Highlights
-
•
Intratumour heterogeneity (ITH) has been demonstrated in various tumour types.
-
•
Distinct clonal subpopulations can exist within different regions of a tumour.
-
•
ITH has evident implications for cancer diagnosis and treatment.
-
•
There is increasing evidence for the association between ITH and drug resistance.
-
•
ITH may allow the effective use of immunotherapeutics against tumour neo-antigens.
Abstract
Recent advances in sequencing technologies have revealed extensive intratumour heterogeneity (ITH) both within individual tumours and between primary and metastatic tumours for different cancer types. Such genetic diversity may have clinical implications for both cancer diagnosis and treatment with increasing evidence linking ITH and therapeutic resistance. Nonetheless, whilst limiting the activity of targeted agents, tumour genetic heterogeneity may provide a new therapeutic opportunity through generation of neo-antigens that could be recognised and targeted by the patient's own immune system in response to immune-modulatory therapies. Longitudinal genomic studies assessing tumour clonal architecture and its correlation with the underlying immune response to cancer in each particular patient are needed to follow tumour evolutionary dynamics over time and through therapy, in order to further understand the mechanisms behind drug resistance and to inform the development of new combinatorial therapeutic strategies.
Current Opinion in Pharmacology 2013, 13:497–503
This review comes from a themed issue on Cancer
Edited by Massimo Santoro and Francesca Carlomagno
For a complete overview see the Issue and the Editorial
Available online 7th May 2013
1471-4892/$ – see front matter, © 2013 Elsevier Ltd. All rights reserved.
Introduction
The existence of distinct subpopulations of cancer cells within a tumour harbouring different behavioural phenotypes, including tumourigenicity, ability to metastasise and evolve resistance to treatment, has been recognised for many years [1]. Recent advances in sequencing technology have given genetic insight into the extent of intratumour heterogeneity (ITH) (for review see [2]), and have contributed to the opinion that ITH is not simply a tumour characteristic, but through the resolution of distinct subclones, may also have the potential to forecast risk of tumour progression and therapeutic outcome. The pattern of genomic instability, and therefore ITH, in tumours can be generated by different processes indicative of clinical outcome. Chromosomal instability (CIN), an initiator of ITH, is associated with poor prognosis in several tumour types [3–5]. Conversely, microsatellite instability (MSI), also a driver of ITH, is associated with good prognosis in colorectal cancers [6]. Therefore, the relationship between ITH and outcome is likely to be complex and dependent not only on the mechanisms generating ITH in individual tumours but also on tumour extrinsic factors such as the potential indirect impact that different forms of ITH may have on the host immune response [7••].
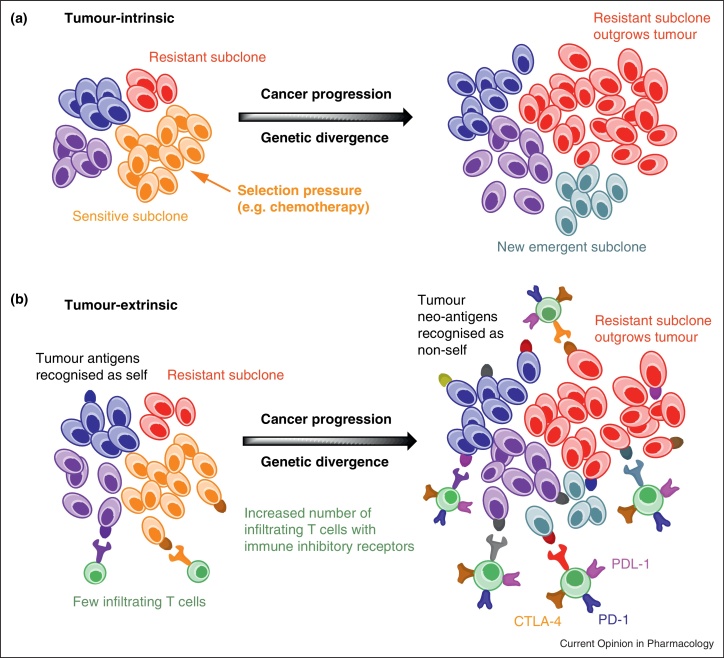
In this article we review the clinical implications of ITH for the genetic stratification of tumours, the emerging evidence that suggests the need to investigate the changing nature of tumour subclonal architecture through therapy and the potential impact of such diversity on anti-tumour immunity. We argue that an in-depth understanding of tumour evolution over time, the mechanisms driving tumour diversity and its impact on immunity may lead to the improved management of cancer patients (Figure 1).
Figure 1.
(a) Tumour-intrinsic representation of clonal evolution with eventual outgrowth of resistant subclones due to selection pressures, such as cancer treatment, and the emergence of new subclones with continued tumour progression. (b) Tumour-extrinsic representation of potential immunological aspects of clonal evolution. With continued clonal evolution, there is the potential for a broader repertoire of tumour-associated neo-antigens recognised as non-self leading to increased T cell infiltration with higher T cell receptor binding affinity. As a consequence, the expression of immune inhibitory receptors, such as PD-1, PDL-1 and CTLA-4, may also be higher. Antibody blockade of such receptors may allow therapeutic intervention that takes advantage of such neo-antigen heterogeneity within a tumour.
Intratumour heterogeneity and clonal evolution
Phenotypic heterogeneity observed in tumours results from both genetic and non-genetic causes of heterogeneity. Spontaneous tumours are known to arise through Darwinian-like somatic clonal evolution involving the acquisition of ‘driver’ events, such as genetic mutations or copy number variations, believed to affect cancer cell proliferation or survival, along with ‘passenger’ events, assumed to be phenotypically silent without a selective fitness advantage [8]. Non-genetic causes of heterogeneity include epigenetic changes [9], differentiation hierarchies as a result of cancer stem cells [10], stochastic biochemical processes within individual cells and heterogeneous tumour microenvironments [8]. Processes of genetic diversification promote tumour progression through clonal evolution so that tumours appear to be composed of evolving cell populations. The linear model of somatic tumour evolution is that of clonal succession, where a series of clonal expansions are triggered by the acquisition of driver events conferring fitness gain, outcompeting and outgrowing other clones [11]. This model implies that tumours are homogenous for functionally significant mutations, and whilst some tumours are found to evolve through linear steps [12••], there is increasing evidence for the existence of genetically distinct clonal subpopulations with substantial genetic divergence coexisting within different regions of the same primary tumour, between primary and secondary tumours, and within metastases [13••]. An alternative model of cancer evolution, distinct from the stepwise accumulation of somatic genetic alterations, is that of chromothripsis, in which a cataclysmic one-off genomic event causes massive DNA alterations acting as a driving force for cancer development and progression [14].
Evidence for intratumour heterogeneity
Recent advances in massively parallel sequencing technologies have enabled the analysis of the complex clonal architecture of both primary and metastatic tumours [15]. Patterns of clonal composition indicate tumour evolutionary paths that underlie tumour progression. An understanding of such evolutionary dynamics is essential in deciphering the clonal origins of metastases and therefore the metastatic process in general, as well as eliciting the mechanisms underlying therapeutic resistance. Several studies have demonstrated genetic diversity within tumours and inferred tumour progression by comparing the mutations and clonal composition between primary and metastatic tumours in different cancer types, including breast, renal, pancreatic, brain and ovarian (for a review see [2,16,17•]).
Intratumour heterogeneity and clinical diagnosis
The validation of predictive biomarkers may be simpler and less subject to tumour sampling bias when present in all regions of a tumour and sustained during disease progression. However, ITH for the expression of genetic and phenotypic biomarkers has been shown in several tumour types. In breast cancer, the amplification of HER2 predicts response to trastuzumab but its distribution can be heterogeneous in primary tumours and associated with shorter disease-free survival times compared to patients with homogenous HER2 amplification [18,19]. Yoon et al. [20] showed that heterogeneous HER2 amplification in oesophageal adenocarcinoma independently predicted worse disease-specific survival and overall survival compared to non-heterogeneous HER2 amplified tumours.
Primary and metastatic tumours can evolve independently and acquire different phenotypes leading to significant genetic divergence, and therefore discordance, between primary and metastatic tumours in terms of biomarkers detected in the diagnostic biopsy [21]. In non-small cell lung cancer (NSCLC), activating mutations in EGFR predict response to gefitinib, but discordance for the EGFR mutation has been shown between primary and metastatic tumours [22,23]. In primary gastric cancers, heterogeneity of HER2 amplification and HER2 protein overexpression has been shown within the same tumour, and between diagnostic biopsies and resected tumours [24]. Discordance in HER2 amplification between primary and metastatic tumours has also been shown in breast cancer [25,26]. In colorectal cancer, Vakiani et al. [27] found mutational concordance between primary and metastatic tumours for KRAS, NRAS, BRAF, PIK3CA and TP53 genes. However, in patients with a history of more than one colorectal primary tumour and interval treatment, there was evidence for discordance in TP53. These examples demonstrate that relying on a single tumour biopsy may lead to sampling bias in some cases and risk missing potentially therapeutically relevant lesions or contribute to the allocation of a mutation as actionable without establishing clonal dominance [28]. Furthermore, distinct subclonal populations appear to be unequally distributed over space and time, indicating that existing biomarkers are subject to change during disease progression [29]. This may pose a challenge for therapeutic strategies if chosen based on an archival primary tumour biopsy.
Intratumour heterogeneity and therapeutic outcome
Most advanced cancers still remain incurable despite significant progress in the fields of cancer research and therapy. Response to therapy is generally of limited duration. This may be due to the inevitable evolution and proliferation of resistant subclonal populations, which may exist before the onset of treatment, under the selective pressure of therapies [30,31••]. In NSCLC, resistance to the EGFR TKI gefitinib is associated with the positive selection of cells harbouring the gatekeeper T790M mutation known to confer insensitivity to gefitinib [32••]. Su et al. [33] demonstrated that in patients with EGFR mutations treated with EGFR TKIs, the presence of low frequency subclones harbouring T790M mutations before the onset of treatment was associated with shorter progression-free survival, and Turke et al. [34•] showed that the presence of subclones with MET amplification was associated with EGFR TKI resistance. In colorectal cancer, wild-type KRAS predicts sensitivity to anti-EGFR antibody therapies such as panitumumab. Diaz et al. [31••] showed that by monitoring circulating tumour DNA in patients treated with panitumumab for initially KRAS wild-type tumours, the emergence of mutations in KRAS could be detected during the course of therapy resulting in acquired resistance. They concluded that subclonal populations harbouring KRAS mutations existed before commencing treatment, and that under the selective pressure of anti-EGFR blockade, resistant subclones rapidly expand and repopulate the tumour. In chronic myeloid leukaemia and gastrointestinal tumours, resistance to imatinib due to mutations in the BCR-ABL fusion protein [35] and KIT [36] respectively, has also been demonstrated in the context of clonal evolution. It should be noted that not all cases of therapeutic resistance are necessarily the result of genetic heterogeneity and that non-genetic causes, such as stochastic epigenetic heterogeneity, may also allow the emergence of resistant clones under selection [37]. These examples demonstrate that relapsed clones in metastatic tumours can often be traced back to low frequency subclones before the start of treatment, hence indicating that the extent of ITH is a likely important determinant of therapeutic outcome.
In light of increasing evidence in support of ITH and its role in treatment resistance, there is a need for alternative therapeutic approaches. Gillies et al. [38] argue that subclonal populations that respond to initial therapy pass through an evolutionary bottleneck rendering them highly susceptible to a second therapy [39••], and that drug resistance in this instance, and the choice of this second therapy, could be anticipated. For example, combined therapy in EGFR mutant NSCLC with an EGFR TKI and EGFR-specific antibody could prevent resistance associated with the expansion of a subclone harbouring a T790M mutation. Approaches like this would require the development of biomarkers predicting likely resistance mechanisms in different patients, and such mechanisms could be targeted either in combination, or alternating, with standard treatment regimens [40]. Treatment dosing schedules could be adapted to prolong the suppression of resistant subpopulations, for example, drug holidays in androgen-dependent prostate cancer [41] and melanoma [42]. Other adaptive approaches could involve combining standard treatment regimens with drugs targeting phenotypes known to contribute to tumour heterogeneity, such as altered tumour vasculature and altered glucose metabolism [38].
Intratumour heterogeneity and anti-tumour immunity
Whilst emerging evidence supports the notion that ITH limits the efficacy of conventional and targeted therapeutics, its overall effect on the immune response to cancer may still be of potential benefit for the patient since intratumoural mutational diversity can provide neo-antigens that may be perceived by the immune system as non-self, producing unique opportunities for the generation of anti-tumour immunity. The wealth of data now being generated through whole genome sequencing of tumour samples provides further support for this concept. In silico-based computer algorithms combined with high-throughput post hoc analyses of data originally generated by Sjoblom et al. [43] revealed that a significant number of candidate tumour neo-antigens arise as a consequence of the multiple gene mutations occurring in breast and colorectal cancers [44]. Furthermore, previous studies in colorectal cancers have shown an association between MSI and good clinical outcomes [6]. MSI is caused by defects in the DNA mismatch-repair system leading to progressive accumulation of mutations, in particular frame-shift mutations that could positively impact immunity. In keeping with this, colorectal tumours with MSI have distinct pathological features, including increased tumour-infiltrating lymphocytes, which have also been associated with better prognosis [45]. One potential explanation is the greater mutational load in tumours with MSI in comparison to CIN tumours, which could result in a higher load of mutated self-peptides or neo-antigens seen as non-self by the immune system, increasing tumour immunogenicity and promoting enhanced T-cell activation and tumour infiltration [46]. Based on this, conventional or targeted agents capable of inducing substantial tumour cell death might produce an in vivo ‘vaccine’ or priming effect which could be further enhanced by interference with immune-modulatory pathways. Whilst the neo-antigenic repertoire generated by ITH could be seen as non-self by the immune system, the type of tumour cell death and inflammatory environment within the tumour will define their immunogenicity and the final outcome of the immune response (i.e. tumour progression versus regression). Importantly, immunity to tumour-associated antigens can be potentiated given proper identification and manipulation of immune-regulatory checkpoints restricting T cell function [47,48]. This has been recently illustrated by several high profile clinical trials in which antibody blockade of the immune inhibitory receptors PD-1, PD-L1 or CTLA-4 produced significant clinical benefits against a variety of cancers, including metastatic melanoma [49–52]. In addition to CTLA-4 and PD-1, a large number of trials are currently investigating the anti-tumour activity of monoclonal antibodies against related inhibitory receptors (Lag-3 and B7-H3), as well as of agonistic antibodies against immune-stimulatory receptors. In this particular group, antibodies against different members of the tumour necrosis receptor (TNFR) family (such as OX40, GITR, CD40, CD27 and 4-1BB) are under active investigation either as single agents or in combination with chemotherapies and targeted-therapies (for a review see [53•]).
Future directions and conclusion
Although ITH may complicate diagnostic and treatment decisions, it can be clinically useful in predicting clinical outcome. In Barrett's oesophagus [54•] and breast cancer [55,56], ITH has been shown to predict invasive progression. Conversely, extreme CIN, an initiator of ITH, has been shown to be associated with improved long-term survival in oestrogen receptor (ER)-negative breast cancer [57•,58]. Extensive ITH in tumours provides greater opportunity for adaptive responses to selective pressures such as hypoxia, chemotherapy and radiotherapy [39••] and therefore, measurements of genetic and phenotypic heterogeneity may be of significant value in patient risk stratification [59]. Reliable methods to interrogate tumours and elicit their underlying clonal architecture need to be developed in order to test the association between distinct mechanisms of ITH and clinical outcome. ITH poses a challenge for effective cancer therapy, and the resulting heterogeneous expression of biomarkers may have implications in terms of accurate diagnosis and treatment outcome [17•]. Longitudinal genomic analysis of tumours at diagnosis, during treatment and at relapse may inform new approaches and shine a light upon tumour adaptive mechanisms through therapy. Clinical trials and biomarker studies should consider such designs to demonstrate the potential benefit of adaptive therapy in response to tumour evolution through the disease course. Obtaining multiple tumour biopsies to study tumour clonal architecture in such trials may be clinically challenging but should at least be considered. Potential non-invasive alternatives to re-biopsy in patients with multiple or inaccessible metastases may include molecular imaging and circulating tumour DNA [60].
With the development of improved technologies allowing the interrogation of ITH, our understanding of tumours and their evolutionary trajectories may lead to better design of clinical trials in search of improved therapeutic interventions to anticipate the emergence of drug resistance mechanisms and generate improved predictive and prognostic biomarkers [61,62]. Whilst cancer cells cannot anticipate future evolutionary events or the selective pressures they may encounter, we should prepare for, and proactively manage, such changes and use our acquired knowledge of tumour evolutionary dynamics to predict and guide treatment strategies in order to attempt to improve patient outcomes. Underlying mechanisms of ITH that result in increased mutational diversity may in theory result in the generation of neo-antigens recognised by the immune system as non-self. The pipeline of new immunotherapeutic drugs offers a newer and larger window of opportunity through which tumour sensitivity could be enhanced via the rational combination of targeted and immune-therapies where targeted therapies will promote tumour destruction and neo-antigen exposure (generated through ITH) to the immune system, whilst manipulation of immune-regulatory pathways will potentially enable a powerful, diverse and durable response against the tumour.
Conflict of interest
None declared.
Role of the funding source
None.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
CSw is funded by the Medical Research Council, Cancer Research UK and the Breast Cancer Research Foundation.
Contributor Information
Sergio A Quezada, Email: s.quezada@ucl.ac.uk.
Charles Swanton, Email: Charles.Swanton@cancer.org.uk.
References
- 1.Heppner G.H., Miller B.E. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2:5–23. doi: 10.1007/BF00046903. [DOI] [PubMed] [Google Scholar]
- 2.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carter S.L., Eklund A.C., Kohane I.S., Harris L.N., Szallasi Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat Genet. 2006;38:1043–1048. doi: 10.1038/ng1861. [DOI] [PubMed] [Google Scholar]
- 4.Habermann J.K., Doering J., Hautaniemi S., Roblick U.J., Bundgen N.K., Nicorici D., Kronenwett U., Rathnagiriswaran S., Mettu R.K., Ma Y. The gene expression signature of genomic instability in breast cancer is an independent predictor of clinical outcome. Int J Cancer. 2009;124:1552–1564. doi: 10.1002/ijc.24017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walther A., Houlston R., Tomlinson I. Association between chromosomal instability and prognosis in colorectal cancer: a meta-analysis. Gut. 2008;57:941–950. doi: 10.1136/gut.2007.135004. [DOI] [PubMed] [Google Scholar]
- 6.Wright C.M., Dent O.F., Barker M., Newland R.C., Chapuis P.H., Bokey E.L., Young J.P., Leggett B.A., Jass J.R., Macdonald G.A. Prognostic significance of extensive microsatellite instability in sporadic clinicopathological stage C colorectal cancer. Br J Surg. 2000;87:1197–1202. doi: 10.1046/j.1365-2168.2000.01508.x. [DOI] [PubMed] [Google Scholar]
- Peggs K.S., Segal N.H., Allison J.P. Targeting immunosupportive cancer therapies: accentuate the positive, eliminate the negative. Cancer Cell. 2007;12:192–199. doi: 10.1016/j.ccr.2007.08.023. [DOI] [PubMed] [Google Scholar]; This paper discusses the impact that ITH may have on the host immune response to cancer and therefore the potential for immunotherapeutic strategies to take advantage of such tumour heterogeneity.
- 8.Marusyk A., Almendro V., Polyak K. Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer. 2012;12:323–334. doi: 10.1038/nrc3261. [DOI] [PubMed] [Google Scholar]
- 9.Dick J.E. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 10.Shipitsin M., Campbell L.L., Argani P., Weremowicz S., Bloushtain-Qimron N., Yao J., Nikolskaya T., Serebryiskaya T., Beroukhim R., Hu M. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 11.Nowell P.C. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]; This study demonstrated complex patterns of clonal evolution and significant genetic variability between leukaemic stem cells.
- Navin N., Krasnitz A., Rodgers L., Cook K., Meth J., Kendall J., Riggs M., Eberling Y., Troge J., Grubor V. Inferring tumor progression from genomic heterogeneity. Genome Res. 2010;20:68–80. doi: 10.1101/gr.099622.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that clonal heterogeneity in breast cancer is common using a method involving macro-dissection of solid tumours, flow-sorting cell populations by DNA content and analysing their clonal structure using aCGH and showed that subclones within the same tumour may be anatomically separate or intermixed.
- 14.Stephens P.J., Greenman C.D., Fu B., Yang F., Bignell G.R., Mudie L.J., Pleasance E.D., Lau K.W., Beare D., Stebbings L.A. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Russnes H.G., Navin N., Hicks J., Borresen-Dale A.L. Insight into the heterogeneity of breast cancer through next-generation sequencing. J Clin Invest. 2011;121:3810–3818. doi: 10.1172/JCI57088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horswell S., Matthews N., Swanton C. Cancer heterogeneity and ‘the struggle for existence’: diagnostic and analytical challenges. Cancer Lett. 2012 doi: 10.1016/j.canlet.2012.10.031. [DOI] [PubMed] [Google Scholar]
- Fisher R., Pusztai L., Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br J Cancer. 2013;108:479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]; This review discusses the exiting evidence for ITH in several tumour types.
- 18.Davila E., Amazon K. The clinical importance of the heterogeneity of HER2 neu. Case Rep Oncol. 2010;3:268–271. doi: 10.1159/000319020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seol H., Lee H.J., Choi Y., Lee H.E., Kim Y.J., Kim J.H., Kang E., Kim S.W., Park S.Y. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol. 2012;25:938–948. doi: 10.1038/modpathol.2012.36. [DOI] [PubMed] [Google Scholar]
- 20.Yoon H.H., Shi Q., Sukov W.R., Lewis M.A., Sattler C.A., Wiktor A.E., Wu T.T., Diasio R.B., Jenkins R.B., Sinicrope F.A. Adverse prognostic impact of intratumor heterogeneous HER2 gene amplification in patients with esophageal adenocarcinoma. J Clin Oncol. 2012;30:3932–3938. doi: 10.1200/JCO.2012.43.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoecklein N.H., Klein C.A. Genetic disparity between primary tumours, disseminated tumour cells, and manifest metastasis. Int J Cancer. 2010;126:589–598. doi: 10.1002/ijc.24916. [DOI] [PubMed] [Google Scholar]
- 22.Gomez-Roca C., Raynaud C.M., Penault-Llorca F., Mercier O., Commo F., Morat L., Sabatier L., Dartevelle P., Taranchon E., Besse B. Differential expression of biomarkers in primary non-small cell lung cancer and metastatic sites. J Thorac Oncol. 2009;4:1212–1220. doi: 10.1097/JTO.0b013e3181b44321. [DOI] [PubMed] [Google Scholar]
- 23.Kalikaki A., Koutsopoulos A., Trypaki M., Souglakos J., Stathopoulos E., Georgoulias V., Mavroudis D., Voutsina A. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–929. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Luo H., Li Y., Li J., Cai Z., Su X., Dai D., Du W., Chen T., Chen M. Intratumoral heterogeneity determines discordant results of diagnostic tests for human epidermal growth factor receptor (HER) 2 in gastric cancer specimens. Cell Biochem Biophys. 2012;62:221–228. doi: 10.1007/s12013-011-9286-1. [DOI] [PubMed] [Google Scholar]
- 25.Gancberg D., Di Leo A., Cardoso F., Rouas G., Pedrocchi M., Paesmans M., Verhest A., Bernard-Marty C., Piccart M.J., Larsimont D. Comparison of HER-2 status between primary breast cancer and corresponding distant metastatic sites. Ann Oncol. 2002;13:1036–1043. doi: 10.1093/annonc/mdf252. [DOI] [PubMed] [Google Scholar]
- 26.Cottu P.H., Asselah J., Lae M., Pierga J.Y., Dieras V., Mignot L., Sigal-Zafrani B., Vincent-Salomon A. Intratumoral heterogeneity of HER2/neu expression and its consequences for the management of advanced breast cancer. Ann Oncol. 2008;19:595–597. doi: 10.1093/annonc/mdn021. [DOI] [PubMed] [Google Scholar]
- 27.Vakiani E., Janakiraman M., Shen R., Sinha R., Zeng Z., Shia J., Cercek A., Kemeny N., D’Angelica M., Viale A. Comparative genomic analysis of primary versus metastatic colorectal carcinomas. J Clin Oncol. 2012;30:2956–2962. doi: 10.1200/JCO.2011.38.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yap T.A., Gerlinger M., Futreal P.A., Pusztai L., Swanton C. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003854. 127ps110. [DOI] [PubMed] [Google Scholar]
- 29.Greaves M., Maley C.C. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engelman J.A., Settleman J. Acquired resistance to tyrosine kinase inhibitors during cancer therapy. Curr Opin Genet Dev. 2008;18:73–79. doi: 10.1016/j.gde.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Diaz L.A., Jr., Williams R.T., Wu J., Kinde I., Hecht J.R., Berlin J., Allen B., Bozic I., Reiter J.G., Nowak M.A. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486:537–540. doi: 10.1038/nature11219. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study showed that by monitoring circulating tumour DNA in patients treated with panitumumab for KRAS wild-type tumours, the emergence of KRAS mutations under the selective pressure of anti-EGFR blockade could be detected resulting in acquired resistance. By mathematical modelling they conclude that these resistant subclones were present before commencing therapy.
- Kosaka T., Yatabe Y., Endoh H., Yoshida K., Hida T., Tsuboi M., Tada H., Kuwano H., Mitsudomi T. Analysis of epidermal growth factor receptor gene mutation in patients with non-small cell lung cancer and acquired resistance to gefitinib. Clin Cancer Res. 2006;12:5764–5769. doi: 10.1158/1078-0432.CCR-06-0714. [DOI] [PubMed] [Google Scholar]; This study showed that resistance to the EGFR TKI gefitinib is associated with the positive selection of cells harbouring the gatekeeper T790M mutation known to confer insensitivity to gefitinib.
- 33.Su K.Y., Chen H.Y., Li K.C., Kuo M.L., Yang J.C., Chan W.K., Ho B.C., Chang G.C., Shih J.Y., Yu S.L. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:433–440. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- Turke A.B., Zejnullahu K., Wu Y.L., Song Y., Dias-Santagata D., Lifshits E., Toschi L., Rogers A., Mok T., Sequist L. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study in non-small cell lung cancer showed that the presence of subclones with MET amplification was associated with EGFR TKI resistance.
- 35.Shah N.P., Nicoll J.M., Nagar B., Gorre M.E., Paquette R.L., Kuriyan J., Sawyers C.L. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 36.Liegl B., Kepten I., Le C., Zhu M., Demetri G.D., Heinrich M.C., Fletcher C.D., Corless C.L., Fletcher J.A. Heterogeneity of kinase inhibitor resistance mechanisms in GIST. J Pathol. 2008;216:64–74. doi: 10.1002/path. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janne P.A., Gray N., Settleman J. Factors underlying sensitivity of cancers to small-molecule kinase inhibitors. Nat Rev Drug Discov. 2009;8:709–723. doi: 10.1038/nrd2871. [DOI] [PubMed] [Google Scholar]
- 38.Gillies R.J., Verduzco D., Gatenby R.A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat Rev Cancer. 2012;12:487–493. doi: 10.1038/nrc3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlinger M., Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103:1139–1143. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated spatial ITH within individual clear cell renal cell carcinomas and distinct diagnostic signatures derived from different biopsies of the same tumour.
- 40.Komarova N.L., Wodarz D. Drug resistance in cancer: principles of emergence and prevention. Proc Natl Acad Sci U S A. 2005;102:9714–9719. doi: 10.1073/pnas.0501870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abrahamsson P.A. Potential benefits of intermittent androgen suppression therapy in the treatment of prostate cancer: a systematic review of the literature. Eur Urol. 2010;57:49–59. doi: 10.1016/j.eururo.2009.07.049. [DOI] [PubMed] [Google Scholar]
- 42.Das Thakur M., Salangsang F., Landman A.S., Sellers W.R., Pryer N.K., Levesque M.P., Dummer R., McMahon M., Stuart D.D. Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature. 2013;494:251–255. doi: 10.1038/nature11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sjoblom T., Jones S., Wood L.D., Parsons D.W., Lin J., Barber T.D., Mandelker D., Leary R.J., Ptak J., Silliman N. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 44.Segal N.H., Parsons D.W., Peggs K.S., Velculescu V., Kinzler K.W., Vogelstein B., Allison J.P. Epitope landscape in breast and colorectal cancer. Cancer Res. 2008;68:889–892. doi: 10.1158/0008-5472.CAN-07-3095. [DOI] [PubMed] [Google Scholar]
- 45.Galon J., Costes A., Sanchez-Cabo F., Kirilovsky A., Mlecnik B., Lagorce-Pages C., Tosolini M., Camus M., Berger A., Wind P. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 46.Ishikawa T., Fujita T., Suzuki Y., Okabe S., Yuasa Y., Iwai T., Kawakami Y. Tumor-specific immunological recognition of frameshift-mutated peptides in colon cancer with microsatellite instability. Cancer Res. 2003;63:5564–5572. [PubMed] [Google Scholar]
- 47.Quezada S.A., Peggs K.S., Curran M.A., Allison J.P. CTLA4 blockade and GM-CSF combination immunotherapy alters the intratumor balance of effector and regulatory T cells. J Clin Invest. 2006;116:1935–1945. doi: 10.1172/JCI27745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quezada S.A., Peggs K.S., Simpson T.R., Shen Y., Littman D.R., Allison J.P. Limited tumor infiltration by activated T effector cells restricts the therapeutic activity of regulatory T cell depletion against established melanoma. J Exp Med. 2008;205:2125–2138. doi: 10.1084/jem.20080099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hodi F.S., O’Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B., Gonzalez R., Robert C., Schadendorf D., Hassel J.C. Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med. 2010;363:711–723. doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robert C., Thomas L., Bondarenko I., O’Day S.M., Garbe D.J., Lebbe C., Baurain C., Testori J.F., Grob A. Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med. 2011;364:2517–2526. doi: 10.1056/NEJMoa1104621. [DOI] [PubMed] [Google Scholar]
- 51.Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brahmer J.R., Tykodi S.S., Chow L.Q., Hwu W.J., Topalian S.L., Hwu P., Drake C.G., Camacho L.H., Kauh J., Odunsi K. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N Engl J Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Zhu Y., Chen L. Advances in targeting cell surface signalling molecules for immune modulation. Nat Rev Drug Discov. 2013;12:130–146. doi: 10.1038/nrd3877. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reviews the advances in immuno-modulatory approaches in cancer.
- Maley C.C., Galipeau P.C., Finley J.C., Wongsurawat V.J., Li X., Sanchez C.A., Paulson T.G., Blount P.L., Risques R.A., Rabinovitch P.S. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]; This study demonstrated that clonality diversity measures adapted from ecology and evolution can predict the progression from pre-malignant Barrett's oesophagus to adenocarcinoma.
- 55.Park S.Y., Gonen M., Kim H.J., Michor F., Polyak K. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Invest. 2010;120:636–644. doi: 10.1172/JCI40724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teixeira M.R., Pandis N., Bardi G., Andersen J.A., Heim S. Karyotypic comparisons of multiple tumorous and macroscopically normal surrounding tissue samples from patients with breast cancer. Cancer Res. 1996;56:855–859. [PubMed] [Google Scholar]
- Roylance R., Endesfelder D., Gorman P., Burrell R.A., Sander J., Tomlinson I., Hanby A.M., Speirs V., Richardson A.L., Birkbak N.J. Relationship of extreme chromosomal instability with long-term survival in a retrospective analysis of primary breast cancer. Cancer Epidemiol Biomarkers Prev. 2011;20:2183–2194. doi: 10.1158/1055-9965.EPI-11-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrated that extreme chromosomal instability, a driver of ITH, is associated with improved long-term survival in oestrogen receptor (ER)-negative breast cancer.
- 58.Birkbak N.J., Eklund A.C., Li Q., McClelland S.E., Endesfelder D., Tan P., Tan I.B., Richardson A.L., Szallasi Z., Swanton C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011;71:3447–3452. doi: 10.1158/0008-5472.CAN-10-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almendro V., Marusyk A., Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol. 2013;8:277–302. doi: 10.1146/annurev-pathol-020712-163923. [DOI] [PubMed] [Google Scholar]
- 60.Board R.E., Wardley A.M., Dixon J.M., Armstrong A.C., Howell S., Renshaw L., Donald E., Greystoke A., Ranson M., Hughes A. Detection of PIK3CA mutations in circulating free DNA in patients with breast cancer. Breast Cancer Res Treat. 2010;120:461–467. doi: 10.1007/s10549-010-0747-9. [DOI] [PubMed] [Google Scholar]
- 61.Swanton C., Larkin J.M., Gerlinger M., Eklund A.C., Howell M., Stamp G., Downward J., Gore M., Futreal P.A., Escudier B. Predictive biomarker discovery through the parallel integration of clinical trial and functional genomics datasets. Genome Med. 2010;2:53. doi: 10.1186/gm174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan D.S., Gerlinger M., Teh B.T., Swanton C. Anti-cancer drug resistance: understanding the mechanisms through the use of integrative genomics and functional RNA interference. Eur J Cancer. 2010;46:2166–2177. doi: 10.1016/j.ejca.2010.03.019. [DOI] [PubMed] [Google Scholar]