Abstract
Molecular switches have been instrumental in the development of powerful and versatile genetic tools for probing biochemical processes, such as intracellular signaling, directly within their native contexts. This protocol outlines a method for using a kinase-inducible bimolecular switch, along with live-cell fluorescence microscopy, to directly control and monitor the activity of a specific enzyme in living cells.
Keywords: Live-cell imaging, Biochemical perturbation, Biosensor, Fluorescence
INTRODUCTION
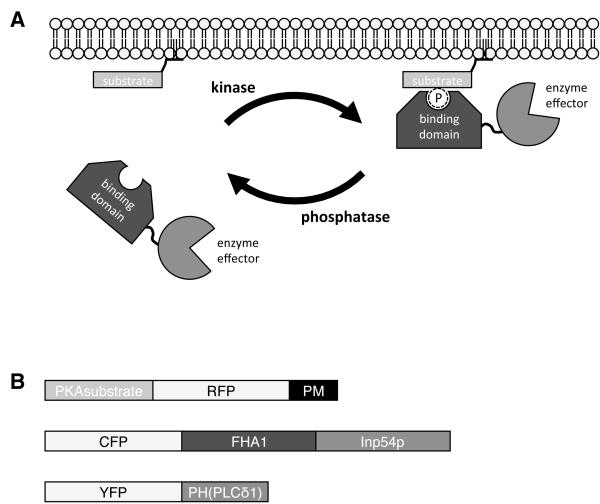
Perturbing a cell’s biochemical processes with ever greater specificity and selectivity is central to the study of intracellular signaling networks. Molecular switches, which couple specific input signals to particular output responses, offer a way for researchers to probe these biochemical networks in more detail. This protocol describes a method for using a kinase-inducible bimolecular switch (KIBS) to selectively and acutely control local enzyme activity in living cells. As illustrated in Figure 1A, the switch consists of a short peptide that acts as a surrogate substrate for a particular kinase, plus a phosphoamino acid-binding domain (PAABD) that will recognize and bind the phosphorylated peptide upon activation of the kinase. By targeting the substrate peptide to a specific subcellular compartment, and tethering the PAABD to an enzymatic effector, activation of the kinase can be used to induce relocalization of the enzyme effector, thus allowing for localized perturbation of the chosen enzyme activity. This system can then be combined with a genetically encoded fluorescent reporter for the activity of interest, for real-time control and monitoring of local enzyme activity in living cells.
Figure 1.
Overview of the KIBS system used in this protocol. (A) A strategy for the control of enzyme activity via translocation using a kinase-inducible bimolecular switch. A substrate peptide corresponding to a specific candidate kinase is anchored to the surface of a particular subcellular compartment (e.g., the plasma membrane). In resting cells, the binding domain, which is fused to an effector enzyme, remains free in the cytosol. Upon stimulation of the kinase, the substrate peptide becomes phosphorylated, recruiting the binding domain to form a complex, and causing translocation of the effector enzyme. This process can be reversed by inhibition of the kinase and dephosphorylation of the substrate, allowing repeated cycles of activation and inactivation of the effector enzyme. (B) Schematic illustration of the constructs used.
STRATEGIC PLANNING
When setting out to selectively control enzyme activity using a KIBS, the specific cellular pathway or process of interest to the investigator should serve as the starting point for all other considerations. Choose a cell type for the experiments that is well-suited for studying the pathway of interest, in particular the activity to be perturbed (e.g., PIP2 degradation), in order to get robust responses. Once the pathway and cell type have been selected, the next major steps are to choose an enyzme effector that will be used to perturb the activity of interest, and the kinase that will be used to power the KIBS.
The goal of this technique is to perturb a specific internal step within a signaling pathway, in contrast to activation of the entire pathway using more tradional pharmacological stimuli. As such, the enzyme effector should be the same enzyme that acts at this particular step in the pathway, or one with a similar catalytic activity. However, before choosing the enzyme effector, it is important to consider how to ensure that the enzyme becomes fully activated in response to the KIBS, and that it is active only when the KIBS is induced. This is critical for minimizing partial or unwanted catalytic activity. One potential solution is to use a constitutively active mutant form of an enzyme whose substrate is only found in a specific subcellular compartment, or is at least not readily accessible from the cytosol, in concert with a translocation-based approach whereby induction of the KIBS results in re-localization of the enzyme effector with its substrate (Fig. 1A).
Re-localization is faciliated by subcellular targeting of the substrate peptide component of the KIBS, via fusion of a targeting sequence to the N- or C-terminus of the peptide. When the peptide is phosphorylated, the PAABD, which is tethered to the enzyme effector, will translocate to the compartment containing the peptide, bringing the effector with it. This ensures that catalytic activity is only observed in response to the KIBS. A benefit of this approach is that it allows for control of local enzyme activity, adding an additional level of selectivity to the perturbation. Unfortunately, this necessarily limits the selection of the enzyme effector, in addition to the activity of interest, as not all enzymes will fit with this approach. Incorporating other enzymes into the KIBS system, such as those that are active throughout the cytosol, will require engineering alternative strategies for satisfying the above parameters. These should be developed based on consultation of the literature and available structural data for the enzyme of interest, along with extensive trial and error.
To construct the KIBS itself, the primary consideration is the kinase that will drive the switch. Most importantly, great care should be taken to identify a kinase that does not directly affect the pathway of interest (see Critical Parameters). Whichever kinase is chosen should have a well-characterized consensus phosphorylation sequence, to be incorporated into the substrate peptide component of the KIBS. The availability of pharmacological agents, such as direct stimulators and inhibitors, should also be taken into account, as these will be necessary to “flip” the switch. Kinase inhibitors are of particular use because they allow for reversal of the switch (provided it can be dephosphorylated by cellular phosphatases) and concomitant deactivation of the effector enzyme, enabling dynamic on- and off-switching of the perturbation. An appropriate PAABD should also be selected, capable of binding specifically and reversibly to the substrate peptide. Multiple such domains, which vary in their binding affinities and precise sequence requirements, have been identified and characterized (Schlessinger and Lemmon, 2003; Yaffe and Elia, 2001; Yaffe and Smerdon, 2001). In some cases, it may be necessary to make slight modifications to the sequence of the substrate peptide to facilitate recognition by the PAABD.
Any effort to perturb a biochemical process should be accompanied by a corresponding technique for measuring the activity being perturbed. Genetically encoded fluorescent reporters are proving to be an invaluable asset in the study of intracellular signaling networks, and many reporters have been generated for tracking a myriad of biochemical activities in living cells (Mehta and Zhang, 2011; Newman et al., 2011). Therefore, it is possible to directly vizualize and monitor KIBS-induced perturbations at the single-cell level by pairing the KIBS with an appropriate reporter specific to the activity of interest. It is also important to visualize the action of the KIBS, in order to ensure that the system works as desired. Directly monitoring the KIBS can be achieved by fusing a different fluorescent protein color variant to each component of the switch.
BASIC PROTOCOL
Stimulating and monitoring enzymatic activity controlled by a kinase-inducible biomolecular switch
The following protocol describes the use of a Protein Kinase A (PKA)-dependent KIBS to perturb plasma membrane PI(4,5)P2 levels by controlling the localization of Inp54p, a constitutively active yeast inositol polyphosphate 5-phosphatase, in Cos7 cells (Sample et al., 2013). Induction of the KIBS leads to translocation of Inp54p to the plasma membrane, resulting in depletion of PI(4,5)P2, which is monitored by a fluorescent biosensor (Fig. 1B). Breifly, cells are first transfected with plasmid DNA encoding the KIBS components and biosensor. The microscope and cells of interest are then readied for the imaging experiment, followed by the acquisition of a time series of fluorescence intensity images. During imaging, the switch is modulated by stimulation and inhibition of PKA, and resulting changes in fluorescence are recorded. Finally, the images are analyzed to quantify the changes in enyzme effector activity. Although epifluorescence microscopy is used in this example, the protocol can be adapted for use with other fluorescence microscopy systems (e.g., confocal fluorescence). The overall experimental procedure can also be generalized to any KIBS-based enzyme perturbation system.
Materials
Cos7 cells plated on 35-mm glass-bottom dishes (see SUPPORT PROTOCOL)
Plasmid DNA: CFP-FHA1-Inp54p, PKAsubstrate-RFP-PM, YFP-PH(PLCδ1) (Sample et al., 2013)
Opti-MEM I Reduced Serum Medium (Gibco)
Lipofectamine 2000 (Invitrogen)
HBSS* imaging buffer (see recipe)
Immersion oil
Fsk stock solution: 50 mM forskolin (Calbiochem) in DMSO
IBMX stock solution: 100 mM 3-isobutyl-1-methylxanthine (Sigma) in DMSO
H89 stock solution: 10 mM H89 (Sigma) in DMSO
Inverted fluorescence microscope with appropriate objective, filters/mirrors, detector, and image acquisition software; for example:
Light source (e.g., XBO 75W xenon arc lamp, Carl Zeiss)
Zeiss Axiovert 200M microscope (Carl Zeiss)
40x/1.3-NA oil-immersion objective lens
Dichroic mirror, excitation filters (CFP, YFP, RFP), emission filters (CFP, YFP, RFP)
Cooled charge-coupled device (CCD) camera (e.g., MicroMAX BFT512, Roper Scientific)
Lambda 10-2 filter changer (Sutter Instruments)
METAFLUOR 7.7 imaging software (Molecular Devices)
PC to run microscope
Spreadsheet application (e.g., Microsoft Excel)
Transfect cells
-
1. In a microcentrifuge tube, prepare a 2:1:1 mixture of the PKAsubstrate-RFP-PM:CFPFHA1-Inp54p:YFP-PH(PLCδ1) plasmid DNA, totaling 1 μg, adding Opti-MEM to bring the total volume to 50 μL.
It is important to thoroughly mix the plasmid DNA after it is added to the microcentrifuge tube, as this will improve the efficiency with which the cells take up all three plasmids during transfection. Opti-MEM should only be opened and handled under sterile conditions, such as in a tissue culture hood.
2. In a separate tube, mix 1 to 2 μL Lipofectamine 2000 with Opti-MEM for a total volume of 50 μL. Incubate both the DNA solution and the Lipofectamine solution for 5 minutes at room temperature.
-
3. Add all (50 μL) of the Lipofectamine solution dropwise to the DNA solution and pipet gently to mix. Incubate for 20 minutes at room temperature.
Be careful to mix gently after adding the Lipofectamine solution to the DNA. Do not vortex.
-
4. In a tissue culture hood, add all (100 μL) of the Lipofectamine/DNA solution dropwise to a 35-mm glass-bottom dish containing Cos7 cells. Rock the dish gently back and forth to evenly disperse the transfection reagents.
The volumes listed are sufficient for transfecting one 35-mm dish. Prepare tubes for each additional dish to be transfected.
Make sure to select a method of transfection that is appropriate for the cell line being used. Efficient transfection increases the number of cells that can be imaged per dish, and also helps increase the signal-to-noise ratio of the fluorescent probes. As such, transfection conditions should be verified and optimized prior to imaging.
Setup microscope and prepare cells for imaging
-
5. Power on the microscope and imaging equipment. Load the imaging software.
Use the imaging software to confirm the filter configuration and acquisition protocol are correct for the experiment. Filters should be verified using control experiments to make sure they are appropriate for imaging the desired fluorescence intensities (e.g., CFP, YFP, RFP). The filter sets needed for this experiment are listed in Table 1.
-
6. Aspirate the culture medium from the 35-mm glass-bottom imaging dish. Wash the cells two times with 1 mL of HBSS* imaging buffer, then add 1 to 2 mL of HBSS* imaging buffer and securely position the imaging dish onto the microscope stage.
Use care when handling the cells; aspiration and addition of solutions should be performed gently so that cells do not detach from the dish. For example, while holding the dish at a slight angle, add or remove solutions from the side of the dish, then rock the dish back and forth to cover the entire surface. Avoid pipetting directly over the cells. Before placing the dish on the microscope stage, add a small drop of immersion oil to the objective. Once the dish is secured, carefully raise the objective until the oil is spread out in a small circle against the bottom of the imaging dish. Make certain that the imaging dish is secured on the stage and does not shift easily, in order to avoid unintentional movements during imaging.
-
7. Select cells of interest based on the appropriate morphology, as well as the desired expression level and cellular distribution of the probes.
Make certain the observed cell morphology is appropriate for the cells being used; Cos7 cells should appear flat and spread out.
Typically, cells with good expression levels are those with average to high fluorescence intensities. Too-high intensities could entail disrutpion of endogenous signaling pathways, as well as potential phototoxicity, whereas too-low intensities may result in reduced signal-to-noise.
Distribution of fluorescence intensity in the cell is determined by the subcellular targeting of the probe, and cells should be chosen that exhibit well-localized fluorescence. For this experiment, select cells that have strong plasma membrane but minimal cytosolic intensity in the RFP and YFP channels, and average or low cytosolic intensity in the CFP channel. This will facilitate visualization of probe translocation.
Table 1.
Filter Sets Used in This Protocola
| Imaging channel | Excitation filter (CWL/BP) |
Dichroic mirror |
Emission filter (CWL/BP) |
Eyepiece filter |
|---|---|---|---|---|
| CFP | 420/20 | 450 | 475/40 | 460LP |
| YFP | 495/10 | 515 | 535/25 | 525LP |
| RFP | 568/55 | 600 | 653/95 | 620LP |
Abbreviations: CWL, center wavelength; BP, band-pass; LP, long-pass.
Acquire images and collect data
-
8. Begin acquiring a time series of images for each channel (e.g., CFP, YFP, RFP).
Use the imaging software to set the time lapse interval between each set of acquisitions, as well as the excitation exposure time for each individual channel. The time lapse should be rapid enough to capture the relevant dynamics in the experiment; 10 to 30 seconds is typically sufficient. In addition, be sure to choose an optimal exposure time for the experiment. A lengthy exposure can lead to photobleaching or toxicity, whereas short exposures reduce the signal-to-noise ratio. Typical exposure times for CFP, YFP, and RFP are 500, 50, and 20 milliseconds, respectively, though these values can be adjusted based on expression levels.
-
9. Stimulate PKA activity to induce the bimolecular switch.
Acquire several sets of images in order to establish a pre-treatment baseline of activity (generally 3 to 5 minutes). Remove 300 to 500 μL of imaging buffer from the imaging dish and add to a microcentrifuge tube containing small volumes of both Fsk and IBMX. Gently add this mixture back to the imaging dish (final concentrations: 50 μM Fsk and 100 μM IBMX) by pipetting down the side of the dish, careful not to disturb the cells. Carefully pipet to evenly mix the drug throughout the dish. Continue acquiring the image series for several additional minutes to capture the stimulated response.
-
10. (Optional) Inhibit PKA activity to reverse the bimolecular switch.
Remove 300 to 500 uL of imaging buffer from the imaging dish and add to a microcentrifuge tube containing a small volume of H89. Gently add this mixture back to the imaging dish (final concentration: 20 μM H89) by pipetting down the side of the dish, careful not to disturb the cells. Carefully pipet to evenly mix the drug throughout the dish. Continue acquiring the image series for several additional minutes to capture the response.
Perform data analysis
-
11. Using the imaging software, select a region of interest (ROI) for each cell that corresponds to a major portion of the cytosol, lacking vacuoles, and log the fluorescence intensities from each channel acquired during the experiment.
In general, the choice of ROIs will depend on the type of probe(s) being imaged, as well as the specific subcellular localization. To cut down on noise, ROIs should be kept in the same cellular region throughout the time series. Individual ROIs may be moved or adjusted to account for slight cell movements. Cell-tracking software may be used, if available. Also, be sure to select a background region (e.g., an untransfected cell or cell-free region) in order to perform background subtraction to correct for contributions from cell autofluorescence or other artifacts.
12. Export the data and generate a plot of the background-subtracted fluorescence intensity versus time for both the CFP and YFP channels using a standard spreadsheet or graphing application.
-
13. (Optional) Use Equation 1 to calculate the membrane-to-cytosol intensity ratio for the CFP and YFP channels, and generate a plot of the ratio versus time:
Equation 1 Repeat Step 11, this time also selecting an ROI for each cell that corresponsds to a clearly defined region of the plasma membrane. Since translocation to or from the plasma membrane produces reciprocal changes in membrane and cytosolic fluorescence intensity, a larger translocation response can sometimes be observed by plotting the ratio.
SUPPORT PROTOCOL
Maintenance and preparation of mammalian cells for live-cell fluorescence microscopy
Successful cell culture is essential to any live-cell imaging application, whether using immoralized cell lines or cells derived from primary tissues. The protocol below outlines the procedure for splitting Cos7 cells, both for prolonged maintenance and for use in imaging experiments. This protocol can be modified for use with other cell lines by consutling and following the appropriate cell culture guidelines for the cells of choice.
Materials
Cos7 cells (American Type Culture Collection)
70% Ethanol (EtOH)
Dulbecco’s Phosphate Buffered Saline (DPBS, Gibco)
Cos7 cell culture medium (see recipe)
0.25% Trypsin/ethylenediamine tetraacetic acid (EDTA) solution (Gibco)
Tissue culture incubator
Tissue culture hood
Tissue culture flasks (e.g., T-25 cm2, BD Falcon)
35-mm glass-bottom imaging dishes (MatTEK)
-
1. Maintain Cos7 cells in T-25 cm2 flasks in a humidified, 37 °C incubator with a 5% CO2 atmosphere.
Passage cells whenever they reach 70 to 80% confluence, which can be determined by breifly examining the cells under a standard dissecting microscope. It is important to verify the doubling time of the cells being used, as splitting times can vary for different cell lines.
-
2. Switch on the white light and blower in the tissue culture hood, and wipe down the interior surface of the hood with 70% EtOH.
Make sure that all necessarly supplies, including pipets, pipet tips, flasks, media aliquots, dishes, etc., are available in the hood before beginning. This minimizes coming and going from inside the hood, reducing the potential for contamination.
3. Carefully transfer the T-25 flask from the incubator to the hood.
4. Remove the culture medium from the flask. Remove all traces of serum by gently washing two times with 2 mL of DPBS.
-
5. Pipet 300 μL of the trypsin/EDTA solution into the flask, and rock gently back and forth to evenly disperse the solution over the cells. Let sit for 2 to 5 minutes, then add 4.7 mL of fresh culture media to the flask and mix the cells by gently pipetting up and down.
Make sure that cells have fully dettached before adding fresh media. Look for rounded cells under a microscope or gently sway the flask from side to side to help cells dettach. When adding media, be careful not to over-pipet, as this could damage the cells.
6. Perform a 1:10 split into a new T-25 flask to maintain the cells.
7. For imaging experiments, perform a 1:10 split of cells into the 35-mm glass-bottom dishes. Cells should reach 60 to 70% confluence after approximately 24 hours, and should be transfected at this confluence.
REAGENTS AND SOLUTIONS
All solutions should be made using water with 18.2 MΩ-cm resistivity, and pepared under sterile conditions in a tissue culture hood.
HBSS* imaging buffer
Hank’s Balanced Salt Solution (Gibco)
20 mM HEPES (Invitrogen)
2.0 g/L D-glucose (Sigma)
Adjust pH to 7.4, then filter sterilize using a 0.22 μm filter. Store a 50 mL aliquot at room temperature near the microscope room and store the remainder at 4 °C.
Cos7 cell culture medium
Dulbecco’s Modified Eagle’s Medium, 4.5 g/L glucose (Gibco)
10% fetal bovine serum (FBS, Sigma)
1% penicillin-streptomycin (Sigma-Aldrich)
Store culture media at 4 °C and warm to 37 °C before using with cells.
COMMENTARY
Background Information
Tools designed to perturb various signaling processes are essential for unraveling the molecular logic of intracellular signaling. Scientists have historically relied on general activators and inhibitors, such as hormones and small-molecular agonists that are still used to this day, to perturb different signaling activities. However, these agents typically act very broadly, affecting multiple steps in a given pathway or network. A more desirable approach is one that enables selective, targeted activation or inhibition of specific enzymes at discrete points within a signaling pathway, thereby giving researchers the ability to tease apart biochemical interactions between select pathway components. In recent years, several techniques have been developed in pursuit of these kinds of targeted perturbations. For the most part, these approaches are all loosely based on the concept of the molecular switch.
A molecular switch can be broadly defined as a molecular system, capable of existing in either of two states (e.g., conformations), which can be converted from one state to the other by a specific input stimulus. For example, Wu et al. (2009) recently generated a light-activated form of the small GTPase Rac by fusing constitutively active Rac with a molecular switch composed of the light-oxygen-voltage (LOV) domain and Jα-helical extension from the Avena sativa phototropin 1 protein. Illumination with 453 nm light triggers a conformational switch within the LOV/Jα complex that allows Rac to act upon its substrates. Molecular switches can also be used to control dimer formation between protein domains, offering another strategy for discretely controling select biochemical processes (Crabtree and Schreiber, 1996). The immunosuppressive compound rapamycin mediates the formation of a dimeric inhibitory complex between FKBP12 and mTORC1, and a commonly used perturbation technique involves incorporating the rapamycin binding domains from these proteins into chimeras that specifically associate upon rapamycin treatment. This type of chemically induced dimerization often involves the rapamicin-induced translocation of an enzyme effector to a specific region of the cell, and has been used to selectively control enzyme activity at both the plasma membrane and various intracellular organelles (Inoue et al., 2005; Inoue and Meyer, 2008; Komatsu et al., 2010; Lin et al., 2012; Phua et al., 2012; Ueno et al., 2011).
Kinase-inducible bimolecular switches also rely on dimer formation to facilitate control of enzyme activity. Like chemically induced dimerization, the kinase-inducible molecular switch is highly modular and can be adapted for a variety of uses. In addition, kinase-inducible bimolecular switches offer a much greater degree of reversibility, which allows for more dynamic control of enzyme activities. Kinase-dependent molecular switches were initially engineered as part of the development of genetically encoded fluorescent kinase activity reporters (Mehta and Zhang, 2011; Newman et al., 2011). The switch is composed of a consensus phosphorylation sequence for the kinase of interest and a phosphoamino acid-binding domain (PAABD), such that phosphorylation of the substrate sequence drives association with the PAABD. In a typical kinase activity reporter, both components are expressed as a single polypeptide, wherein association of the PAABD with the phosphorylated substrate produces a conformational change. However, Herbst et al. (2011) were able to show that the switch also functions in a bimolecular format, wherein the switch controls dimer formation, which was then successfully adapted for use in the targeted perturbation of enzyme activity (Sample et al., 2013).
Critical Parameters
Selection of the kinase
Prior to designing a KIBS, careful consideration must be given to selecting the kinase that will drive the switch. It is essential to select a kinase that does not function in the signaling pathway being studied, and cannot independently induce the enzymatic activity that will eventually be perturbed. Since kinases often participate in multiple signaling pathways, minimizing the potential for unwanted cross-talk is critical for ensuring that any experiments performed using the KIBS produce meaningful results. Once a KIBS has been constructed to control a particular biochemical activity in a given cell type, further consideration is also warranted before transfering the KIBS into a different cell type or modifying it to target a different pathway. Ultimately, it may prove necessary to engineer a specific KIBS, based on a different kinase, for each new application. The proliferation of genetically encoded, fluorescence resonance energy transfer (FRET)-based kinase activity reporters means that substrate/PAABD pairs corresponding to a variety of kinases have already been tested and optimized, potentially simplifying the development of additional KIBS-based perturbation systems for this purpose. However, efforts to generate a KIBS that is essentially orthogonal to the target signaling network(s), by introducing a kinase found only in specialized cells, or in a different organism, into the cells of interest along with the corresponding substrate, may provide researchers with a more universal solution in the future.
Control experiments
It is of critical importance that control experiments be carried out to validate any KIBS-based perturbation system. One generally useful negative control involves performing the KIBS experiments using a modified KIBS featuring a catalytically inactive mutant form of the enzyme effector. This experiment is designed to verify that any observed change in the activity of interest upon KIBS induction is specifically dependent on the activity of the effector enzyme. Correspondingly, this can also serve as a control for whether stimulation of the kinase alone has any effect on the activity of interest. The latter can also be verified using cells that do not express the KIBS.
Another important control experiment is to use a phosphorylation site mutant of the substrate peptide. Since the underyling mechanism of the KIBS is based on association of a PAABD with a phosphorylated peptide, mutating the phosphorylation site to a nonphosphorylatable residue should disable the KIBS and abrogate the enzyme effector response. This control is useful to provide further confirmation that the candidate kinase and the biochemical activity of interest are coupled together exclusively by the action of the KIBS, and not through some endogenous mechanism.
Keep in mind that these examples are simply a starting point for designing control experiments, applicable to all KIBS-based perturbation studies. Care should be taken to design and perform addional experiments tailored to the requirements of the specific KIBS, enzyme effector, pathway, and cell type being used.
Troubleshooting
Table 2 provides a summary of some potential areas where problems may arise during a KIBS experiment, along with possible causes and strategies for performing troubleshooting.
Table 2.
Steps For Troubleshooting KIBS-mediated Control of Enzyme Activity
| Problem | Possible causes | Possible solutions |
|---|---|---|
| Dim cells | Poor transfection | Optimize transfection |
| Incorrect filters | Verify microscope filters | |
| No stable baseline | Cell morphology changes | Optimize imaging conditions (e.g., temperature, buffer) |
| Photobleaching | Reduce exposure time or use neutral density filters |
|
| KIBS response smaller or slower than expected |
Incorrect drug addition | Mix well or use perfusion |
| Drug concentration too low | Verify ammount of drug needed to fully activate kinase |
|
| Too high PAABD expression | Optimize substrate/PAABD ratio during transfection |
|
| Weak substrate/PAABD interaction |
Optimize substrate amino acid sequence for specific PAABD |
|
| High basal kinase activity | Pretreat cells with inhibitor; select a different kinase |
|
| Effector response smaller or slower than expected |
Reporter expression level | Optimize transfection |
| Low effector enzyme activity | Verify enzyme selection using controls |
Anticipated Results
Treatment with Fsk/IBMX should produce a decrease in cytoplasmic CFP fluorescence intensity, along with increased cytoplasmic YFP fluorescence. Subsequent treatment with H89 should reverse these effects (Sample et al., 2013).
Time Considerations
All of the steps outlined above can be completed over a period of 3 days. This includes splitting cells into 35-mm imaging dishes (~30 minutes, day 1), transfecting the cells (~30 minutes, day 2), and acquiring and analyzing the data (~1 hour, day 3). Generally, the precise time required to carry out a KIBS experiment will vary based on multiple factors such as the doubling time of the cells, the time needed for probe expression, and the kinetics of the pathway being investigated.
ACKNOWLEDGEMENT
This work is supported by National Institutes of Health R01 DK073368 and DP1 CA174423.
LITERATURE CITED
- Crabtree GR, Schreiber SL. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem Sci. 1996;21:418–422. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- Herbst KJ, Allen MD, Zhang J. Luminescent kinase activity biosensors based on a versatile bimolecular switch. Journal of the American Chemical Society. 2011;133:5676–5679. doi: 10.1021/ja1117396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Heo WD, Grimley JS, Wandless TJ, Meyer T. An inducible translocation strategy to rapidly activate and inhibit small GTPase signaling pathways. Nat Methods. 2005;2:415–418. doi: 10.1038/nmeth763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Meyer T. Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration. PLoS One. 2008;3:e3068. doi: 10.1371/journal.pone.0003068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T, Kukelyansky I, McCaffery JM, Ueno T, Varela LC, Inoue T. Organelle-specific, rapid induction of molecular activities and membrane tethering. Nat Methods. 2010;7:206–208. doi: 10.1038/nmeth.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin B, Holmes WR, Wang CJ, Ueno T, Harwell A, Edelstein-Keshet L, Inoue T, Levchenko A. Synthetic spatially graded Rac activation drives cell polarization and movement. Proceedings of the National Academy of Sciences. 2012;109:E3668–3677. doi: 10.1073/pnas.1210295109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S, Zhang J. Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Annual Review of Biochemistry. 2011;80:375–401. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RH, Fosbrink MD, Zhang J. Genetically encodable fluorescent biosensors for tracking signaling dynamics in living cells. Chem Rev. 2011;111:3614–3666. doi: 10.1021/cr100002u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phua SC, Pohlmeyer C, Inoue T. Rapidly relocating molecules between organelles to manipulate small GTPase activity. ACS Chemical Biology. 2012;7:1950–1955. doi: 10.1021/cb300280k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sample V, Ni Q, Mehta S, Inoue T, Zhang J. Controlling enzymatic action in living cells with a kinase-inducible bimolecular switch. ACS Chemical Biology. 2013;8:116–121. doi: 10.1021/cb300393w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlessinger J, Lemmon MA. SH2 and PTB domains in tyrosine kinase signaling. Science’s STKE : Signal Transduction Knowledge Environment. 2003;2003:RE12. doi: 10.1126/stke.2003.191.re12. [DOI] [PubMed] [Google Scholar]
- Ueno T, Falkenburger BH, Pohlmeyer C, Inoue T. Triggering actin comets versus membrane ruffles: distinctive effects of phosphoinositides on actin reorganization. Science Signaling. 2011;4:ra87. doi: 10.1126/scisignal.2002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YI, Frey D, Lungu OI, Jaehrig A, Schlichting I, Kuhlman B, Hahn KM. A genetically encoded photoactivatable Rac controls the motility of living cells. Nature. 2009;461:104–108. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MB, Elia AE. Phosphoserine/threonine-binding domains. Current Opinion in Cell Biology. 2001;13:131–138. doi: 10.1016/s0955-0674(00)00189-7. [DOI] [PubMed] [Google Scholar]
- Yaffe MB, Smerdon SJ. PhosphoSerine/threonine binding domains: you can't pSERious? Structure. 2001;9:R33–38. doi: 10.1016/s0969-2126(01)00580-9. [DOI] [PubMed] [Google Scholar]