0077 |
| Glucagon-like peptide 2 receptor | GLP2R | 3.32 | 0.0077 |
| cDNA FLJ41861 fis, clone NTONG2008672 | C1orf170 | 3.27 | 0.0152 |
| Follistatin-like 3 (secreted glycoprotein) | FSTL3 | 3.24 | 0.0077 |
| Syntaxin 1A (brain)] | STX1A | 3.23 | 0.0077 |
| HtrA serine peptidase 1 | HTRA1 | 3.23 | 0.0077 |
| Laminin, gamma 3 | LAMC3 | 3.20 | 0.0077 |
| Noggin | NOG | 3.18 | 0.0077 |
| Bone morphogenetic protein 6 | BMP6 | 3.13 | 0.0109 |
| Metallothionein 3 | MT3 | 3.10 | 0.0152 |
| Decay-accelerating factor 4ab | CD55 | 3.10 | 0.0077 |
| Syntaxin 1A | STX1A | 3.05 | 0.0077 |
| Cytokine receptor-like factor 1 | CRLF1 | 3.04 | 0.0109 |
Table II.
The top 50 down-regulated genes in damaged cartilage compared to undamaged cartilage
| Gene name | Gene symbol | Fold change | P-value |
|---|---|---|---|
| Laminin, beta 1 | LAMB1 | −15.3 | 0.0076 |
| SPARC-like 1 | SPARCL1 | −11.5 | 0.028 |
| Basic helix-loop-helix domain containing, class B, 5 | BHLHB5 | −9.8 | 0.011 |
| Glycoprotein (transmembrane) nmb | GPNMB | −9.5 | 0.0076 |
| Major histocompatibility complex, class II, DM beta | HLA-DMB | −8.7 | 0.015 |
| Major histocompatibility complex, class II, DR alpha | HLA-DRA | −8.3 | 0.021 |
| Collagen, type XIV, alpha 1 | COL14A1 | −8.3 | 0.0077 |
| Chemokine (C-X-C motif) ligand 12 | CXCL12 | −8.0 | 0.038 |
| Phosphodiesterase 1A | PDE1A | −7.9 | 0.0077 |
| NEL-like 1 | NELL1 | −7.7 | 0.021 |
| Secreted frizzled-related protein 4 | SFRP4 | 7.6 | 0.011 |
| Matrix metallopeptidase 2 | MMP2 | 7.2 | 0.0077 |
| Allograft inflammatory factor 1 | AIF1 | 7.2 | 0.021 |
| Metallopeptidase 13 | MMP13 | 7.1 | 0.021 |
| Collagen, type XIV, alpha 1 | COL14A1 | 7.0 | 0.015 |
| Proprotein convertase subtilisin/kexin type 1 | PCSK1 | 6.8 | 0.0077 |
| Secreted frizzled-related protein 1 | SFRP1 | 6.8 | 0.038 |
| Interferon-induced protein 44 | IFI44 | 6.5 | 0.0077 |
| Complement component 3 | C3 | 6.5 | 0.011 |
| Plasminogen activator | PLAU | 6.4 | 0.011 |
| Raftlin, lipid raft linker 1 | RFTN1 | 6.4 | 0.0077 |
| Matrix metallopeptidase 1 | MMP1 | 6.3 | 0.011 |
| Plexin domain containing 1 | PLXDC1 | 6.0 | 0.0077 |
| Serpin peptidase inhibitor, clade F | SERPINF1 | 5.8 | 0.0077 |
| Colony stimulating factor 1 receptor | C51FR | 5.8 | 0.0077 |
| Cadherin 11, type 2 | CDH11 | 5.51 | 0.0077 |
| Olfactomedin-like 3 | OLFML3 | 5.47 | 0.0077 |
| Fibrinogen-like 2 | FGL2 | 5.44 | 0.0152 |
| Matrix metallopeptidase 9 | MMP9 | 5.43 | 0.0077 |
| Major histocompatibility complex, class II, DR beta 4 | HLA-DRB4 | 5.33 | 0.0382 |
| G protein-coupled receptor 124 | GPR124 | 5.28 | 0.0077 |
| Hepatitis A virus cellular receptor 2 | HAVCR2 | 5.27 | 0.0077 |
| Twist homologue 1 | TWIST1 | 5.16 | 0.0077 |
| Insulin-like growth factor 1 | IGF1 | 5.09 | 0.0077 |
| Chemokine (C–C motif) ligand 2 | CCL2 | 5.03 | 0.0077 |
| Bone marrow stromal cell antigen 2 | BST2 | 5.03 | 0.0077 |
| Family with sequence similarity 20, member A | FAM20A | 5.01 | 0.0077 |
| Olfactomedin-like 3 | OLFML3 | 4.98 | 0.0077 |
| V-set and immunoglobulin domain containing 4 | VSIG4 | 4.97 | 0.0382 |
| Acyl-Coenzyme A oxidase 2, branched chain | ACOX2 | 4.95 | 0.0077 |
| Lymphoid enhancer-binding factor 1 | LEF1 | 4.94 | 0.0077 |
| Interleukin 13 receptor, alpha 2 | IL13RA2 | 4.89 | 0.0077 |
| Mannosidase, alpha, class 1C, member 1 | MAN1C1 | 4.78 | 0.0077 |
| Phospholipase C-like 2 | PLCL2 | 4.70 | 0.0077 |
| Leucine rich repeat containing 15 | LRRC15 | 4.67 | 0.0109 |
| Brain abundant, membrane attached signal protein 1 | BASP1 | 4.65 | 0.0109 |
| Q8SPE4_9PRIM | THC2632039 | 4.64 | 0.0209 |
| WNT1 inducible signalling pathway protein 2 | WISP2 | 4.55 | 0.0077 |
| Caspase 1 | CASP1 | 4.54 | 0.0077 |
| Acid phosphatase 5, tartrate resistant | ACP5 | 4.53 | 0.0209 |
| Chemokine (C–C motif) ligand 3 | CCL3 | 4.49 | 0.0077 |
Table III.
Enriched GO groups showing differential expression in damaged compared to undamaged cartilage. ↑ = upregulated in damaged cartilage, ↓ = down-regulated in damaged cartilage
| GO Accession | GO term | P-value | Corrected P-value | Example regulated genes |
|---|---|---|---|---|
| GO:0005576 | Extracellular region | 8.12E-30 | 2.09E-24 | ↑ADAMTS6 CILP FGF18 TNFRSF11B ↓B2M CCL3 COL1A1 |
| GO:0031012 | Extracellular matrix | 6.19E-22 | 5.32E-17 | ↑ADAMTSL2 COMP FMOD LAMB3 ↓CHI3L1 COL1A2 FBLN1 MATN2 |
| GO:0005578 | Proteinaceous extracellular matrix | 2.18E-21 | 1.40E-16 | ↑FGF1 MUC2 NTN1 TGM2 ↓COL12A1 CPZ MATN4 THBS2 |
| GO:0002376 | Immune system process | 3.58E-14 | 1.85E-09 | ↑CXCL4 TNFSF10 CD8B CRHR1 ↓IL4R IL18BP CCL2 FAS C2 CCL4 C1 C3 IFIT3 |
| GO:0007155 | Cell adhesion | 4.55E-11 | 1.47E-06 | ↑LAMC3 SDK2 VWA1 DST ADAM12 SDK1 ↓IBSP COL12A1 CADM1 STAB1 NID2 |
| GO:0005615 | Extracellular space | 9.78E-11 | 2.80E-06 | ↑CXCL14 FGF18 ANGPTL4 HTRA1 ↓PTN PLAU SEPP1 SPON2 |
| GO:0007165 | Signal transduction | 1.89E-08 | 3.48E-04 | ↑TNFAIP6 FGF1 AXIN2 WIF1 ↓CAMK1 NOG IGF1 FRZB SFRP1 WISP2 WNT10A |
| GO:0044254 | Multicellular organismal protein catabolic process | 9.30E-08 | 0.001042428 | ↓MMP13 MMP2 MMP9 |
| GO:0044256 | Protein digestion | 9.30E-08 | 0.001042428 | ↓MMP11 MMP9 MMP3 |
| GO:0044259 | Multicellular organismal macromolecule metabolic process | 9.30E-08 | 0.001042428 | ↓MMP 1 MMP19 MMP11 |
| GO:0032963 | Collagen metabolic process | 9.30E-08 | 0.001042428 | ↓MMP1 MMP2 MMP3 |
| GO:0030574 | Collagen catabolic process | 9.30E-08 | 0.001042428 | ↑PRSS2 ASCL5 ↓MMP 9 MMP11 MMP13 |
| GO009611 | Response to wounding | 2.81E-07 | 0.002584188 | ↑FN1 ↓TLR4 CFHR1 LY96 C4BPA CX3CR1 PTX3 |
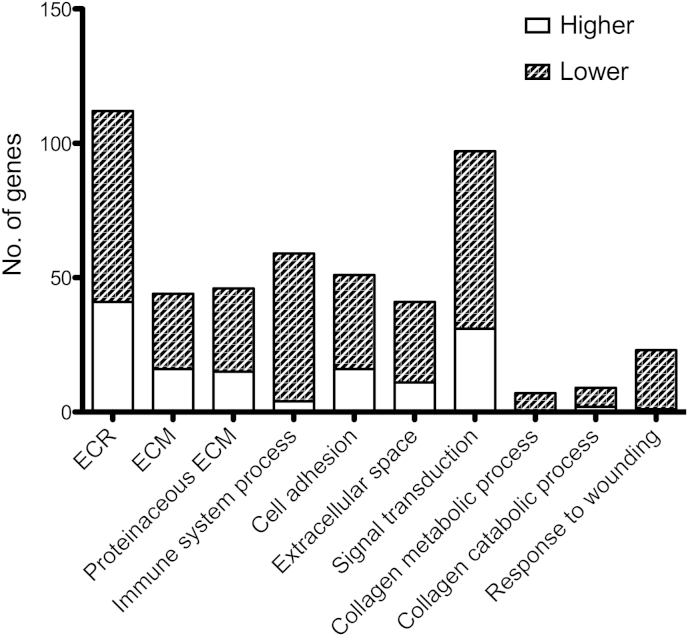
67 Gene ontology (GO) groups were enriched (P < 0.01) Table II, Fig. 2. Regulated GO groups include those related to Extracellular Matrix (ECM), immune system process, signal transduction, collagen metabolic process and response to wounding. Strikingly, ECM-related GO groups were the major groups showing up-regulation of gene expression in damaged cartilage, whilst genes from all significant GO groups showed down-regulation in damaged cartilage.
Fig. 2.
Distribution of up- and down-regulated genes within enriched GO groups. Differentially expressed genes were classified according to their GO group. GO groups that were significantly enriched were selected and the number of genes within each group that were up- or down-regulated in damaged compared to undamaged cartilage was calculated. Genes were included if there expression was showed >2 fold change in expression in all nine individuals with P < 0.05. Unfilled bars represent genes that were expressed at a higher level y in damaged compared to undamaged cartilage. Shaded bars represent those expressed at a lower level in damaged compared to undamaged cartilage. ECR = Extracellular Region. ECM = Extracellular Matrix.
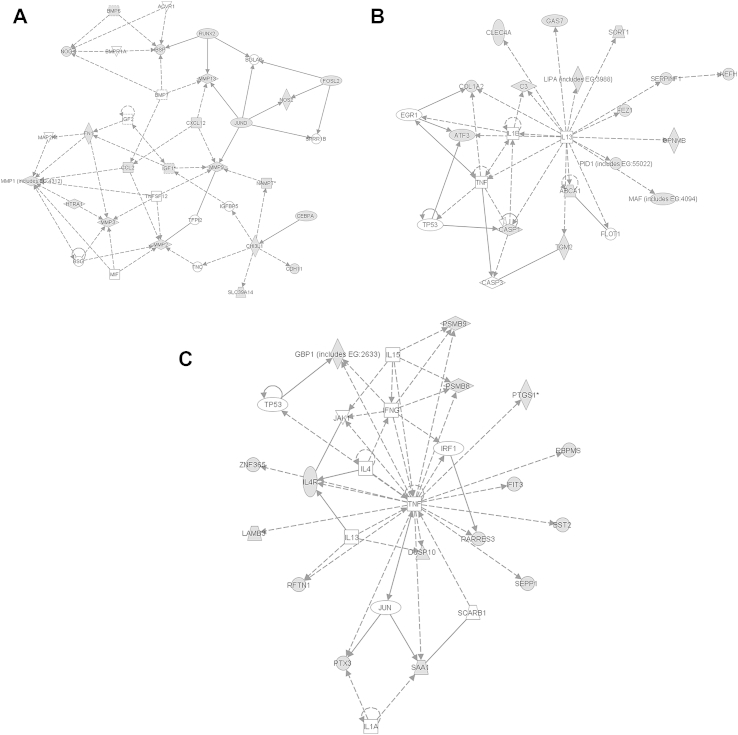
Ingenuity analysis identified three major regulated pathways showing dysregulation between damaged and undamaged cartilage [Fig. 3]. These are:
-
1.
Cellular Development, Skeletal and Muscular System Development and function, Lymphoid tissue structure development
-
2.
Inflammatory response, cell death, haematological disease
-
3.
Skeletal and muscular disorders, cell signalling, small molecule biochemistry
Fig. 3.
Pathway analysis. Ingenuity analysis on differentially expressed genes in damaged and undamaged cartilage revealed three protein–protein interaction networks showing significant association. These were, A. Cellular Development, Skeletal and Muscular System Development and function, Lymphoid tissue structure development. B. Inflammatory response, cell death, haematological disease. C. Skeletal and muscular disorders, cell signalling, small molecule biochemistry Nodes shaded grey are genes with significant expression differences in the microarray.
We carried out further pathway analysis using the DAVID and Webgestalt bioinformatic tools (Supplementary Tables II-IV). These confirmed the marked alteration between damaged and undamaged cartilage of cell signalling, inflammatory and cell matrix related pathways. Significantly associated pathways (P < 0.05) included ECM-receptor interactions, focal adhesion, haematopoietic cell lineage, TGF signalling, and MMPs. Furthermore disease enrichment analysis identified arthritis, joint diseases, inflammation and rheumatoid arthritis (P < 1 × 10–18). microRNA target analysis identified OA- and inflammation relevant miRs including miR200, miR145 miR23a (P < 1 × 10–5).
Comparison with other published microarrays of human knee OA cartilage confirmed OA-related changes in expression of genes including FN1, INHBA and TNFAIP63, 7, 8. However some genes including MMP1, MMP3 and COL1A2 showed an opposite direction of regulation compared to that previously reported3, 6, 7, 8. Genes previously not reported as regulated in OA include RARRES3, ADAMTSL2 and DUSP1. A single study of human hip OA cartilage vs neck of femur fracture control revealed 899 differential expressed genes, 59 of these are also altered in our study; 34 in the same direction (e.g., MMP1, MMP3); 25 in the opposite direction (e.g., BMP6 and TGM2)14. COL12A1 and STAB1 are the only differentially expressed genes within genetic loci marked by SNPs that genome wide association studies have identified as key signals in OA15, 16, 17. Comparison microarray analysis of murine and rat injury models of OA showed quite distinct gene expression profiles, however some common genes including INHBA, TNFAIP6 and TGM2 were identified4, 11, 18, 19.
Real-time PCR
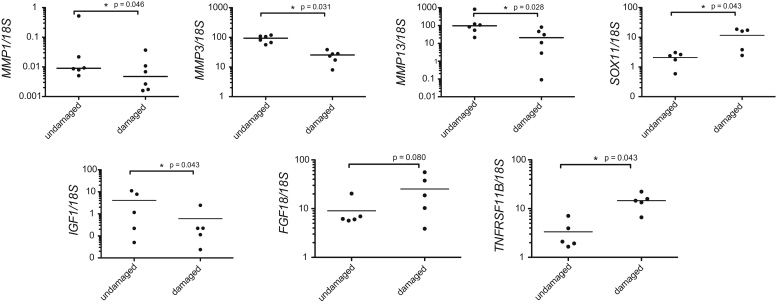
SOX11, IGF1, FGF18, MMP1, MMP3 and MMP13 were selected for confirmation of microarray data. Genes were selected based on their broad spectrum of roles in cartilage biology. SOX11, a transcription factor in embryonic development, was selected as it showed a high fold change in gene expression (8.66 fold higher in damaged cartilage) yet is relatively unstudied in OA. FGF18 (4.69 higher in damaged cartilage) IGF1 (5.10 lower in damaged cartilage), MMP1 (6.28 fold lower in damaged cartilage), MMP3 (3.19 fold lower in damaged cartilage) and MMP13 (7.05 lower in damaged cartilage) were selected due to their known functions in cartilage development and homoeostasis, and to confirm the direction of MMP expression changes reported in our microarray data20, 21, 22. TNFRSF11B (3.69 higher in damaged cartilage), has previously shown increased expression in OA cartilage but is generally regarded as a mediator of bone rather than cartilage metabolism23, 24.
Significant differences in gene expression were detected for SOX11: 6.7 fold higher in damaged cartilage (P = 0.043), IGF1: 5.2 fold lower in damaged (P = 0.043), TNFRSF11B: 6.9 fold higher in damaged (P = 0.043), MMP1: 1.88 lower in damaged (P = 0.046) MMP3: 3.68 fold lower in damaged (P = 0.031), MMP13: 4.62 fold lower in damaged (P = 0.028) [Fig. 4]. These expression changes confirmed the direction of gene expression changes detected in our microarray analysis. There was no significant difference in FGF18 expression (P = 0.08) although the pattern of expression suggested an increase in damaged cartilage and expression differences were seen with immunohistochemistry (data not shown).
Fig. 4.
Real-time PCR analysis. RNA was extracted from damaged and undamaged cartilage to confirm expression changes seen in the microarray analysis. Real-time PCR was carried out to assess gene expression of MMP1, MMP3, MMP13, SOX11, IGF1, FGF18 and TNFRSF11B.
Discussion
This study is the first to detail gene expression changes between undamaged and damaged cartilage taken from the same joint compartment of individuals with a common and specific phenotype of OA (AMG). The pattern of cartilage damage in AMG is reproducible across individuals. We show clear gene expression differences between damaged and undamaged cartilage and provides important clues to dysregulated pathways in OA. This study highlights the complexity of the OA joint and the variation in gene expression phenotype of chondrocytes from different locations within the same joint. Thus our study emphasizes the requirement to understand the behaviour of different cartilage regions in order to successfully study OA and target therapeutics.
We have identified changes in ECM, immune system processes and signal transduction between damaged and undamaged cartilage. Protein–protein interaction networks related to cellular, skeletal and muscular system development also showed significant gene expression changes. The identification of immune system process and response to wounding are particularly interesting as they support the growing notion that inflammatory processes are relevant to OA pathogenesis25, 26. Although only a small percentage of genes show altered expression between damaged and undamaged cartilage these potentially have significant biological implications on tissue activity due to their aggregate effects and impact on chondrocyte behaviour.
Matrix degrading enzymes MMP1, MMP2, MMP3, MMP9 and MMP13 showed higher levels of expression in undamaged cartilage. Our study highlights that different regions of cartilage in an OA joint may have differing catabolic and anabolic activity. The higher expression of MMPs in undamaged cartilage adjacent to a lesion may be an attempt to actively remodel undamaged cartilage in response to an altered cellular environment. This in turn may reflect the changes in expression of ECM components. These expression changes may be driven by changes in weight-bearing as a result of cartilage damage elsewhere in the joint and by inherent loading differences due to location within the joint. Studies of hip OA show decreased MMP expression in hip OA cartilage, perhaps suggesting a similar mechanical environment of the hip cartilage sampled. Decreased load has been shown to increase MMP expression in tendon, cartilage and bone cells and thus mechanical off-loading may be driving the MMP expression changes seen in our study27, 28, 29, interestingly, no differential Tissue inhibitor of metalloproteinase (TIMP) expression was detected in our study. This suggests that MMP activity may be further reduced in damaged compared to undamaged cartilage due to greater availability of TIMP to inhibit MMP activity. Future work should assess MMP activity across damaged and undamaged cartilage regions.
Inflammatory processes are known to regulate ECM production and remodelling. Our study shows up-regulation of inflammatory genes (e.g., C2, CCL2, CCL3) in undamaged cartilage as well as alteration in MMPs and matrix genes suggesting a distinct but necessary inflammatory component drives OA. Inflammatory mediators expressed in undamaged cartilage as well as bone and synovial tissue may impact chondrocyte phenotype in damaged regions. The lower expression of inflammatory genes in damaged regions may also indicate a completely dysregulated responsiveness in this highly degraded cartilage where chondrocytes previously in deep zones are now exposed to the biochemical milieu of the joint.
Genes down-regulated in damaged cartilage include members of the complement system e.g., C2 and C4BPA. The membrane attack arm of the complement pathway has previously been reported to be involved in OA30. In support of this, the relative up-regulation of complement in undamaged cartilage in AMG may act to provide low-level complement activation and contribute to OA pathogenesis.
Our study also supports the hypothesis that disrupted chondrocyte signal transduction is a key modulator of OA pathogenesis. The Bone Morphogenetic Protein (BMP) and Wnt pathways are key for development and homoeostasis of cartilage and showed significant alterations in gene expression between damaged and undamaged cartilage.
A number of BMP antagonists show differential expression including NOG, FSTL3 CHRDL2 and BMPER. NOG is expressed in articular cartilage, is required for articular joint formation, and NOG ± mice are protected from inflammation-drive joint destruction31, 32. FSTL3 is an inhibitor of Activin signalling and knockout mice show severe articular cartilage lesions when compared to wild-type controls33. CHRDL2 is increased in the midzone of OA cartilage and can inhibit chondrocyte mineralization34. BMPER can antagonize BMP-induced chondrogenesis by ATDC5 chondroprogenitor cells, however it has also been reported to have pro-BMP activity through binding to chordin35, 36. Up- and down-regulation of BMP antagonists and modulators in our study suggest the balance of BMP signalling is altered in distinct cartilage regions in OA and that Smad signalling in general is disrupted. In support of this, HTRA1 and ADAMTSL2, antagonists of TGFβ signalling are increased in damaged cartilage in our study. BMP mainly signals through Smad1/5/8 and TGFβ through Smad2/3. However TGFβ-mediated Smad1/5/8 signalling is reported to become dominant in OA with a concurrent decrease in both TGFβ and Smad2/3 signalling, Smad3 null mice also show increased OA development37, 38, 39. In our study, INHBA, an activator of Smad2/3 signalling is upregulated in damaged cartilage, and its up-regulation may suggest an attempt at reactivation of Smad2/3 signalling in a feedback loop. Our data suggests that not only is there a potential for a change in Smad2/3 and Smad1/5/8 balance in OA, but also that the fine-tuning of these Smad-pathways and their interactions is disrupted.
Under- and over-activation of the Wnt pathway results in OA in transgenic mice and Wnt pathway members are regulated by articular cartilage injury in vitro [ref] and show altered expression in human OA40, 41. The Wnt antagonist, SFRP3 is down-regulated in damaged cartilage in our study and SFRP3 knockout mice show increased proteoglycan loss in OA models42. Expression of a number of Wnt – inducible genes including LEF1, WISP2 and TWIST1 were decreased in damaged cartilage in our study, as was the Wnt receptor FZD5. Furthermore FGF18, RUNX2, IGF1, FMOD, COL5A1, FN1 are also Wnt target genes showing altered expression. Our study not only confirms considerable dysregulation of Wnt signalling in OA but also suggests dysregulation within the same joint dependent on cartilage degradation status and anatomical location. The mechanism underlying this dysregulation are unknown and may result from biological and mechanical upstream regulators of both Wnt and TGF/BMP pathways and cross-talk between these and other signal transduction pathways in the chondrocyte.
Other regulated genes include DUSP10, a phosphatase that blocks MAPK activation and can protect from oxidative stress, and PSMB9 a proteosomal component; suggesting a role for stress-induced signalling in OA pathogenesis43, 44. RARRES3 a retinoic acid receptor showed decreased expression in damaged cartilage and may indicate a role of retinoic acid signalling, a key developmental signalling pathway, in OA.
Altered expression of signal transduction components will markedly impact chondrocyte behaviour between damaged and undamaged cartilage. These differences in signalling activity pose obvious complications when targeting pathways with OA therapies. Altered activities within damaged and undamaged regions could potentially cause deleterious effects in one cartilage zone despite positive effects in the other. This highlights the importance of discriminating the microanatomical location and degradation status of cartilage so that achievable therapeutic targets are identified. This can only be achieved by studying cartilage with known damage status and location and not indiscriminately harvesting entire joint surfaces.
Developmental- and skeletal injury-related networks also showed altered expression between damaged and undamaged cartilage. Regulated genes include ATF3 and TNFRSF11B. ATF3 is upregulated during chondrocyte terminal differentiation and its overexpression decreases SOX9 promoter activity and increases RUNX2 expression, thus altering the direction of chondrogenic differentiation45. TNFRSF11B (OPG) is an osteoclastogenesis inhibitor and shows increased expression in damaged cartilage in our study. The altered expression of bone-related genes may impact not only the chondrocytes but also the underlying subchondral bone, and also suggests regulation of pathways more akin to endochondral development. The significant alteration in developmental gene expression supports the hypothesis that chondrocytes in OA may assume a more developmental phenotype as an attempt at tissue repair.
A number of genes showing differential expression in our study corroborate those seen in other microarray studies of human and murine OA. However several genes show opposite changes in gene expression. This may be accounted for by the distinct phenotype of AMG and our comparison of specific microanatomical regions, whilst in other reports, OA cartilage has been compared to non-OA or early-OA samples from different individuals. Thus OA cartilage in other studies may consist of varying proportions of damaged and undamaged cartilage. Although predominant gene expression changes can be captured, the proportion of each region present in these pooled sample will impact the directionality of gene expression changes. Additionally variation due to confounding factors such as drug treatment are removed by comparing cartilage from within the same joint in AMG patients vs between OA patients and controls. It is also likely that temporal changes in gene expression occur during progression from AMG, in our study, to end-stage tricompartmental OA, in other studies.
Only a limited number of genes were found in common when we compared the gene expression changes in the present study with those previously reported in the DMM murine model of OA. This may reflect the different mechanisms and speed of disease onset, time of sampling and the inclusion of whole joint or entire cartilage (both damaged and undamaged) when analysing expression in animal studies. Genes in common include TNFAIP6 and INHBA (upregulated in OA and damaged cartilage) and may be representative of major OA-related pathways and expression changes – as these changes are also seen in other published studies of gene expression in human knee OA7, 8.
Studies, including that by Sato et al. do compare damaged and undamaged cartilage from within the same knee in human OA8. Our work confirmed the directional regulation of a number of genes identified by Sato et al. (e.g.,. ETS1, SOX11, TNFAIP6) emphasising that defining cartilage degradation status does result in more consistent identification of gene expression changes in OA. The cartilage used in the work by Sato et al. is taken from end-stage tricompartmental OA. The early disease phenotype of these patients, and thus the mechanisms underlying disease pathogenesis is likely to be heterogenous compared to the more well-defined natural history of AMG. This may account for differences between this and our study and may provide clues to genes involved in driving AMG vs those driving other OA phenotypes.
It has been postulated that AMG cartilage lesions may be exacerbated by heel strike force46 and future studies should seek to model and integrate topographical differences in joint loading over time with disease-mediated changes in regional gene expression. Chondrocyte phenotype will undoubtedly be influenced by the alteration in weight-bearing regions secondary to failing and eroded cartilage, with changes in MMP expression in our study potentially reflecting mechanical off-loading. It will also be important to assess gene expression across microanatomical regions in non-OA controls to allow further delineation, by subtractive analysis, of OA- and loading-associated genes. This will build upon the limitations of our study and provide further understanding of OA pathogenesis and potential targets for therapeutic intervention.
Through using a defined phenotype of OA we have captured and compared a temporal and spatial stage of cartilage degeneration and identified key genes and pathways regulated in OA. We hypothesize that in AMG and in OA the signalling environment of chondrocytes in distinct microanatomical regions is altered. This altered signalling environment results in an altered cellular phenotype specific to the chondrocyte location within the tibial plateau. Chondrocytes undergo repair attempts leading to alteration in ECM components and the attempted activation of developmental pathways as well as the production of soluble factors such as inflammatory cytokines. Behaviour of chondrocytes in distinct regions may influence, and be influenced by secreted proteins from chondrocytes in other regions as well as underlying subchondral bone and synovium. We have shown distinct gene expression profiles between damaged and undamaged cartilage taken from the same joint of individuals with a defined and common phenotype of knee OA. This emphasises the importance of using phenotypically and microanatomically well-defined regions of cartilage when analysing OA gene expression and chondrocyte behaviour and when developing therapeutics.
Author contributions
SS and RR drafted and wrote the manuscript and carried out laboratory work and data analysis. RD carried out MMP expression confirmation. PAH and IMC contributed to the experimental design and drafting of the manuscript. AJC provided critical revision of the article. AJP provided patient samples, input to the design and analysis of data and the initial conception of the study.
Conflict of Interest
No conflicts of interest.
Role of funding sources
We would like to thank the NIHR Musculoskeletal BRU, Royal College of Surgeons, England and Arthritis Research UK (19,222) for funding this work. There were no sponsors to this project.
Acknowledgements
We would like to thank Zhidao Xia for help with RNA extraction and members of the Oxford Musculsoskeletal Biobank for their help in collecting samples. We would also like to thank the patients of the NOC for participating in this study.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.joca.2013.12.009.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Dillon C.F., Rasch E.K., Gu Q., Hirsch R. Prevalence of knee osteoarthritis in the United States: arthritis data from the Third National Health and Nutrition Examination Survey 1991–94. J Rheumatol. 2006;33:2271–2279. [PubMed] [Google Scholar]
- 2.Goldring M.B., Goldring S.R. Osteoarthritis. J Cell Physiol. 2007;213:626–634. doi: 10.1002/jcp.21258. [DOI] [PubMed] [Google Scholar]
- 3.Aigner T., Fundel K., Saas J., Gebhard P.M., Haag J., Weiss T. Large-scale gene expression profiling reveals major pathogenetic pathways of cartilage degeneration in osteoarthritis. Arthritis Rheum. 2006;54:3533–3544. doi: 10.1002/art.22174. [DOI] [PubMed] [Google Scholar]
- 4.Appleton C.T., Pitelka V., Henry J., Beier F. Global analyses of gene expression in early experimental osteoarthritis. Arthritis Rheum. 2007;56:1854–1868. doi: 10.1002/art.22711. [DOI] [PubMed] [Google Scholar]
- 5.Fukui N., Miyamoto Y., Nakajima M., Ikeda Y., Hikita A., Furukawa H. Zonal gene expression of chondrocytes in osteoarthritic cartilage. Arthritis Rheum. 2008;58:3843–3853. doi: 10.1002/art.24036. [DOI] [PubMed] [Google Scholar]
- 6.Ijiri K., Zerbini L.F., Peng H., Otu H.H., Tsuchimochi K., Otero M. Differential expression of GADD45beta in normal and osteoarthritic cartilage: potential role in homeostasis of articular chondrocytes. Arthritis Rheum. 2008;58:2075–2087. doi: 10.1002/art.23504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karlsson C., Dehne T., Lindahl A., Brittberg M., Pruss A., Sittinger M. Genome-wide expression profiling reveals new candidate genes associated with osteoarthritis. Osteoarthritis Cartilage. 2010;18:581–592. doi: 10.1016/j.joca.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Sato T., Konomi K., Yamasaki S., Aratani S., Tsuchimochi K., Yokouchi M. Comparative analysis of gene expression profiles in intact and damaged regions of human osteoarthritic cartilage. Arthritis Rheum. 2006;54:808–817. doi: 10.1002/art.21638. [DOI] [PubMed] [Google Scholar]
- 9.Davidson R.K., Waters J.G., Kevorkian L., Darrah C., Cooper A., Donell S.T. Expression profiling of metalloproteinases and their inhibitors in synovium and cartilage. Arthritis Res Ther. 2006;8:R124. doi: 10.1186/ar2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Longo U.G., Loppini M., Fumo C., Rizzello G., Khan W.S., Maffulli N. Osteoarthritis: new insights in animal models. Open Orthop J. 2012;6:558–563. doi: 10.2174/1874325001206010558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeser R.F., Olex A.L., McNulty M.A., Carlson C.S., Callahan M.F., Ferguson C.M. Microarray analysis reveals age-related differences in gene expression during the development of osteoarthritis in mice. Arthritis Rheum. 2012;64:705–717. doi: 10.1002/art.33388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rout R., McDonnell S., Hulley P., Jayadev C., Khan T., Carr A. The pattern of cartilage damage in antero-medial osteoarthritis of the knee and its relationship to the anterior cruciate ligament. J Orthop Res. 2013;31:908–913. doi: 10.1002/jor.22253. [DOI] [PubMed] [Google Scholar]
- 13.Rout R., McDonnell S., Benson R., Athanasou N., Carr A., Doll H. The histological features of anteromedial gonarthrosis–the comparison of two grading systems in a human phenotype of osteoarthritis. Knee. 2011;18:172–176. doi: 10.1016/j.knee.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 14.Xu Y., Barter M.J., Swan D.C., Rankin K.S., Rowan A.D., Santibanez-Koref M. Identification of the pathogenic pathways in osteoarthritic hip cartilage: commonality and discord between hip and knee OA. Osteoarthritis Cartilage. 2012;20:1029–1038. doi: 10.1016/j.joca.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 15.Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380:815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerkhof H.J., Lories R.J., Meulenbelt I., Jonsdottir I., Valdes A.M., Arp P. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010;62:499–510. doi: 10.1002/art.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima M., Takahashi A., Kou I., Rodriguez-Fontenla C., Gomez-Reino J.J., Furuichi T. New sequence variants in HLA class II/III region associated with susceptibility to knee osteoarthritis identified by genome-wide association study. PLoS One. 2010;5:e9723. doi: 10.1371/journal.pone.0009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burleigh A., Chanalaris A., Gardiner M.D., Driscoll C., Boruc O., Saklatvala J. Joint immobilization prevents murine osteoarthritis and reveals the highly mechanosensitive nature of protease expression in vivo. Arthritis Rheum. 2012;64:2278–2288. doi: 10.1002/art.34420. [DOI] [PubMed] [Google Scholar]
- 19.Wei T., Kulkarni N.H., Zeng Q.Q., Helvering L.M., Lin X., Lawrence F. Analysis of early changes in the articular cartilage transcriptisome in the rat meniscal tear model of osteoarthritis: pathway comparisons with the rat anterior cruciate transection model and with human osteoarthritic cartilage. Osteoarthritis Cartilage. 2010;18:992–1000. doi: 10.1016/j.joca.2010.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Jenniskens Y.M., Koevoet W., de Bart A.C., Weinans H., Jahr H., Verhaar J.A. Biochemical and functional modulation of the cartilage collagen network by IGF1, TGFbeta2 and FGF2. Osteoarthritis Cartilage. 2006;14:1136–1146. doi: 10.1016/j.joca.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 21.Burrage P.S., Mix K.S., Brinckerhoff C.E. Matrix metalloproteinases: role in arthritis. Front Biosci. 2006;11:529–543. doi: 10.2741/1817. [DOI] [PubMed] [Google Scholar]
- 22.Moore E.E., Bendele A.M., Thompson D.L., Littau A., Waggie K.S., Reardon B. Fibroblast growth factor-18 stimulates chondrogenesis and cartilage repair in a rat model of injury-induced osteoarthritis. Osteoarthritis Cartilage. 2005;13:623–631. doi: 10.1016/j.joca.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 23.Upton A.R., Holding C.A., Dharmapatni A.A., Haynes D.R. The expression of RANKL and OPG in the various grades of osteoarthritic cartilage. Rheumatol Int. 2012;32:535–540. doi: 10.1007/s00296-010-1733-6. [DOI] [PubMed] [Google Scholar]
- 24.Kadri A., Ea H.K., Bazille C., Hannouche D., Liote F., Cohen-Solal M.E. Osteoprotegerin inhibits cartilage degradation through an effect on trabecular bone in murine experimental osteoarthritis. Arthritis Rheum. 2008;58:2379–2386. doi: 10.1002/art.23638. [DOI] [PubMed] [Google Scholar]
- 25.Berenbaum F., Eymard F., Houard X. Osteoarthritis, inflammation and obesity. Curr Opin Rheumatol. 2013;25:114–118. doi: 10.1097/BOR.0b013e32835a9414. [DOI] [PubMed] [Google Scholar]
- 26.Haseeb A., Haqqi T.M. Immunopathogenesis of osteoarthritis. Clin Immunol. 2013;146:185–196. doi: 10.1016/j.clim.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leigh D.R., Abreu E.L., Derwin K.A. Changes in gene expression of individual matrix metalloproteinases differ in response to mechanical unloading of tendon fascicles in explant culture. J Orthop Res. 2008;26:1306–1312. doi: 10.1002/jor.20650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun H.B. CITED2 mechanoregulation of matrix metalloproteinases. Ann N Y Acad Sci. 2010;1192:429–436. doi: 10.1111/j.1749-6632.2009.05305.x. [DOI] [PubMed] [Google Scholar]
- 29.Visigalli D., Strangio A., Palmieri D., Manduca P. Hind limb unloading of mice modulates gene expression at the protein and mRNA level in mesenchymal bone cells. BMC Musculoskelet Disord. 2010;11:147. doi: 10.1186/1471-2474-11-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q., Rozelle A.L., Lepus C.M., Scanzello C.R., Song J.J., Larsen D.M. Identification of a central role for complement in osteoarthritis. Nat Med. 2011;17:1674–1679. doi: 10.1038/nm.2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brunet L.J., McMahon J.A., McMahon A.P., Harland R.M. Noggin, cartilage morphogenesis, and joint formation in the mammalian skeleton. Science. 1998;280:1455–1457. doi: 10.1126/science.280.5368.1455. [DOI] [PubMed] [Google Scholar]
- 32.Lories R.J., Daans M., Derese I., Matthys P., Kasran A., Tylzanowski P. Noggin haploinsufficiency differentially affects tissue responses in destructive and remodeling arthritis. Arthritis Rheum. 2006;54:1736–1746. doi: 10.1002/art.21897. [DOI] [PubMed] [Google Scholar]
- 33.Oldknow K.J., Seebacher J., Goswami T., Villen J., Pitsillides A.A., O'Shaughnessy P.J. Follistatin-like 3 (FSTL3) mediated silencing of transforming growth factor beta (TGFbeta) signaling is essential for testicular aging and regulating testis size. Endocrinology. 2013;154:1310–1320. doi: 10.1210/en.2012-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakayama N., Han C.Y., Cam L., Lee J.I., Pretorius J., Fisher S. A novel chordin-like BMP inhibitor, CHL2, expressed preferentially in chondrocytes of developing cartilage and osteoarthritic joint cartilage. Development. 2004;131:229–240. doi: 10.1242/dev.00901. [DOI] [PubMed] [Google Scholar]
- 35.Binnerts M.E., Wen X., Cante-Barrett K., Bright J., Chen H.T., Asundi V. Human Crossveinless-2 is a novel inhibitor of bone morphogenetic proteins. Biochem Biophys Res Commun. 2004;315:272–280. doi: 10.1016/j.bbrc.2004.01.048. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J.L., Patterson L.J., Qiu L.Y., Graziussi D., Sebald W., Hammerschmidt M. Binding between Crossveinless-2 and Chordin von Willebrand factor type C domains promotes BMP signaling by blocking Chordin activity. PLoS One. 2010;5:e12846. doi: 10.1371/journal.pone.0012846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blaney Davidson E.N., Remst D.F., Vitters E.L., van Beuningen H.M., Blom A.B., Goumans M.J. Increase in ALK1/ALK5 ratio as a cause for elevated MMP-13 expression in osteoarthritis in humans and mice. J Immunol. 2009;182:7937–7945. doi: 10.4049/jimmunol.0803991. [DOI] [PubMed] [Google Scholar]
- 38.Blaney Davidson E.N., Vitters E.L., van der Kraan P.M., van den Berg W.B. Expression of transforming growth factor-beta (TGFbeta) and the TGFbeta signalling molecule SMAD-2P in spontaneous and instability-induced osteoarthritis: role in cartilage degradation, chondrogenesis and osteophyte formation. Ann Rheum Dis. 2006;65:1414–1421. doi: 10.1136/ard.2005.045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang X., Chen L., Xu X., Li C., Huang C., Deng C.X. TGF-beta/Smad3 signals repress chondrocyte hypertrophic differentiation and are required for maintaining articular cartilage. J Cell Biol. 2001;153:35–46. doi: 10.1083/jcb.153.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu M., Chen M., Zuscik M., Wu Q., Wang Y.J., Rosier R.N. Inhibition of beta-catenin signaling in articular chondrocytes results in articular cartilage destruction. Arthritis Rheum. 2008;58:2053–2064. doi: 10.1002/art.23614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu M., Tang D., Wu Q., Hao S., Chen M., Xie C. Activation of β-catenin signaling in articular chondrocytes leads to osteoarthritis-like phenotype in adult β-catenin conditional activation mice. J Bone Miner Res. 2009;24:12–21. doi: 10.1359/JBMR.080901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lories R.J., Peeters J., Bakker A., Tylzanowski P., Derese I., Schrooten J. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice. Arthritis Rheum. 2007;56:4095–4103. doi: 10.1002/art.23137. [DOI] [PubMed] [Google Scholar]
- 43.Theodosiou A., Smith A., Gillieron C., Arkinstall S., Ashworth A. MKP5, a new member of the MAP kinase phosphatase family, which selectively dephosphorylates stress-activated kinases. Oncogene. 1999;18:6981–6988. doi: 10.1038/sj.onc.1203185. [DOI] [PubMed] [Google Scholar]
- 44.Wu J., Mei C., Vlassara H., Striker G.E., Zheng F. Oxidative stress-induced JNK activation contributes to proinflammatory phenotype of aging diabetic mesangial cells. Am J Physiol Renal Physiol. 2009;297:F1622–F1631. doi: 10.1152/ajprenal.00078.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.James C.G., Woods A., Underhill T.M., Beier F. The transcription factor ATF3 is upregulated during chondrocyte differentiation and represses cyclin D1 and A gene transcription. BMC Mol Biol. 2006;7:30. doi: 10.1186/1471-2199-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gulati A., Chau R., Beard D.J., Price A.J., Gill H.S., Murray D.W. Localization of the full-thickness cartilage lesions in medial and lateral unicompartmental knee osteoarthritis. J Orthop Res. 2009;27:1339–1346. doi: 10.1002/jor.20880. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.