Abstract
Drug resistance acquired by cancer cells is a significant challenge in the clinic and requires impairing the responsible pathological pathway. Administering chemotherapeutics along with silencing resistance-basis activity using RNA interference (RNAi) is expected to restore the activity of the chemotherapeutic. generate synergistic cancer eradication. This study attempted to reverse tamoxifen (TAM)-resistance in breast cancer by silencing a mitochondrial enzyme, manganese superoxide dismutase (MnSOD), which dismutates TAM-induced reactive oxygen species (ROS) (i.e., superoxide) to less harmful hydrogen peroxide and hampers therapeutic effects. Breast cancer cells were co-treated with TAM and MnSOD siRNA-delivering nanoparticles (NPs) made of a siRNA/poly(amidoamine) (PAMAM) dendriplex core and an acid-degradable polyketal (PK) shell. The (siRNA/PAMAM)-PK NPs were designed for the PK shell to shield siRNA from nucleases, minimize detrimental aggregation in serum, and facilitate cytosolic release of siRNA from endosomal compartments. This method of forming the PK shell around the siRNA/PAMAM core via surface-initiated photo-polymerization enables ease of tuning NPs’ size for readily controlled siRNA release kinetics. The resulting NPs were notably homogenous in size, resistant to aggregation in serum, and invulnerable to heparan sulfate-mediated disassembly, compared to siRNA/PAMAM dendriplexes. Gel electrophoresis and confocal microscopy confirmed efficient siRNA release from the (siRNA/PAMAM)-PK NPs upon stimuli-responsive hydrolysis of the PK shell. Sensitization of TAM-resistant MCF7-BK-TR breast cancer cells with (MnSOD siRNA/PAMAM)-PK NPs restored TAM-induced cellular apoptosis in vitro and significantly suppressed tumor growth in vivo, as confirmed by biochemical assays and histological observations. This study implies that combined gene silencing and chemotherapy is a promising strategy to overcoming a significant challenge in cancer therapy.
Keywords: RNA interference, drug-resistant tumor model, apoptosis, gene therapy, stimuli-responsive nanoparticles
1. Introduction
One of the immediate utilities of RNAi in biomedical applications is to sensitize target cells to therapy by silencing an antagonistic pathological pathway or resistance against chemotherapeutic agents [1–3]. For example, approximately 50% of breast cancer patients undergoing chemotherapy develop resistance against tamoxifen (TAM), leading to relapse and increased mortality [4]. When effective, TAM engages mitochondrial estrogen receptor (ER)-β as an antagonist, inducing the intrinsic apoptotic program that damages mitochondrial DNA of breast cancer cells by stimulating reactive oxygen species (ROS) (i.e., superoxide) formation [5]. This cytotoxic effect is offset when the expression of manganese superoxide dismutase (MnSOD) is up-regulated in TAM-resistant breast cancer cells and superoxide is catalyzed to less harmful hydrogen peroxide [6, 7]. Therefore, it is important to interfere with MnSOD activity in order to achieve effective therapy for TAM-resistant breast cancer. Re-sensitizing cancer cells to chemotherapy by silencing the pathological pathway may result in a lower necessary dose of chemotherapeutic agents administered, reducing adverse side effects.
Delivery of small interfering RNA (siRNA) has been a pivotal technological challenge in applying RNAi in biomedical applications [8, 9]. siRNA delivery carriers must protect the cargo from degradation and clearance, efficiently enter cells, and release siRNA into the cytoplasm [10, 11]. Cationic polymers, such as polyethylenimine (PEI) and poly-L-lysine (PLL), are capable of complexing nucleic acids for shielding and charge compensation, and facilitating escape from the endosome via hypothetic proton sponge effect [12–14]. Cationic dendrimers, such as poly(amidoamine) (PAMAM), are promising nucleic acid delivery carriers for their highly controlled molecular weight (size), homogeneity, and low cytotoxicity compared to other cationic polymers [15]. PAMAM is known to efficiently complex anionic siRNA (dendriplex) via electrostatic interaction with cationic primary amines on its surface, while tertiary amines inside buffer protons at an acidic pH [16–18]. PAMAM is known to deliver large nucleic acids such as plasmid DNA [19]. However, very few reports have been made on the delivery of small nucleotides (e.g., siRNA) by PAMAM, and studies indicate that higher generation PAMAM is required to ensure efficient complexation and hence protection of siRNA from serum proteins and nucleases [20, 21]. In this study, siRNA/PAMAM dendriplexes were shelled with acid-degradable polyketal (PK) layer, resulting in (siRNA/PAMAM)-PK nanoparticles (NPs). The PK shell 1) reinforces stable complexation of siRNA with PAMAM, 2) shields the siRNA/PAMAM core from nucleases and serum proteins, and 3) facilitates siRNA release into the cytoplasm upon rapid hydrolysis in the mildly acidic endosome. The surface-initiated photo-polymerization employed in synthesizing (siRNA/PAMAM)-PK NPs also enables precise control over size, hence, stimuli-triggered release kinetics. Shelling polyplex and virus cores with a PK shell via surface-initiated photo-polymerization has demonstrated enhanced gene delivery [22, 23].
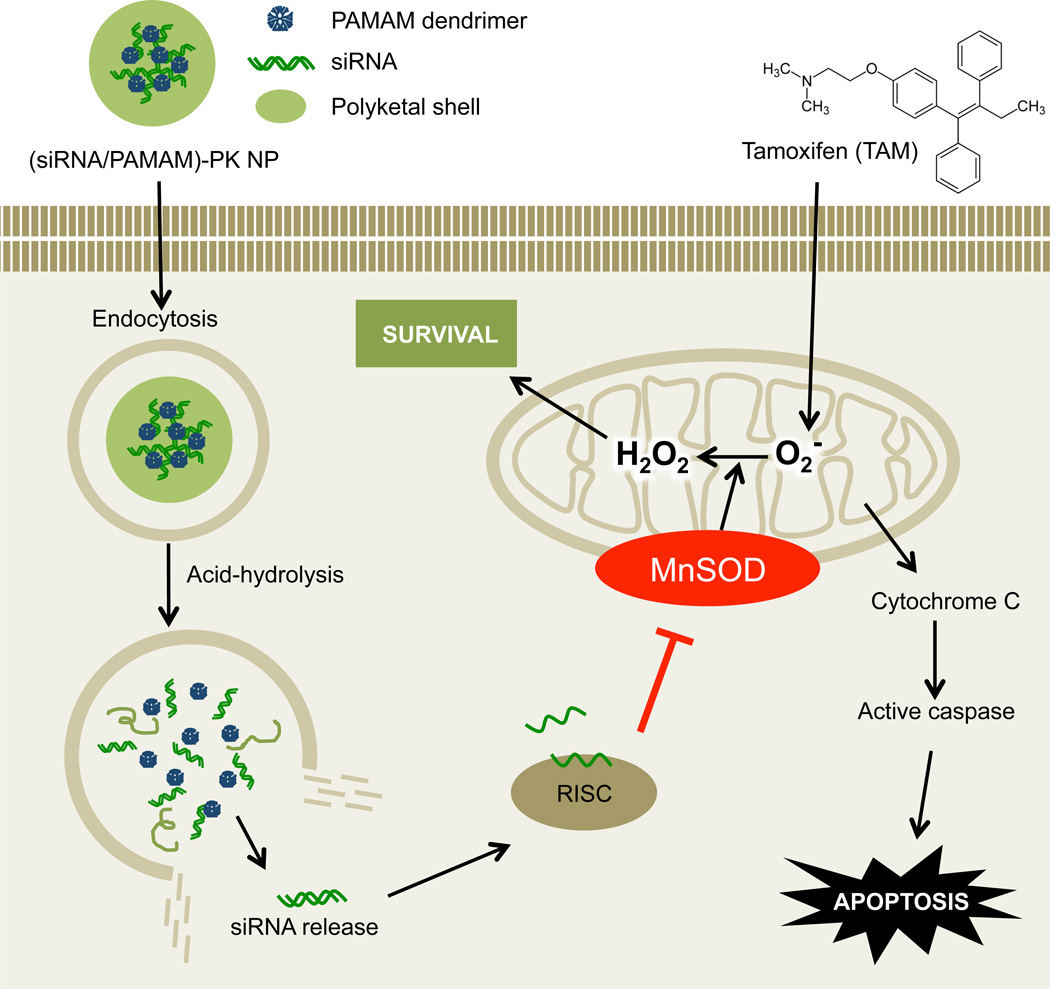
This study aimed to demonstrate the feasibility of restoring TAM-sensitivity by administering MnSOD siRNA into in vivo tumors, which initially show resistance to the selective estrogen receptor modulator (SERMs) (e.g., TAM) (Figure 1). The (siRNA/PAMAM)-PK NPs developed in this study were also fully characterized for their high stability in the presence of serum proteins, stimuli-triggered degradation, and efficient intracellular release of siRNA.
Fig. 1.
Reversed TAM-resistance in breast cancer by delivery of MnSOD siRNA encapsulated in (siRNA/PAMAM)-PK NPs. MnSOD-mediated conversion of TAM-generated superoxide to hydrogen peroxide is blocked when MnSOD siRNA is released into the cytoplasm from the stimuli-responsive core-shell NPs, resulting in enhanced apoptosis of TAM-resistant breast cancer cells.
2. Materials and methods
2.1. Materials
PAMAM dendrimer (G5; 28 kDa) was purchased from Sigma-Aldrich (St. Louis, MO). MnSOD siRNA was purchased from Santa Cruz Biotechnology (Dallas, TX). siRNA with a scrambled sequence (scr siRNA) and carboxyfluorescein (FAM)-labeled scr siRNA were purchased from Ambion (Foster City, CA). PEG-NHS (Mw 5,000 Da) was purchased from Jenchem (Walnut Creek, CA). Eosin-5-isothiocyanate, nucBlue Live Cell Stain, CellLight® early endosomes-RFP and LysoTracker Red DND-99 were purchased from Invitrogen (Carlsbad, CA). Ascorbic acid was purchased from Acros Organics (Geel, Belgium). Heparan sulfate (HS) was purchased from Sigma-Aldrich (St. Louis, MO). Acid-degradable cationic ketal monomer and cross-linker were synthesized as previously published [15]. 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 3,3-diaminobenzidine, and protease inhibitors cocktail were purchased from Sigma-Aldrich.
Tamoxifen-resistant breast cancer cell line, MCF7-BK-TR (kindly provided from Dr. Suzanne Fuqua, Baylor University), were cultured in Dulbecco’s modified Eagle’s medium/nutrient mixture F12 (DMEM/F12) medium (Sigma, St. Louis, MO) supplemented with sodium bicarbonate (1.2 mg/mL) (Hyclone, Logan, UT), 10 % fetal bovine serum (FBS) (Hyclone), 1 % antibiotics and antimycotic (Gibco, Grand Island, NY), and 100 nM tamoxifen (Sigma).
Apoptosis of the breast cancer cells was measured by DeadEnd™ Fluorometric TUNEL system (Promega, Madison, WI) and Annexin V apoptosis kit (Calbiochem, La Jolla, CA). MnSOD expression and caspase-7 in the cells were compared by gel electrophoresis and immunoblotting using caspase-7 mouse monoclonal antibody (Cell Signalling Technology, Danvers, MA) and MnSOD antibody (Santa Cruz Biotechnology). E2 and TAM pellets were purchased from Innovative Research of America (Sarasota, FL). The breast cancer cells were grafted in nu/nu mice in matrigel purchased from Becton Dickinson (Franklin Lakes, NJ).
2.2. Synthesis and characterization of (siRNA/PAMAM)-PK NPs
Photo-initiator, eosin, was conjugated to the amines on the surface of PAMAM dendrimer. Briefly, 10 mg of PAMAM dendrimer dissolved in methanol was evaporated under vacuum and re-dispersed in 610 µL of 10 mM sodium bicarbonate buffer (pH 8.0). Eosin-5-isothiocyanate (0.2 mg in 20 µL DMSO) was added to 500 µL of the PAMAM dendrimer-containing solution (8.2 mg of PAMAM dendrimer), followed by stirring for 3 h at room temperature without exposure to light. After 3 h, eosin-conjugated PAMAM dendrimer was purified from unreacted eosin-5-isothiocyanate using a PD Mini size-exclusion column (GE Healthcare, Pittsburgh, PA) with 10 mM HEPES buffer (pH 7.4). Then a mixture of 6.1 µL of eosin-conjugated PAMAM dendrimer (50 µg) and 1 mL of 10 mM HEPES buffer (pH 7.4) in a glass vial was briefly stirred on ice, followed by adding 10 mg of acid-degradable cationic ketal monomers in 50 µL 10 mM HEPES buffer and 10 µg of MnSOD siRNA in 50 µL of the same buffer. After 5 min of stirring, 10 mg of ascorbic acid in 50 µL of 10 mM HEPES buffer was added and photo-polymerization was initiated by irradiating the resulting mixture with 700 klux halogen light. At 10 min of photo-polymerization, 10 mg of acid-degradable cationic ketal monomer and 4 mg of acid-degradable ketal cross-linker (both in 50 µL of 10 mM HEPES buffer) were added and the polymerization was continued for additional 5 min. (siRNA/PAMAM)-PK core-shell NPs were obtained after centrifuging the polymerized solution twice using Amicon ultracentrifugal filter (MWCO 10 kDa; Millipore, Billerica, MA), at 3500 rpm (2100 g) at 4 °C for 30 min to remove unreacted monomer, cross-linker, and ascorbic acid. The NPs were finally re-dispersed in 1 mL of de-ionized (DI) water. Smaller NPs were prepared by the same method using lowered monomer and cross-linker amounts to 10 mg and 2 mg, respectively. Control nanoparticles with scrambled sequence siRNA (scr siRNA) were also prepared in the same way. For the in vivo study, both (scr siRNA/PAMAM)-PK and (MnSOD siRNA/PAMAM-PK NPs were PEGylated. Briefly, the NPs in 1 mL of DI water were reacted separately with 0.5 mg of PEG-NHS (Mw 5,000 Da) in 0.5 mL of DI water for 2 h at 4 °C. Unreacted PEG-NHS was removed by centrifugal filtration (MWCO 10 kDa) at 3000 rpm for 30 min at 4 °C. The purified PEGylated NPs and dendriplexes were re-dispersed in 1 mL DI water for size and surface charge measurements. siRNA/PAMAM dendriplexes were also PEGylated using same method.
The size and surface charge of the NPs and dendriplexes in 1 mL of DI water were measured by dynamic light scattering (DLS) and zeta-potential analysis using a Malvern Zetasizer Nano ZS (Malvern Instruments, Westborough, MA) at 25 °C. To investigate serum protein-induced aggregation, 10 % FBS was added to NP- and dendriplex-containing solutions in 1 mL DI water and incubated for 1 h at room temperature. The size changes of NPs and dendriplexes before and after adding FBS were measured by DLS.
2.3. Transmission electron microscopy (TEM) and gel electrophoresis
(siRNA/PAMAM)-PK NPs were incubated in 100 mM acetate buffer (pH 5.0) at 37 °C for 6 h to hydrolyze the acid-degradable PK shell. In order to dissociate the siRNA-PAMAM dendriplex core, heparan sulfate (HS, Sigma-Aldrich, St. Louis, MO) was further added at a final concentration of 1 % (w/v). The aliquots (10 µL) of both siRNA/PAMAM dendriplexes and (siRNA/PAMAM)-PK NPs with and without acid-hydrolysis were dropped on carbon-coated grids (Ted Pella, Redding, CA), followed by uranyl acetate staining, and dried in vacuo overnight at room temperature. The morphology and size of the nanoparticles were then examined under a Philips CM20 transmission electron microscope (Philips Electronic Instruments, Mahwah, NJ) at 80 kV. The aliquots (containing 1 µg siRNA) of (siRNA/PAMAM)-PK NPs treated under different conditions (i.e., with and without acid-hydrolysis; with and without incubation with HS) were loaded in 1 % agarose gel containing 1 µg/mL of ethidium bromide in Tris-acetate-EDTA (TAE) buffer. The gel was electrophoresed at 110 V for 30 min and the siRNA bands were visualized on a UV transilluminator.
2.4. Confocal microscopy
(siRNA/PAMAM)-PK NPs encapsulating FAM-labeled scr siRNA were prepared as described earlier. TAM-resistant MCF7-MK-TR cells were plated at a density of 5 × 104 cells/well in a 8-well culture slide (BD Biosciences, Franklin Lakes, NJ), 24 h prior to incubation with (siRNA/PAMAM)-PK NPs. (siRNA/PAMAM)-PK NP-containing fresh growth media (0.5 mL) replaced the existing media and the cells were incubated for 2 h or 10 h at 37 °C. Then 1 drop of nucBlue Live Cell Stain was added to the media and cells were incubated at room temperature for additional 20 min, followed by rinsing twice with Hank’s balanced salt solution (HBSS). Lysosomes were stained by adding 200 µL of 100 nM LysoTracker Red DND-99 in HBSS to each well and incubating the cells for 1 min at room temperature, followed by rinsing twice with HBSS. To visualize early endosomes, another group of the cells were incubated with 10 µL of CellLight® early endosomes-RFP per well (30 particles per cell as recommended by the supplier) for 24 h prior to incubation with NPs. After 2 or 10 h, the nuclei of the cells were stained and rinsed as previously described. The cells were observed under an Olympus IX2 inverted microscope (Olympus America, Inc., Melville, NY) coupled with a Fluoview 1000 confocal scanning microscopy setup (FV10-ASW) (Olympus America, Inc.) and a 60×/1.2 NA water immersion plan apochromat objective. The confocal microscopy set up was equipped to visualize FAM-labeled scr siRNA (a 488 nm multiple argon laser, a 488 nm dichroic mirror, and a 505–540 nm band pass barrier filter), lysosomes stained with LysoTracker Red DND-99 and early endosomes stained with CellLight® early endosomes-RFP (a 559 nm helium-neon laser, a SMD640 dichroic mirror, and a 575–675 nm band-pass barrier filter), and nucBlue-stained nuclei (a 405 nm diode laser, a 405 nm dichroic mirror, and a 430–470 nm band pass barrier filter). Fluorescence images were captured and processed using FV10-ASW 1.6 viewer (Olympus America, Inc.). The cells were scanned in three dimensions as a z-stack of two-dimensional images (1024 × 1024 pixels). Images cutting horizontally through approximately the middle of the cellular height were selected to exclude the fluorescence signals on the cellular surface from the intracellularly localized ones. Each emission light was separately scanned for individual excitations to eliminate cross-talk of the dyes. FAM-labeled scr siRNA localized in early endosomes or lysosomes were presented in yellow and its relative intracellular distribution was quantitatively compared using Image J.
2.5. In vitro transfection for cellular apoptosis
MCF7-BK-TR cells plated at a density of 5 × 106 cells/well in a 6-well plate were transfected with (siRNA/PAMAM)-PK NPs at a concentration of 2.5 µg of siRNA (Scr or MnSOD siRNA). After 48 h, the cells were washed and the cells undergoing apoptosis were identified using the DeadEnd™ Fluorometric TUNEL system, according to the manufacturer's instruction. Briefly, cells in culture were washed with PBS after treatments, fixed with 4% formaldehyde in phosphate-buffered saline, permeabilized with Triton® X-100, labeled with flurorescein-12-dUTP, and stained with propidium iodide for nuclei, and green fluorescence of apoptotic cells was analyzed by fluorescence microscopy. MnSOD knockdown in the cells incubated with (siRNA/PAMAM)-PK NPs was determined by immunoprecipitation/western blot. The cells were grown to subconfluency in a 100 mm petri dish, incubated with (siRNA/PAMAM)-PK NPs for 48 h, prior to protein expression. After the culture medium was removed, the cell monolayer was rinsed with PBS at room temperature. The adherent cells were removed using a cell scraper and further microcentrifuged at 10,000 rpm for10 min at 4 °C). To the resulting pellet, 0.5 mL of lysis buffer with protease inhibitor was added and the mixture was passed through a 21-gauge needle to shear the DNA. The cell lysate was centrifuged at 10,000 rpm for 10 min at 4 °C. Then the supernatant was transferred to a new microcentrifuge tube. In order to pre-clear whole cell lysate, 0.25 µg of proteinA/G-agarose beads was added to cell lysate and incubated for 30 min at 4 °C then the lysate was centrifuged at 3,000 rpm for one min. The supernatants were mixed in 40 µL of 2× electrophoresis sample buffer. The samples were boiled for 5 min and electrophoresed, transblotted to the nitrocellulose membrane. Then non-specific binding of membrane was blocked by incubation with bovine serum albumin (BSA) (Sigma-Aldrich) in PBS without Tween 20 (Sigma-Aldrich) at room temperature in a covered container for 2 h. The membrane was washed with Tris buffered saline three times and then incubated with primary antibody overnight at 4°C The membrane was washed three times for 5 min with Tris buffered saline with Tween 20 (TBST). The membrane was incubated for 45 min at room temperature with horseradish peroxidase (HRP)-conjugated secondary antibody and washed three times for 5 min each with TBST and once for 5 min with Tris buffered saline. The protein was visualized using ECL reagents. Cell viability was quantified by the colorimetric MTT assay, which measures mitochondrial dehydrogenase activity in viable cells. This method is based on the conversion of the MTT to MTT-formazan crystal by mitochondrial enzyme. In brief, MCF7 cells were incubated with (siRNA/PAMAM)-PK NPs for 24 h. At the end of incubation the cells were washed with phenol red-free medium and then incubated with medium containing 1 mg/mL MTT for 4 h at 37 °C. MTT/formazan was extracted by overnight incubation at 37 °C with 100 µL extraction buffer, consisting of 20% sodium dodecyl sulfate (SDS) and 50% formamide adjusted to pH 4.7 with 0.02% acetic acid and 0.025 N HCl. Optical densities at 570 nm were measured using extraction buffer as a blank.
2.6. TAM-resistant tumor xenograft model co-transplanted with (siRNA/PAMAM)-PK NPs
Athymic (nu/nu) female mice (4–6 week old, Jackson laboratory, Bar Harbor, ME) were housed under pathogen-free conditions with a 12 h light/12 h dark schedule. Each mouse received one 21-day release E2 pellet for 3 days before replacing with TAM pellets for 3 days (Innovative Research of America, Sarasota, FL). MCF7-BK-TR cells growing in log phase were harvested by trypsinization and washed twice with PBS. The cells were resuspended.in 50/50 PBS/matrigel (Becton Dickinson, Franklin Lakes, NJ) along with (MnSOD siRNA/PAMAM)-PK NPs (right) and (scr siRNA/PAMAM)-PK NPs (left) to a final concentration of 5×106 cells in 200 µL. To establish tumor xenografts, the cells in PBS/matrigel/(siRNA/PAMAM)-PK NPs were injected subcutaneously (s.c.) into both flanks of (nu/nu) female mice (6 mice/group). All procedures were carried out following the protocol approved by The Institutional Animal Care and Use Committee. Tumors were allowed to grow for 7 weeks after cell injection. The mice were sacrificed, and the tumors were harvested. The skin and connective tissues were dissected from the tumors, and the tumor weight was measured and calculated. The tissues were divided into two parts. One part was fixed in 10% formalin, paraffin-embedded, cut in 5-µm sections, stained with hematoxylin and eosin, and evaluated. For immunostaining, the formalin-fixed, paraffin-embedded tissue sections were de-paraffinized, dehydrated, and incubated in 0.3% H2O2 in absolute methanol for 30 min to block endogenous peroxidase. Nonspecific staining was blocked using normal serum at a 1:50 dilution for 30 min, followed by incubation overnight at 4°C with specific primary antibodies against Annexin V (goat polyclonal IgG) (Santa Cruz Biotechnology, Dallas, TX). Sections were then rinsed in PBS and immunostained using an Annexin V apoptosis kit (Calbiochem, La Jolla, CA). A positive reaction was visualized as brown stain after incubating the slides with 3,3-diaminobenzidine for 5 min. The other section stored in liquid nitrogen was rinsed with distilled water, counterstained with Gill’s hematoxylin for 1 min, and mounted with Universal Mount (Research Genetics, Huntsville, AL). Apoptotic cells (brown stained) were counted under Nikon Epifluorescnce microscope (Nikon, Melvillen NY) using the Optimas 6 software program (Optimas Corp., Bothell, WA). The apoptotic index was calculated by dividing the number of apoptotic cells by the total number of cells counted per sample.
Caspase-7 activity and MnSOD knockdown were determined from frozen tumors, pulverized by mortar and pestle, and the proteins were extracted with buffer containing protease inhibitors cocktail (Sigma). Proteins were further homogenized by polytron(homogenizer) (Brinkmann, Westbury, NY) and the solutions were centrifuged at 14,000 g for 10 min at 4°C. Proteins were separated by 10 % SDS-polyacrylamide gel electrophoresis (PAGE), followed by immunoblot using caspase-7 mouse monoclonal antibody (Cell Signaling Technology, Danvers, MA) and MnSOD antibody (Santa Cruz Biotechnology) to detect endogenous uncleaved and cleaved 30 and 20 kDa fragments of caspase-7 and MnSOD, respectively.
3. Results
3.1. Surface-initiated photo-polymerization of an acid-degradable PK layer onto siRNA/PAMAM dendriplexes for enhanced siRNA delivery
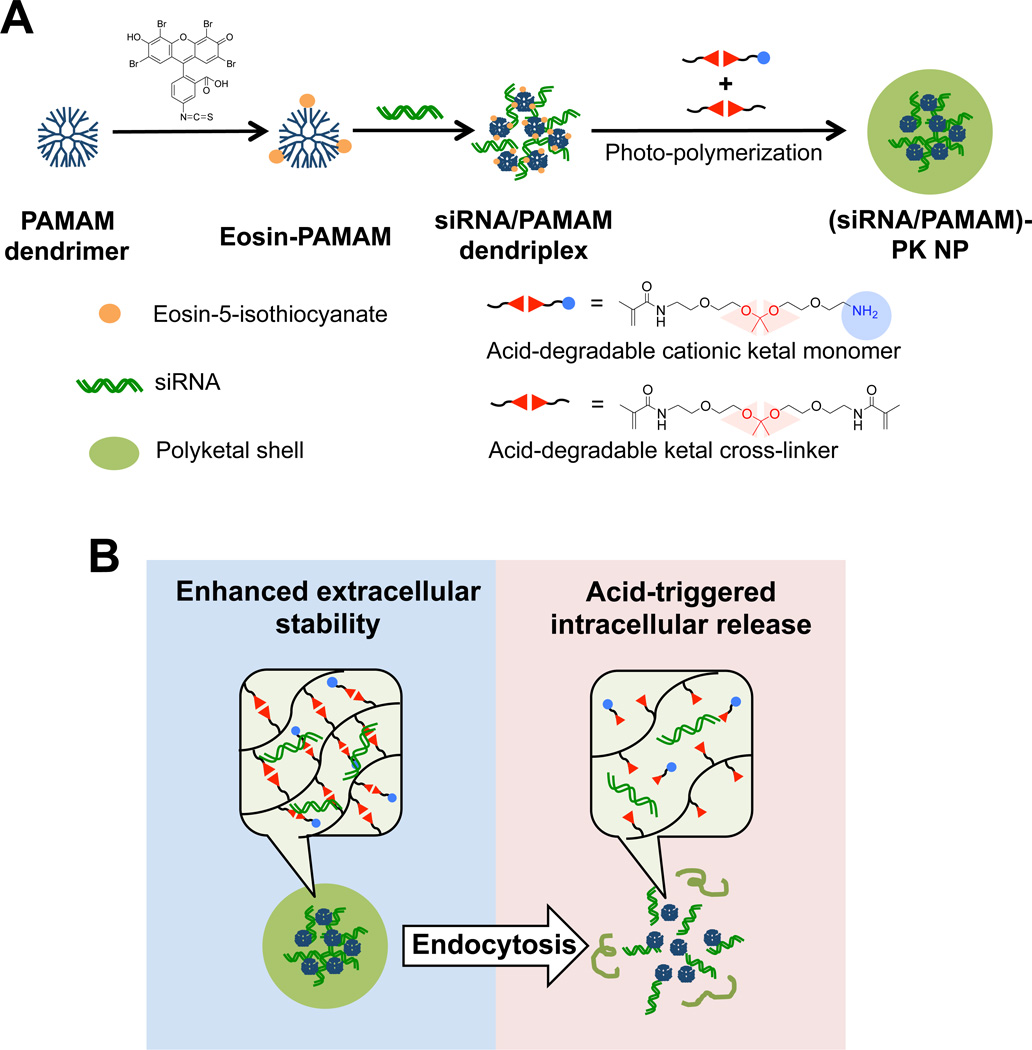
The limited extracellular stability and intracellular siRNA release capability of siRNA/PAMAM dendriplexes were addressed by grafting an acid-degradable PK shell around the dendriplex core via surface-initiated photo-polymerization (Figure 2a). In order to ensure the formation of PK shell around siRNA/PAMAM dedriplexes without generating empty PK particles (i.e., seedless particles), photo-initiator eosin was conjugated only onto PAMAM dendrimers. Using the facile tunability of PK shell’s thickness by surface-initiated polymerization, (siRNA-PAMAM)-PK NPs in two different sizes, hence varied acid-degradability, were prepared (Table 1). They were synthesized using the same amount of siRNA but varying monomer and cross-linker concentrations during the polymerization. The 132 nm (siRNA-PAMAM)-PK NPs are supposed to generate greater acid-responsive siRNA release upon hydrolysis than 72 nm ones. The surface charges of both NPs were comparable.
Fig. 2.
(A) Synthesis of (siRNA/PAMAM)-PK NPs with siRNA/PAMAM dendriplex core and acid-degradable PK shell. (B) Chemically tuned transformation of (siRNA/PAMAM)-PK NPs before and after cellular uptake. Cross-linked PK shell protects and stabilizes siRNA/PAMAM dedriplex core from serum proteins and nucleases, while acid-hydrolysis in the endosome releases siRNA/PAMAM dendriplex into the cytoplasm, playing stimuli-mediated dual roles.
Table 1.
(siRNA/PAMAM)-PK NPs with varying acid-degradability.
| Size (nm) | PDI | Zeta potential (mV) |
|
|---|---|---|---|
| (siRNA/PAMAM)-PK NP 1 | 72.1 ± 7.1 | 0.15 | 18.1 ± 0.4 |
| (siRNA/PAMAM)-PK NP 2 | 131.8 ± 3.7 | 0.17 | 22.9 ± 0.4 |
The PK shell is designed to shield siRNA from nucleases, re-enforce the structural integrity of the dendriplex core (avoidance of disassembly and aggregation under biological conditions), and enhance intracellular trafficking (cytosolic release of siRNA), which comprehensively contribute to successful siRNA delivery in vivo. Upon endocytosis, the PK shell hydrolyzes in the mildly acidic endosomes, destabilizes the endosome via NP swelling and proton buffering by amines of the PK shell and PAMAM dendrimer in the core [24], and releases siRNA into the cytoplasm (Figure 2b).
3.2. Improved structural homogeneity and enhanced stability of (siRNA/PAMAM)-PK NPs in serum, and stimuli-triggered siRNA release
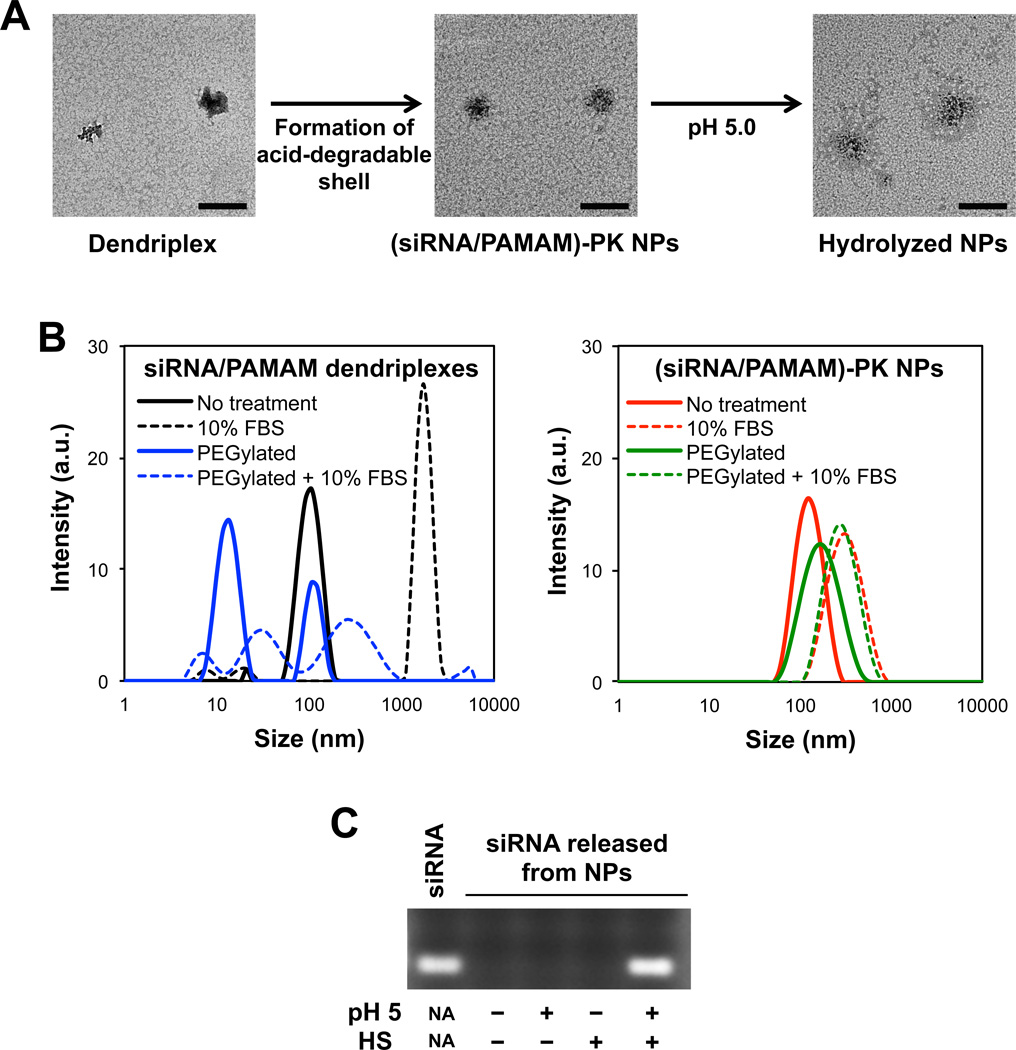
Chemical and structural homogeneity, high-density surface groups, and relatively low cytotoxicity are often described as advantages of employing dendrimers for biomedical applications. However, when nucleic acids are complexed with dendrimers, the resulting dendriplexes show a high polydispersity in size, which is exacerbated over time [25]. Moreover, complexation of small nucleotides (e.g., siRNA) with lower generation PAMAM (< generation 7) has been reported to be inefficient, due to limited molecular interactions [14], which also increases size and polydispersity. Immediate formation of cross-linked polymeric shell around a siRNA/PAMAM dendriplex is hypothesized to supplement the limited molecular interactions between PAMAM dendrimer and siRNA, and stabilize the siRNA/PAMAM dendriplexes, especially under biological conditions. The results shown in Figure 3 verify the hypothesis. The polydispersity index (PDI) of siRNA/PAMAM dendriplexes dropped from 0.27 (represented by the left image in Figure 3a) to 0.16 when they were shelled by a PK layer (represented by the middle image in Figure 3b). The irregular morphology and polydispersed sizes of siRNA/PAMAM dendriplexes, which has also been reported by others [18], were greatly improved in (siRNA/PAMAM)-PK NPs with spherical morphology and relatively uniform size distribution. The dendriplexs’ tendency to aggregate and prematurely release siRNA in serum, which are detrimental to in vivo applications, are also clearly corrected (Figure 3b). After 1 h incubation in 10% serum, the size of siRNA/PAMAM dendriplexes increased more than 20-fold (PDI jumped from 0.26 to 0.75), while the size of (siRNA/PAMAM)-PK NP increased about two-fold (PDI change from 0.17 to 0.31). The significant aggregation of siRNA/PAMAM dendriplexes can lead to rapid clearance by the reticuloendothelial (RES) system, poor access to tissues (e.g., tumor), and inefficient cellular uptake. Appearance of a new peak slightly larger than 10 nm in size when siRNA/PAMAM dendriplexes were incubated with serum also indicates their structural instability (e.g., premature siRNA release) under a biologically relevant condition.
Fig. 3.
(A) Morphology of dendriplexes, (siRNA/PAMAM)-PK NPs before acid-hydrolsysis, and acid-hydrolyzed (siRNA/PAMAM)-PK NPs, observed by TEM (bar = 200 nm). (B) Size analysis of dendriplexes (left) and (siRNA/PAMAM)-PK NPs (right) incubated with (dashed lines) and without (solid lines) 10 % serum for 1 h. PEGylated dendriplexes and (siRNA/PAMAM)-PK NPs under the same conditions were also analyzed. (C) Release of siRNA from differentially treated (siRNA/PAMAM)-PK NPs. siRNA was released only when (siRNA/PAMAM)-PK NPs were incubated at an acidic pH followed by subsequent HS treatment.
When siRNA/PAMAM dendriplexes were PEGylated, a large peak around 10 nm appeared, indicating that PEGylation onto the amines on the surface of PAMAM disrupted the electrostatic interactions with siRNA (Figure 3b). It is also speculated that conjugating targeting moieties using the same NHS-mediated conjugation may also destabilized the siRNA/PAMAM dendriplexes. Further incubation of siRNA/PAMAM dendriplexes in 10 % FBS induced the complete loss of their structural integrity, as indicated by a number of peaks in a wide range (Figure 3b). In contrast, when (siRNA/PAMAM)-PK NPs were PEGylated, only slight increase in size was observed without the generation of any additional peaks (Figure 3b). This confirms the enhanced structural stability of the siRNA/PAMAM dendriplexes by the PK shell, and facile conjugation of additional functional moieties such as targeting molecules onto the (siRNA/PAMAM)-PK NPs is also expected. Incubation in 10% FBS resulted in the same sizes of both PEGylated and unPEGylated (siRNA/PAMAM)-PK NPs.
The acid-degradable PK shell is designed to play dual roles: protection of siRNA/PAMAM dendriplexes from nucleases and serum proteins during circulation and stimuli-triggered intracellular release of siRNA upon acid-hydrolysis (Figure 2a). The later hypothesis was tested by gel electrophoresis of differentially treated (siRNA/PAMAM)-PK NPs (Figure 2c). siRNA was released only when (siRNA/PAMAM)-PK NPs were incubated at an acidic pH and subsequently with HS, an anionic biomacromolecule known to displace nucleic acids and disassemble complexes of nucleic acids and cationic polymers. The premature release of siRNA by HS was also effectively prevented by the PK shell, represented by the observation of no siRNA release when (siRNA/PAMAM)-PK NPs were incubated with HS, without acid-hydrolysis. This result also infers that siRNA in (siRNA/PAMAM)-PK NPs would be efficiently protected from other biomacromolecules such as nucleases. Acid-hydrolysis alone was unable to release free siRNA, implying that siRNA would be released into the cytoplasm from the acidic endosome as siRNA/PAMAM dendriplexes.
3.3. Facilitated intracellular siRNA release from endosomal compartments
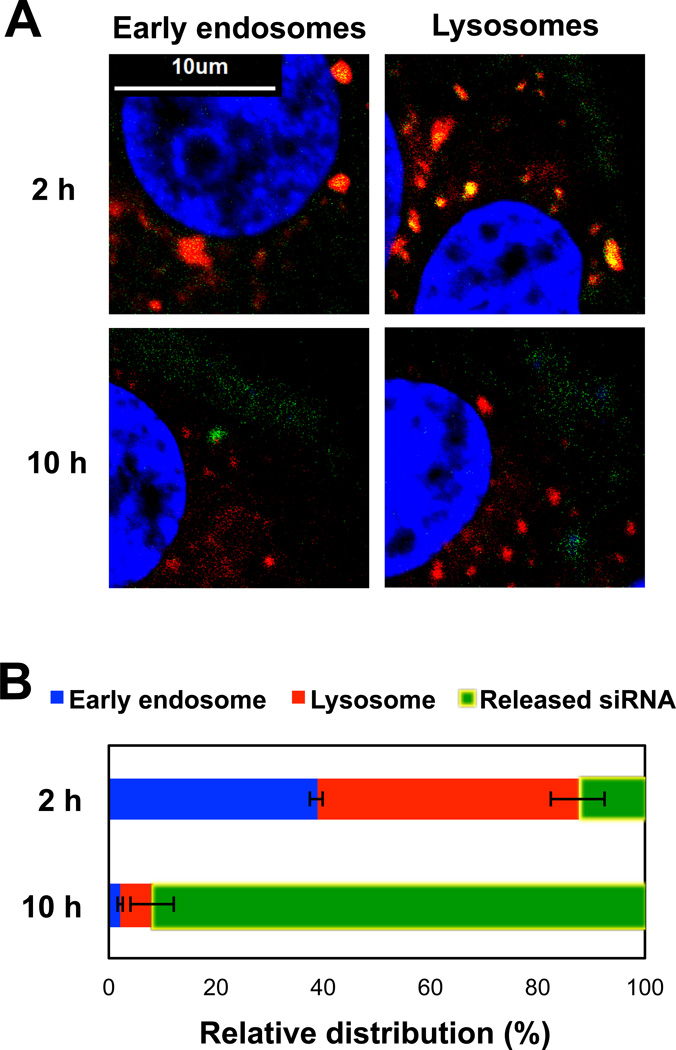
One of the significant hurdles in nucleic acid delivery is cytosolic release from endosomal compartments. FAM-labeled siRNA was used to prepare (siRNA/PAMAM)-PK NPs and its time-dependent intracellular trafficking in MCF7-BK-TR cells was observed (Figure 4). Early-endosomes and lysosomes were separately labeled as red, and nuclei were stained blue in order to ensure correct vertical depth and distinguish cytoplasm from nucleus. At 2 h of incubation, 40 and 50% siRNA was localized in the early-endosomes and the lysosomes, respectively (Figures 4a and b), Therefore, most siRNA (90%) was entrapped in the endosomal compartments at an early intracellular process. At 10 h of incubation, however, only a negligible amount of internalized siRNA (2%) was found in the early-endosomes (2%) and slightly more were entrapped in the lysosomes (6%), while releasing more than 90% of siRNA into the cytoplasm (Figure 4a and b).
Fig. 4.
(A) Intracellular distribution of FAM-labeled siRNA (green) after (siRNA/PAMAM)-PK NPs were incubated with MCF7-BK-TR cells incubated for 2 and 10 h. Early endosome (red, left) and lysosomes (red, right) were separately labeled, and siRNA co-localized in early endosomes or lysosomes is represented as yellow. (B) Quantified intracellular distributions of siRNA at 2 and 10 h of incubation with MCF7-BK-TR cells.
3.4. Reversed TAM-resistance by MnSOD knock-down in breast cancer cells
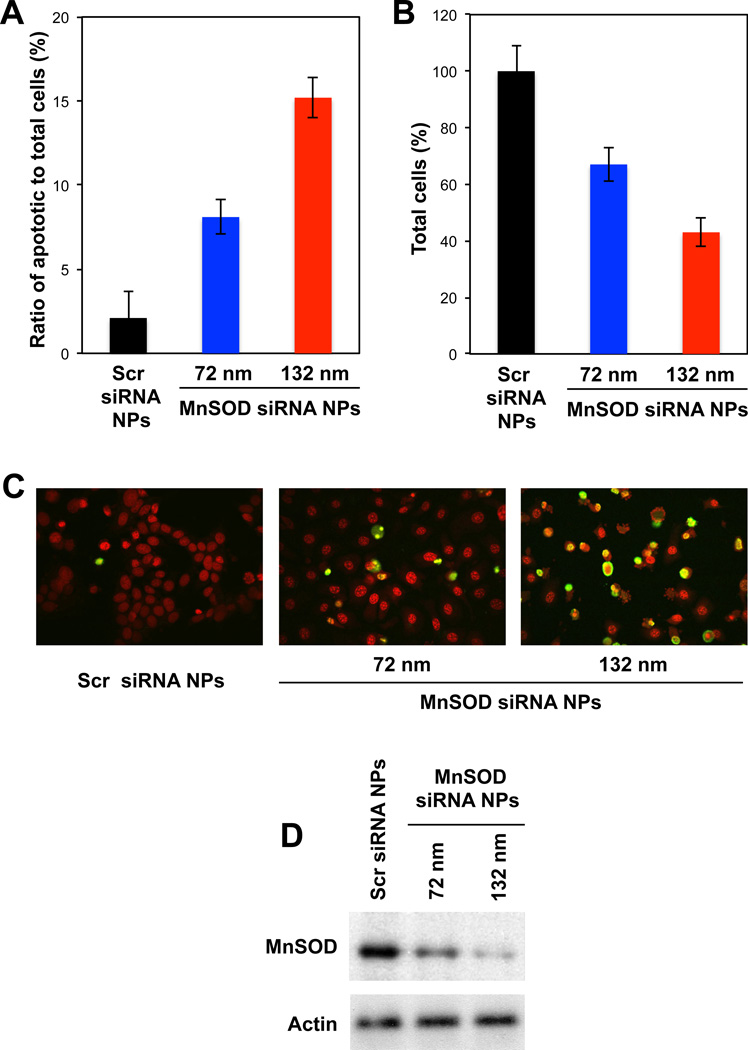
TAM-resistance occurs in nearly half of breast cancer patients undergoing this therapy and has been suggested to occur due to a high agonistic activity of mitochondrial ER-β [6]. MnSOD, a key enzyme in ER-β pathway, dismutates superoxide formation and thereby quenches the cytotoxic effects of TAM. As shown in Figures 5a, b, and c, TAM-resistant MCF7-BK-TR breast cancer cells were re-sensitized to TAM and the cellular apoptosis was significantly enhanced when MnSOD siRNA was delivered by (siRNA/PAMAM)-PK NPs (MnSOD siRNA NPs herein). In contrast, (siRNA/PAMAM)-PK NPs containing scr siRNA (scr siRNA NPs herein) resulted in minimal apoptosis and eradication of the cells. It was also confirmed that the NP-enhanced cellular apoptosis and eradication was due to silenced MnSOD expression by MnSOD siRNA NPs (Figure 5d). Noticeably, 130 nm MnSOD siRNA NPs exhibited higher cellular apoptosis (~ two-fold) (Figures 5a and c) and eradication (~ 1.3 fold) (Figure 5b) as well as silencing of MnSOD expression (Figure 5c) compared to 70 nm MnSOD siRNA NPs. Larger (siRNA/PAMAM)-PK NPs are synthesized with more acid-degradable monomers and cross-linkers than smaller ones, therefore, possess stronger acid-responsiveness. The results in Figure 5 suggest that rapid release of siRNA from the endosomal compartments is required to achieve efficient gene silencing.
Fig. 5.
(A) Apoptosis of MCF7-BK-TR cells incubated with TAM and with Scr siRNA NPs (129 nm) or MnSOD siRNA NPs (72 and 132 nm) for 48 h, confirmed by TUNEL staining (C). (B) Cell viability measured by MTT assay after 48 h of incubation with Scr siRNA NPs and MnSOD siRNA NPs (72 and 132 nm) in the presence of TAM. (D) Western blot of MnSOD expression in MCF7-BK-TR cells incubated with Scr siRNA NPs and MnSOD siRNA NPs (72 and 132 nm).
3.5. Recovered apoptosis of TAM-resistant breast tumor co-transplanted with MnSOD siRNA NPs in vivo
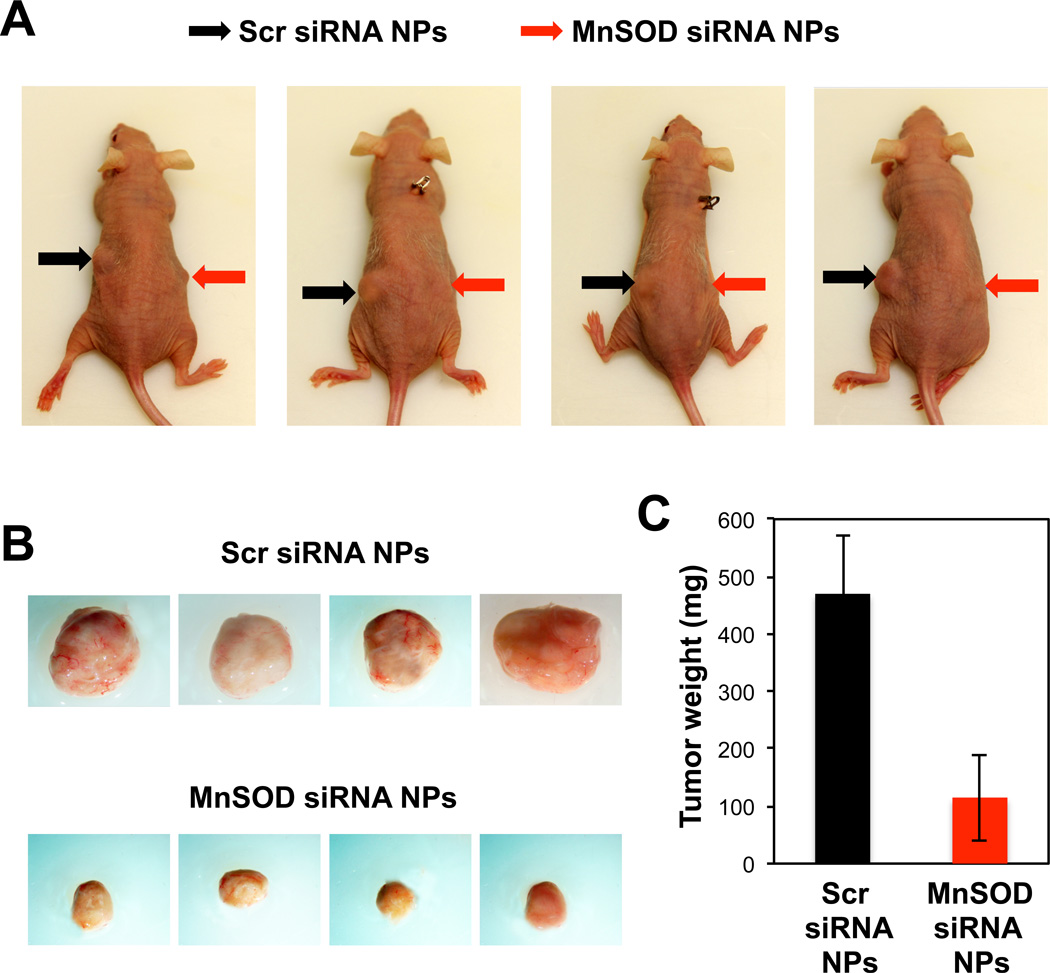
Sensitization of drug-resistant cancer cells to a chemotherapeutic agent in vivo is often not as effective as in vitro [26, 27]. TAM-resistant MCF7-BK-TR cells were mixed in Matrigel along with MnSOD siRNA or scr siRNA-containing NPs and injected into the flanks of nude mice that also had TAM pellets inserted subcutaneously under the skin of the back to maintain relevant TAM concentrations. This in vivo model allows a rapid formation of chemotherapy-resistant solid tumor environment with a constantly high TAM concentration in the tumor area, demonstrating the potential for clinical translation of this study. The results shown in Figure 6 clearly demonstrate effectively reversed TAM-resistance in breast tumor in vivo by MnSOD siRNA NPs. After 7 weeks, the growth of TAM-resistant tumors co-transplanted with scr siRNA NPs in the mice was much more obvious than those co-transplanted with MnSOD siRNA NPs (Figure 6a). The NP-mediated reversal of TAM-resistance in breast tumor was evident when the tumors were harvested from euthanized mice (Figure 6b). The tumors co-transplanted with scr siRNA NPs were approximately 5 times larger in weight (Figure 6c) than the tumors co-transplanted with MnSOD siRNA NPs.
Fig. 6.
(A) Mice with MnSOD siRNA NP-carrying (right flank) and scr siRNA NP-carrying (left flank) TAM-resistant breast tumor at 7 weeks post-transplantation. (B) Harvested tumors from the mice shown in (A). The tumors are presented at the same scale and they were also compared by average weight (C).
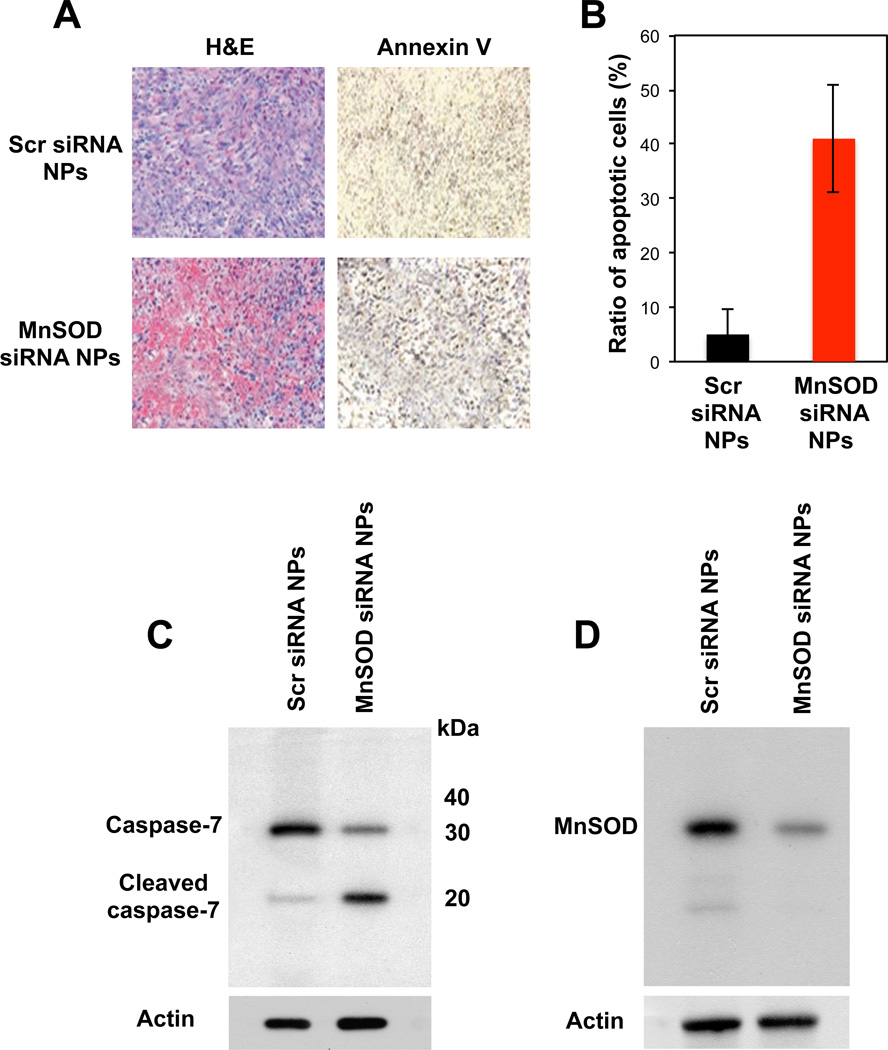
The effectively reversed TAM-resistance to apoptosis in breast tumor in vivo by MnSOD siRNA NPs was further histo-pathologically and biochemically confirmed (Figure 7). Annexin-V staining revealed that about 40% of the cells in TAM-resistant breast tumors underwent apoptosis at the time of assessment when co-transplanted with MnSOD siRNA NPs (Figure 7a and b) as marked by activation of a caspase (e.g., capsase-7) (Figure 7c). Caspase-7 is a cysteine peptidase that is cleaved to its active subunits by other caspases and induces apoptosis [28]. This suggests that a continual process of apoptosis occurred, resulting from MnSOD siRNAs NP in the presence of TAM, leading to the much smaller tumors at 7 weeks. In contrast, only a small fraction of the cells (less than 5%) in the scr siRNA NP-co-transplanted, TAM-resistant breast cancer underwent apoptosis (Figure 7a and b) and the caspase-7 in such tumor cells were found uncleaved (Figure 7c). It was also confirmed that the apoptosis of TAM-resistant breast cancer cells in vivo was triggered by NP-mediated MnSOD silencing. The results shown in Figures 6 and 7 strongly imply that drug-resistance of cancers can effectively be reversed in vivo by silencing the responsible molecular pathway.
Fig. 7.
(A) Histo-phatological confirmation of apoptosis of TAM-resistant tumor in vivo co-transplanted with MnSOD siRNA NP, by Annexin V staining. (B) A relative number of apoptotic to total cells in the randomly selected TAM-resistant tumor areas. (C and D) Enhanced caspase-activated apoptosis of TAM-resistant tumor in vivo (C) via NP-mediated MnSOD silencing.
4. Discussion
It is a general consensus in a number of reports that the promise for RNAi as a molecular therapeutichas been significantly hampered by inefficient means of administration [29]. Unlike delivering cDNA to express a protein (e.g., cytokines) for bystander effects, silencing a target gene expression at a desirable level in target cells is a daunting task. Employing RNAi as a synergistic supplement to other modes of therapy, however, can be an immediately applicable alternative use of this potentially revolutionary technology. Drug-resistant cancer cells that survive the first round of chemotherapy lead to increased failure of succeeding therapies and morbidity, requiring re-sensitization of the cells to therapy. In this study, a resistance in breast cancer cells to a commonly used anti-endocrine chemotherapeutic agent, TAM, was reversed in vitro and in vivo by silencing a key antagonistic protein, MnSOD, using nanoparticles consisting of an siRNA carrying core and an intracellular stimuli-responsive polymeric shell. The in vivo tumor model used in this study mimics clinically relevant conditions where breast tumors grow in the presence of TAM. The results in this study show that the MnSOD siRNA NPs enhanced intrinsic apoptotic activity of TAM-resistant breast cancer cells, implicating the high potential for clinical translation of the study.
The primary aim of the study was to validate the concept of reversing TAM-resistance in breast cancer in vitro and in vivo using siRNA-carrying NPs, therefore, the NPs were co-transplanted rather than being separately administered after tumor establishment. Although the results are promising, further refinement and optimization are needed to bring this exciting technology a step closer to clinic. Biodistribution of the NPs upon a systemic administration (e.g., intravenous injection) and targeted accumulation of the NPs in the tumor need to be explored in a subsequent study. Systemic administration of the NPs resulting in comparably reversed TAM-resistance in the breast tumor in vivo would conclusively confirm the clinical utility of the strategy investigated in this study. According to a number of literatures [10, 30], the size of the NPs used in this study (~ 130 nm) is in an appropriate range to achieve size-dependent passive tumor targeting via enhanced permeation and retention (EPR) effect. The size of the NPs can be easily tuned by controlling the thickness of PK shell around the siRNA/PAMAM core simply by varying surface-initiated photo-polymerization conditions (e.g., monomer and cross-linker concentrations) as demonstrated in this study. Smaller NPs with thinner PK shell may have higher access to tumors. However, thinner PK shell also results in lowered gene silencing efficiency due to limited cytosolic release of siRNA, as shown in Figure 5. On the other hand, too thick NPs may have poor pharmacokinetic properties, over-responsiveness to an endosomal condition (i.e., rapid endosomal destabilization), and may be toxic to cells. The abundantly available amines on the surface of the NPs can serve as conjugation handles for tumor targeting. Therefore, the NPs should be optimized for rapid accumulation in tumors upon systemic administration and efficient gene silencing.
PAMAM dendrimers have been a promising polymeric nanomaterial for biomedical applications. The primary amines on their surface can complex anionic nucleic acids, and the tertiary amines inside broaden the proton buffering capacity for endosomal escape in a cell. However, unlike linear cationic polymers, dendrimers complex nucleic acids on their surface, exposing their cargos to inactivating conditions. This results in aggregation and disassembly of nucleic acid/PAMAM dendriplexes by serum proteins and degradation of nucleic acids by nucleases. Overcoming these limitations is indispensable in using PAMAM dendrimers for in vivo applications. In this study, an acid-degradable polymeric PK shell was formed around a siRNA/PAMAM dendriplex core. It was demonstrated that the dendriplex core was fully protected by the PK shell from disassembly and aggregation in serum (Figure 3), in addition to efficient intracellular release of siRNA (Figure 4). As recently reported, shelling the nucleic acid/polymer core with a cross-linked polymeric layer enables lyophilization for easy handling and long-term storage. The PK shell can be further engineered for extended applications. Including other functional molecules in the PK shell (e.g., inorganic NPs) that are responsive to external triggers (e.g., laser-irradiation) can enable multi-modal sensitization (e.g., gene silencing and hyperthermia) of drug-resistance cancer cells. Co-encapsulation of chemotherapeutic drugs is anticipated to achieve synergistic therapy via normalized pharmacokinetic windows, which reduce required doses of the therapeutic molecules (i.e., lowered side effects). Inorganic NPs and chemoagents can be co-encapsulated in the PK shell by adding them to monomers and cross-linkers during the photo-polymerization process. Therefore, the PK shell not only improves the siRNA/PAMAM dendriplex core’s properties for in vivo gene silencing but also offers high functional versatility for expanded biomedical applications.
5. Conclusion
The limited capability of siRNA/PAMAM dendriplexes for gene silencing was greatly ameliorated by forming acid-degradable PK shell via surface-initiated photo-polymerization. The resulting (siRNA/PAMAM)-PK NPs showed improved stability in serum, facilitated siRNA release into the cytoplasm, and efficient gene silencing. The TAM-resistance of breast cancer cells was reversed when an antagonistic MnSOD activity was efficiently silenced by MnSOD siRNA NPs both in vitro and in vivo. The promising results of this study and high versatility of the NPs developed in this study indicates high possibility of clinical translation in a variety of biomedical applications upon further structural and functional optimizations.
Acknowledgment
The project described was partially supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant UL1 TR000153. The authors thank Jessica Kemp for proofreading and making comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Chen AM, Zhang M, Wei D, Stueber D, Taratula O, Minko T, et al. Co-delivery of doxorubicin and Bcl-2 siRNA by mesoporous silica nanoparticles enhances the efficacy of chemotherapy in multidrug-resistant cancer cells. Small. 2009;5:2673–2677. doi: 10.1002/smll.200900621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kamada M, So A, Muramaki M, Rocchi P, Beraldi E, Gleave M. Hsp27 knockdown using nucleotide-based therapies inhibit tumor growth and enhance chemotherapy in human bladder cancer cells. Mol Cancer Ther. 2007;6:299–308. doi: 10.1158/1535-7163.MCT-06-0417. [DOI] [PubMed] [Google Scholar]
- 3.Meng H, Liong M, Xia T, Li Z, Ji Z, Zink JI, et al. Engineered design of mesoporous silica nanoparticles to deliver doxorubicin and P-glycoprotein siRNA to overcome drug resistance in a cancer cell line. ACS Nano. 2010;4:4539–4550. doi: 10.1021/nn100690m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Early Breast Cancer Trialists' Collaborative Group. Tamoxifen for early breast cancer: an overview of the randomised trials. The Lancet. 1998;351:1451–1467. [PubMed] [Google Scholar]
- 5.Pedram A, Razandi M, Wallace DC, Levin ER. Functional estrogen receptors in the mitochondria of breast cancer cells. Mol Biol Cell. 2006;17:2125–2137. doi: 10.1091/mbc.E05-11-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Razandi M, Pedram A, Jordan V, Fuqua S, Levin ER. Tamoxifen regulates cell fate through mitochondrial estrogen receptor beta in breast cancer. Oncogene. 2012:1–12. doi: 10.1038/onc.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matés JM, Sánchez-Jiménez FM. Role of reactive oxygen species in apoptosis: implications for cancer therapy. Int J Biochem Cell B. 2000;32:157–170. doi: 10.1016/s1357-2725(99)00088-6. [DOI] [PubMed] [Google Scholar]
- 8.Shim MS, Kwon YJ. Efficient and targeted delivery of siRNA In Vivo. FEBS J. 2010;277:4814–4827. doi: 10.1111/j.1742-4658.2010.07904.x. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen J, Szoka FC. Nucleic acid delivery: the missing pieces of the puzzle? Acc Chem Res. 2012;45:1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alexis F, Pridgen E, Molnar LK, Farokhzad OC. Factors affecting the clearance and biodistribution of polymeric nanoparticles. Mol Pharm. 2008;5:505–515. doi: 10.1021/mp800051m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon YJ. Before and after endosomal escape: roles of stimuli-converting siRNA/polymer interactions in determining gene silencing efficiency. Acc Chem Res. 2012;45:1077–1088. doi: 10.1021/ar200241v. [DOI] [PubMed] [Google Scholar]
- 12.Boussif O, Lezoualc'h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, et al. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. P Natl Aca Sci USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benjaminsen RV, Mattebjerg MA, Henriksen JR, Moghimi SM, Andresen TL. The possible "proton sponge" effect of polyethylenimine (PEI) does not include change in lysosomal pH. Mol Ther. 2013;21:149–157. doi: 10.1038/mt.2012.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richard I, Thibault M, De Crescenzo G, Buschmann MD, Lavertu M. Ionization behavior of chitosan and chitosan-DNA polyplexes indicate that chitosan has a similar capability to induce a proton-sponge effect as PEI. Biomacromolecules. 2013;14:1732–1740. doi: 10.1021/bm4000713. [DOI] [PubMed] [Google Scholar]
- 15.Dufès C, Uchegbu IF, Schätzlein AG. Dendrimers in gene delivery. Adv Drug Del Rev. 2005;57:2177–2202. doi: 10.1016/j.addr.2005.09.017. [DOI] [PubMed] [Google Scholar]
- 16.Nandy B, Maiti PK. DNA compaction by a dendrimer. J Phys Chem B. 2011;115:217–230. doi: 10.1021/jp106776v. [DOI] [PubMed] [Google Scholar]
- 17.Froehlich E, Mandeville JS, Weinert CM, Kreplak L, Tajmir-Riahi HA. Bundling and aggregation of DNA by cationic dendrimers. Biomacromolecules. 2011;12:511–517. doi: 10.1021/bm1013102. [DOI] [PubMed] [Google Scholar]
- 18.Ouyang D, Zhang H, Parekh HS, Smith SC. The effect of pH on PAMAM dendrimer-siRNA complexation: endosomal considerations as determined by molecular dynamics simulation. Biophys Chem. 2011;158:126–133. doi: 10.1016/j.bpc.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 19.Kukowska-Latallo JF, Bielinska AU, Johnson J, Spindler R, Tomalia DA, Baker JR. Efficient transfer of genetic material into mammalian cells using Starburst polyamidoamine dendrimers. Proc Natl Aca Sci USA. 1996;93:4897–4902. doi: 10.1073/pnas.93.10.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang H, DeLong R, Fisher M, Juliano R. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharm Res. 2005;22:2099–2106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- 21.Shen X-C, Zhou J, Liu X, Wu J, Qu F, Zhang Z-L, et al. Importance of size-to-charge ratio in construction of stable and uniform nanoscale RNA/dendrimer complexes. Org Biomol Chem. 2007;5:3674–3681. doi: 10.1039/b711242d. [DOI] [PubMed] [Google Scholar]
- 22.Cho SK, Kwon YJ. Polyamine/DNA polyplexes with acid-degradable polymeric shell as structurally and functionally virus-mimicking nonviral vectors. J Control Release. 2011;150:287–297. doi: 10.1016/j.jconrel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 23.Cho SK, Kwon YJ. Simultaneous gene transduction and silencing using stimuli-responsive viral/nonviral chimeric nanoparticles. Biomaterials. 2012;33:3316–3323. doi: 10.1016/j.biomaterials.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Rocchi P, Peng L. Dendrimers as non-viral vectors for siRNA delivery. New J Chem. 2012;36:256–263. [Google Scholar]
- 25.Eichman JD, Bielinska AU, Kukowska-Latallo JF, Baker JR., Jr The use of PAMAM dendrimers in the efficient transfer of genetic material into cells. Pharm Sci Technol Today. 2000;3:232–245. doi: 10.1016/s1461-5347(00)00273-x. [DOI] [PubMed] [Google Scholar]
- 26.van Zuylen L, Nooter K, Sparreboom A, Verweij J. Development of multidrug-resistance convertors: sense or nonsense? Invest New Drugs. 2000;18:205–220. doi: 10.1023/a:1006487003814. [DOI] [PubMed] [Google Scholar]
- 27.Thomas H, Coley HM. Overcoming multidrug resistance in cancer: an update on the clinical strategy of inhibiting p-glycoprotein. Cancer control. 2003;10 doi: 10.1177/107327480301000207. 159-. [DOI] [PubMed] [Google Scholar]
- 28.Lakhani SA, Masud A, Kuida K, Porter GA, Booth CJ, Mehal WZ, et al. Caspases 3 and 7: Key mediators of mitochondrial events of apoptosis. Science. 2006;311:847–851. doi: 10.1126/science.1115035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mello C. A conversation with Craig C Mello on the discovery of RNAi. Cell Death Differ. 2007;14:1981–1984. doi: 10.1038/sj.cdd.4402249. [DOI] [PubMed] [Google Scholar]
- 30.Morille M, Passirani C, Vonarbourg A, Clavreul A, Benoit J-P. Progress in developing cationic vectors for non-viral systemic gene therapy against cancer. Biomaterials. 2008;29:3477–3496. doi: 10.1016/j.biomaterials.2008.04.036. [DOI] [PubMed] [Google Scholar]