Abstract
Hereditary myopathy with early respiratory failure is an autosomal dominant myopathy caused by mutations in the 119th fibronectin-3 domain of titin. To date all reported patients with the most common mutation in this domain (p.C30071R) appear to share ancestral disease alleles. We undertook this study of two families with the p.C30071R mutation to determine whether they share the same haplotype as previously reported British families or whether the mutation arose as a de novo event. We sequenced the 119th fibronectin-3 domain in these two probands and flanking polymorphisms associated with the British haplotype in hereditary myopathy with early respiratory failure. A family of Indian descent had a haplotype that was not compatible with the British shared haplotype. Cloning of the 119th fibronectin-3 domain in this patient demonstrated polymorphisms rs191484894 and novel noncoding variant c.90225C>T on the same allele as the mutation, which is distinct from previously reported British families. This proves that the p.C30071R mutation itself (rather than the haplotype containing this mutation) causes hereditary myopathy with early respiratory failure and suggests its independent origin in different ethnic groups.
Keywords: Hereditary myopathy with early respiratory failure, Titin, Titinopathy, Myofibrillar myopathy, Haplotype
1. Introduction
Hereditary myopathy with early respiratory failure (HMERF) is an autosomal dominant myopathy causing skeletal muscle weakness (proximal and/or distal) with early involvement of respiratory muscles. It is caused by mutations in the 119th fibronectin 3 (FN3) domain of titin (TTN) [1,2]. The clinical presentation can be quite variable, although the condition should be particularly suspected in patients with disease onset in mid-life, with preferential involvement of distal leg muscles, and symptoms of respiratory muscle weakness such as orthopnoea [2]. Creatine kinase is often mildly elevated (up to 1000 U/ml) and muscle MRI shows a characteristic pattern which includes fatty infiltration of semitendinosus, sartorius and gracilis at the thigh and anterior compartment lower leg muscles [2]. The condition meets diagnostic criteria for myofibrillar myopathy on muscle pathology, and has been identified in approximately 5% of undiagnosed myofibrillar myopathy cohorts from two separate studies [3,4], although muscle pathology may also be nonspecific in some patients [2]. The first mutation to be associated with HMERF was the p.R32450W mutation in the kinase domain [5]. However, no further HMERF cases with kinase domain mutations have since been identified [3,6], and this variant is now reported as a low frequency polymorphism in controls of European descent (rs140319117), and patients from this family also have a mutation in the 119th FN3 domain of TTN [7].
The most common mutation associated with this condition is the p.C30071R mutation in TTN, present in 20 of 30 reported HMERF families [1–4,8,9]. However, to date all families characterised with this mutation share common haplotypes, implying that all have inherited an ancestral allele: 8 UK-based families and one Canadian family (with British ancestry) share a single haplotype [3,9]. All families with this mutation reported by Palmio and colleagues also share a common haplotype, which includes a British family (Family E) [8], and three previously reported Scandinavian families [1]. Given that British families are involved in both studies, it is probable that all families share the same haplotype, although this is not confirmed given the different methodologies employed in defining the haplotypes between studies. This leaves only two non-UK families with this mutation whose haplotypes have not been characterised, from the work of Toro et al. [4]. We studied these two families to determine whether the p.C30071R mutation in these patients is on the same ancestral allele, or whether it occurred as a de novo event.
2. Materials and methods
The probands of two previously reported families (Family B and Family C from Toro et al. [4]) were studied in this report. Family B resides in Canada although is of Indian ancestry. Family C is of Spanish descent. Neither of these families is known to have British ancestors. Clinical details are described in the work by Toro et al. [4]. Sequencing of leucocyte DNA was performed using previously described methods for the 119th FN3 domain of TTN and flanking polymorphisms that define the UK haplotype [3]. Haplotypes were manually reconstructed assuming the minimum number of recombinations. The PCR-amplified 119th FN3 domain from Family B was gel purified using Qiaquick gel extraction kit (Qiagen, Manchester UK), ligated into pGEM-T vector system (Promega, Southampton, UK), heat-shock transfected into JM109 competent bacteria (Promega, Southampton, UK) and cloned on selective ampicillin/X-gal LB agar plates. Clones were amplified with PCR using M13 primers. Sequencing was performed using BigDye (Applied Biosystems) according to the manufacturer’s protocol with an ABI 3130XL sequencer.
3. Results
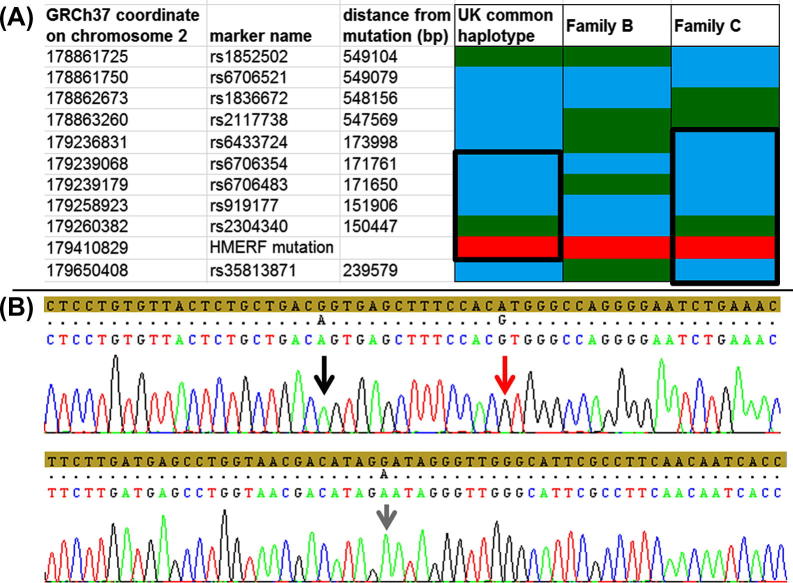
Family B (India) does not share the UK haplotype. Sequencing results of SNP markers was incompatible with the shared haplotype of the previously reported UK families (Fig. 1a). Upstream from the mutation in the 119th FN3 domain were two heterozygous variants, c.90374C>T (listed as rs191484894 in dbSNP), and a novel synonymous variant, c.90225C>T (using NM_001256850 as the reference sequence). Sequencing of clones of the PCR fragment demonstrated that both variants are on the same allele as the p.C30071R mutation at c.90211T>C (Fig. 1b).
Fig. 1.
(A) The shared haplotypes are demonstrated, assuming the minimum number of recombinations. Green boxes indicate reference sequence, whereas blue boxes indicate the polymorphic variant. The HMERF mutation is indicated by red boxes. The UK common haplotype represented here is the shared haplotype in 5 of 8 reported UK families, although a black box in this column indicates the core haplotype of 171 kbp which is shared by all 8 families. Family B (India) does not share the haplotype. The column for Family C (Spain) indicates the compatible shared haplotype with UK families (outlined by a black box). (B) Sequencing chromatograph demonstrating the cloned disease allele from the 119th FN3 domain in Family B. The amber band indicates the reference sequence. The upper trace is the region immediately surrounding the HMERF mutation c.90211T>C (p.C30071R) (indicated by a red arrow). The novel noncoding variant c.90225C>T is on the same allele as the mutation and indicated by a black arrow. The lower trace is the sequence immediately surrounding the other heterozygous variant, c.90374C>T (a.k.a: rs191484894) which also segregated with the disease mutation on the same allele.
Sequencing results from the proband of Family C (Spain) was compatible with the shared haplotype of the UK families of a minimum size of 673 kbp (defined by rs6433724, rs6706354, rs6706483, rs919177, the mutation at position chr2.179410829A>G (reference sequence GRCh37), and rs35813871) (Fig. 1a). Within this shared region is the 171 kbp haplotype which is shared between all previously reported UK families with the p.C30071R mutation.
4. Discussion
This study provides definitive proof of the pathogenicity of the p.C30071R mutation associated with HMERF. To date, haplotype studies suggest that all patients with this mutation originated from a very remote common founder [3,8]. Therefore, in the absence of functional evidence of pathogenicity for this single amino acid substitution, it would only be accurate to state that the haplotype containing the p.C30071R mutation is associated with HMERF. However, Family B from India has a different genetic background, with two polymorphisms within the same FN3 domain that are not present in the UK patients, and which we show to be in cis with the p.C30071R mutation. This indicates that the p.C30071R mutation itself causes HMERF.
HMERF is already considered to be an under-recognised condition that is more common than expected and is distributed internationally [3,4,8]. Its true prevalence is unknown, although the presence of the p.C30071R mutation on a different haplotype in Family B indicates the possibility that this position in TTN may be a mutational hotspot and supports a higher than expected prevalence, and perhaps a worldwide distribution of this condition [4,10].
The shared haplotype of Spanish Family C with the UK families is consistent with our findings suggesting the p.C30071R mutation was an ancient occurrence, and it is possible that the other European and Scandinavian patients share the same common founder. The fact that this is a late-onset disease thus evading reproductive selection is probably what allowed the ancestral allele to have broadly permeated an international population. We therefore reiterate that titinopathy is an important diagnostic consideration in patients with autosomal dominant adult-onset myopathy.
Acknowledgements
GP is the recipient of a Bisby Fellowship from the Canadian Institutes of Health Research. PFC is an Honorary Consultant Neurologist at Newcastle upon Tyne Foundation Hospitals NHS Trust, is a Wellcome Trust Senior Fellow in Clinical Science (084980/Z/08/Z), and a UK NIHR Senior Investigator. PFC receives additional support from the Wellcome Trust Centre for Mitochondrial Research (096919Z/11/Z), the Medical Research Council (UK) Centre for Translational Research in Neuromuscular Diseases, and EU FP7 TIRCON, and the National Institute for Health Research (NIHR) Newcastle Biomedical Research Centre based at Newcastle upon Tyne Hospitals NHS Foundation Trust and Newcastle University. The research was supported in part by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke.
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Ohlsson M., Hedberg C., Bradvik B. Hereditary myopathy with early respiratory failure associated with a mutation in A-band titin. Brain. 2012;135(Pt 6 (June)):1682–1694. doi: 10.1093/brain/aws103. [DOI] [PubMed] [Google Scholar]
- 2.Pfeffer G., Elliott H.R., Griffin H. Titin mutation segregates with hereditary myopathy with early respiratory failure. Brain. 2012;135(Pt 6 (June)):1695–1713. doi: 10.1093/brain/aws102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeffer G., Barresi R., Wilson I.J. Titin founder mutation is a common cause of myofibrillar myopathy with early respiratory failure. J Neurol Neurosurg Psychiatry. 2013 doi: 10.1136/jnnp-2012-304728. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Toro C., Olive M., Dalakas M.C. Exome sequencing identifies titin mutations causing hereditary myopathy with early respiratory failure (HMERF) in families of diverse ethnic origins. BMC Neurol. 2013;13 doi: 10.1186/1471-2377-13-29. 29,2377-13-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lange S., Xiang F., Yakovenko A. The kinase domain of titin controls muscle gene expression and protein turnover. Science. 2005;308(5728 (June)):1599–1603. doi: 10.1126/science.1110463. [DOI] [PubMed] [Google Scholar]
- 6.Pfeffer G., Griffin H., Pyle A., Horvath R., Chinnery P.F. Reply: Hereditary myopathy with early respiratory failure is caused by mutations in the titin FN3 119 domain. Brain. 2013 doi: 10.1093/brain/awt306. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedberg C., Melberg A., Dahlbom K., Oldfors A. Hereditary myopathy with early respiratory failure is caused by mutations in the titin FN3 119 domain. Brain. 2013 doi: 10.1093/brain/awt305. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Palmio J., Evila A., Chapon F. Hereditary myopathy with early respiratory failure: Occurrence in various populations. J Neurol Neurosurg Psychiatry. 2013 doi: 10.1136/jnnp-2013-304965. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 9.Pfeffer G, Joseph JT, Innes AM, et al. Titinopathy in a Canadian family sharing the British founder haplotype. Can J Neurol Sci 2014; 41(1) [in press]. [DOI] [PMC free article] [PubMed]
- 10.Izumi R., Niihori T., Aoki Y. Exome sequencing identifies a novel TTN mutation in a family with hereditary myopathy with early respiratory failure. J Hum Genet. 2013;58(5 (May)):259–266. doi: 10.1038/jhg.2013.9. [DOI] [PubMed] [Google Scholar]