Abstract
Objective
Testing for high risk human papillomavirus (HR-HPV) is increasing; however due to limitations in specificity there remains a need for better triage tests. Research efforts have focused recently on methylation of human genes which show promise as diagnostic classifiers.
Methods
Methylation of 26 genes: APC, CADM1, CCND2, CDH13, CDKN2A, CTNNB1, DAPK1, DPYS, EDNRB, EPB41L3, ESR1, GSTP1, HIN1, JAM3, LMX1, MAL, MDR1, PAX1, PTGS2, RARB, RASSF1, SLIT2, SOX1, SPARC, TERT and TWIST1 was measured by pyrosequencing in cytology specimens from a pilot set of women with normal or cervical intraepithelial neoplasia grade 3 (CIN3) histology. Six genes were selected for testing in Predictors 1, a colposcopy referral study comprising 799 women. The three genes EPB41L3, DPYS and MAL were further tested in a second colposcopy referral study, Predictors 2, comprising 884 women.
Results
The six genes selected from the pilot: EPB41L3, EDNRB, LMX1, DPYS, MAL and CADM1 showed significantly elevated methylation in CIN2 and CIN3 (CIN2/3) versus ≤CIN1 in Predictors 1 (p < 0.01). Highest methylation was observed in cancer tissues. EPB41L3 methylation was the best single classifier of CIN2/3 in both HR-HPV positive (p < 0.0001) and negative samples (p = 0.02). Logistic regression modeling showed that other genes did not add significantly to EPB41L3 and in Predictors 2, its classifier value was validated with AUC 0.69 (95% CI 0.65–0.73).
Conclusion
Several methylated genes show promise for detecting CIN2/3 of which EPB41L3 seems the best. Methylated human gene biomarkers used in combination may be clinically useful for triage of women with HR-HPV infections.
Keywords: Cervical intraepithelial neoplasia, Human gene methylation, EPB41L3, Diagnostic biomarker
Highlights
-
•
Methylation of 26 human genes assessed by pyrosequencing in 40 cytology specimens
-
•
Several methylated genes show promise for detecting CIN2/3.
-
•
EBP41L3 shows reproducible biomarker potential in high risk HPV positive women.
Introduction
Cervical cancer is the third most common type of cancer in women worldwide and persistent infection with high-risk human papillomavirus (HR-HPV) is the primary etiological agent of the disease [1,2]. Although HR-HPV infections are very common, only a small proportion (approximately 10%) become persistent, of which a subset drive the progression of occult precursors to cervical intraepithelial neoplasia grades 2 and 3 (CIN2/3) and eventually cervical cancer [3]. Due to high sensitivity, no requirements for preservation of cell morphology and relatively low costs, HR-HPV testing is becoming the preferred method for CIN screening. However, as only a fraction of HR-HPV positive women have clinically relevant lesions, use of cytology as the primary screening test still predominates. Novel diagnostic biomarkers with high specificity are desirable as a compliment to HR-HPV testing. Development of a simple molecular triage test is particularly awaited because such a test may be more easily implemented in less developed countries and in vaginal self-collected specimens, thus leading to considerable reduction in cervical cancer deaths worldwide.
HR-HPV infection is a prerequisite for carcinogenesis but is not sufficient alone. Genetic and epigenetic changes in the host and/or viral genome affect the outcome of HR-HPV infection and identifying these key molecular changes is likely to reveal biomarkers for use in cancer prevention programs. DNA methylation has been extensively investigated as a potential biomarker of human cancers [4]. Carefully orchestrated, DNA methylation plays a crucial role for activating and silencing genes during normal development; however, its disruption contributes to carcinogenesis [5]. In cervical cancer, aberrant methylation can be detected in cervical smears up to 7 years prior to diagnosis indicating promise as a biomarker [6]. Although many human genes exhibit elevated methylation in cervical cancer, an effective testing approach exploiting these changes for triage of HR-HPV positive women requires a robust assay that can separate CIN2/3 from ≤CIN1 with an adequate and consistent differential. Quantitative methylation-specific polymerase chain reaction (QMSP) assays for CADM1 and MAL [7–9] have shown promise as triage tools. Other combination QMSP assays of genes sets such as EPB41L3, JAM3, TERT, C13ORF18 [10,11], LMX1, PAX1, SOX1, NKX6-1 [12], and APC, HIN-1, RARB, RASSF1A, and TWIST1 [13] are also of interest. Furthermore, some studies have identified elevated methylation of single or groups of genes, using different technologies, in both cytology samples as well as biopsy specimens [14]. In the current study, we aimed to evaluate methylation of 26 human genes in cytology samples by pyrosequencing. Genes APC, CADM1, CCND2, CDH13, CDKN2A, CTNNB1, DAPK1, EPB41L3, ESR1, GSTP1, HIN1, JAM3, LMX1, MAL, PAX1, PTGS2, RARB, RASSF1, SLIT2, SOX1, SPARC, TERT and TWIST1 were selected as they were part of earlier described promising QMSP assays or reported as aberrant in at least two separate reports [7–14]. In addition, genes previously not investigated in cervical cancer: DPYS, EDNRB and MDR1 were included as they showed interesting aberrant methylation patterns in other cancers of mucosal tissue [15–17]. Pyrosequencing was chosen as it gives absolute quantification of methylation [18] in contrast to the relative quantification obtained by the most commonly used method QMSP. Our aim was to measure the methylation levels of a set of promising genes on an absolute scale and to directly compare their methylation, initially in a selected set of CIN3 and normal samples. Genes that showed the best potential to separate CIN3 from normal were tested in Predictors 1 (P1) a large colposcopy referral population. The final goal was to investigate the performance of the most informative genes in a second large study Predictors 2 (P2) and to assess their potential as future triage biomarkers of HR-HPV positive women. The results of our investigations may be regarded as independent validation of the biomarkers by virtue of the use of different specimens and assays in a separate and independent laboratory.
Material and methods
Patient cohort
Archived patient materials collected in two earlier studies, Predictors 1 (P1) [19] and Predictors 2 (P2) [20] were used for methylation testing. P1 and P2 comprised 1000 and 1099 women respectively attending colposcopy at Hammersmith or St Mary Hospital in London UK, during 2005–2007 (P1) or 2007–2009 (P2) because of a cytology finding of “mild dyskaryosis or worse”. Women were eligible if they had been referred as a result of one or more abnormal cervical smears, were not pregnant, had not been treated previously for CIN, nor had a hysterectomy. Immediately before colposcopy, cervical specimens were obtained in PreservCyt transport medium. All analyses were based on a centrally reviewed histopathology and took the highest grade of abnormality seen in the biopsy or treatment specimen within 9 months of the initial base-line visit as the final diagnosis. The number of women with different histology findings for P1 and P2 is presented in Supplemental table S1. 201 patients from P1 were excluded due to insufficient material for the methylation study. For identification of HR-HPV positive samples, Linear Array (Roche Molecular Systems, Inc., Pleasanton, CA, USA) results for P1 and BD HPV test (BD Diagnostics, Burlington, NC, USA) results for P2 were used.
Archived patient materials collected in two earlier studies, Predictors 1 (P1) [19] and Predictors 2 (P2) [20] were used for methylation testing. P1 and P2 comprised 1000 and 1099 women respectively attending colposcopy at Hammersmith or St Mary Hospital in London UK, during 2005–2007 (P1) or 2007–2009 (P2) because of a cytology finding of “mild dyskaryosis or worse”. Women were eligible if they had been referred as a result of one or more abnormal cervical smears, were not pregnant, had not been treated previously for CIN, nor had a hysterectomy. Immediately before colposcopy, cervical specimens were obtained in PreservCyt transport medium. All analyses were based on a centrally reviewed histopathology and took the highest grade of abnormality seen in the biopsy or treatment specimen within 9 months of the initial base-line visit as the final diagnosis. The number of women with different histology findings for P1 and P2 is presented in Supplemental table S1. 201 patients from P1 were excluded due to insufficient material for the methylation study. For identification of HR-HPV positive samples, Linear Array (Roche Molecular Systems, Inc., Pleasanton, CA, USA) results for P1 and BD HPV test (BD Diagnostics, Burlington, NC, USA) results for P2 were used.
All women received a patient information sheet explaining the study and provided written consent. Approvals were obtained from the relevant local research ethics committees.
DNA isolation and bisulfite conversion
300 μl of PreservCyt was centrifuged at 13 200 rpm for 2 min and the pellet was resuspended in 200 μl PBS. The genomic DNA was extracted with QIAamp DNA Mini Kit (Qiagen Inc., Hilden, Germany) following the spin protocol recommended by the manufacturer except that DNA was eluted in 60 μl AE buffer. 250 ng of DNA was used in the bisulfite conversion reactions where unmethylated cytosines were converted to uracil with the EZ DNA methylation kit (Zymo research, Irvine, CA) according to manufacturer's instructions.
The methylation assays
The methylation assays for APC, CCND2, CDH13, CDKN2A, DAPK1, DPYS, EDNRB, ESR1, GSTP1, HIN1, MAL, MDR1, PTGS2, RARB, RASSF1, SLIT2 and TWIST1 were previously described [21]. The remaining assays are presented in Supplemental table S2. All primers were designed using PyroMark Assay Design software version 2.0.1.15 (Qiagen). The assays were designed to avoid any overlapping of CpG positions and aimed to investigate the same CpG positions or those in close proximity to CpG sites previously investigated for a gene. To provide the internal control for total bisulfite conversion, a non CG cytosine was included. Although genes C13ORF18 and NKX6-1 were part of combination QMSP assays of interest [10] we were not able to design corresponding pyrosequencing assays due to high CpG density. Polymerase chain reaction (PCR) assays were performed as described in the footnote to Supplemental table 2. The presence of the correct amplicon was confirmed on Qiaxel (Qiagen) and pyrosequencing was done as previously described [21]. All runs included standard curves as positive controls, of 0%, 50% and 100% methylated human DNA and a non-template control.
The methylation assays for APC, CCND2, CDH13, CDKN2A, DAPK1, DPYS, EDNRB, ESR1, GSTP1, HIN1, MAL, MDR1, PTGS2, RARB, RASSF1, SLIT2 and TWIST1 were previously described [21]. The remaining assays are presented in Supplemental table S2. All primers were designed using PyroMark Assay Design software version 2.0.1.15 (Qiagen). The assays were designed to avoid any overlapping of CpG positions and aimed to investigate the same CpG positions or those in close proximity to CpG sites previously investigated for a gene. To provide the internal control for total bisulfite conversion, a non CpG cytosine was included. Although genes C13ORF18 and NKX6-1 were part of combination QMSP assays of interest [10] we were not able to design corresponding pyrosequencing assays due to high CpG density. Polymerase chain reaction (PCR) assays were performed as described in the footnote to Supplemental table 2. The presence of the correct amplicon was confirmed on Qiaxel (Qiagen) and pyrosequencing was done as previously described [21]. All runs included standard curves as positive controls, of 0%, 50% and 100% methylated human DNA and a non-template control.
Data analysis
Mean methylation of the investigated CpG positions, 3–6 per each gene, was used for all the analyses. Median methylation of genes in CIN2/3 and ≤CIN1 tissues was compared using the Mann–Whitney U test. For random selection of 40 samples from P1, a computer random number generator was used. To account for the high number of genes analyzed on the initial sample set, the Benjamin and Hochberg step-up procedure for controlling false discovery rate (FDR) was applied with an FDR limit of 5% [22]. Any failed assays were excluded from the analysis. Raw p-values are presented in the text and tables with footnotes indicating p-values significant at 5% FDR. Spearman's rho correlation coefficient was estimated for methylation levels of the 6 genes in the entire P1 data set. Each gene was evaluated by receiver operating characteristics (ROCs) analyses, area under the curve (AUC) was the main measure used to assess the ability of genes to classify high grade disease. Methylation data from cancers were excluded from all analysis, but are presented in the supplemental data. Univariate and multivariate analysis was done using logistic regression models and compared by likelihood ratio (LR) and χ2 statistics. STATA v12 and GraphPad Prism v5.03 were used for statistical analysis and illustrations.
Results
Methylation of 26 genes in 40 patients from the Pilot Set
The methylation of 26 genes was measured in the pilot set comprising 20 CIN3 and 20 normal controls, all were HR-HPV positive and randomly selected from the P1 study. No assays failed in the pilot set. Overall the methylation of genes was low with 14 genes—JAM3, GSTP1, CDKN2A, APC, HIN1, RARB, RASSF1A, TWIST1, ESR1, CDH13, SPARC, PTGS2, CTNNB1 and CCND2 showing median methylation ≤ 1% in both groups (Supplemental table 3), these were excluded from further consideration. Eight genes: EPB41L3, EDNRB, DPYS, LMX1, CADM1, TERT, PAX1 and MDR1 showed elevated methylation in CIN3 and were individually significant at α = 0.05 (Supplemental table 3). However, after correction for multiple comparisons, only EPB41L3 (p < 0.0001) and EDNRB (p = 0.0004) remained significant and consequently they were tested in the entire P1 study. In addition, LMX1 and DPYS showed relatively high methylation (3–6%) with good ability to separate normal from CIN3 with AUC > 0.7 (Table 1). Thus, these genes were also investigated in the entire P1 cohort. Additionally, MAL and CADM1 were tested in the P1 study because these two genes were extensively evaluated in earlier reports and shown as particularly promising methylation biomarkers. In total, 6 genes were tested in all remaining (759) patients from the P1 cohort.
Table 1.
Median methylation % and interquartile range (IQR), area under the curve (AUC) and 95% confidence intervals (CI), and Mann–Whitney U test P values for the 6 top genes measured in the HR-HPV positive P1 specimens, comparing women diagnosed with histological CIN2/3 to women diagnosed with <CIN1 (includes CIN1 and normal).
| GENE |
<CIN1 |
CIN2/3 |
AUC | 95%CI | Pa | ||
|---|---|---|---|---|---|---|---|
| Medianb (n)c |
IQR | Medianb (n)c |
IQR | ||||
| EPB41L3 | 1.4 (363) |
1.0–2.5 | 3.3 (208) |
1.4–7.6 | 0.73 | 0.68–0.77 | 2.8*10− 19 |
| EDNRB | 1.5 (363) |
0.8–2.6 | 3.3 (208) |
1.5–6.1 | 0.67 | 0.62–0.72 | 3.4*10− 12 |
| DPYS | 4.6 (364) |
3.2–5.8 | 5.4 (208) |
4.2–6.6 | 0.65 | 0.61–0.70 | 7.4*10− 10 |
| LMX1 | 3.9 (348) |
3.3–4.6 | 4.6 (193) |
3.6–7.1 | 0.63 | 0.58–0.68 | 3.4*10− 07 |
| MAL | 2.4 (363) |
1.6–3.5 | 3.2 (206) |
2.1–4.8 | 0.62 | 0.57–0.67 | 2.8*10− 06 |
| CADM1 | 1.6 (357) |
0.9–3.0 | 2.6 (205) |
1.1–5.3 | 0.58 | 0.53–0.62 | 1.1*10− 03 |
Indicates P values not adjusted for multiple testing, all genes remained significantly associated with CIN2/3 (P < 0.05) at a false discovery rate of 5%.
The medians and IQR exclude specimens where the methylation assay failed or gave a value of 0%. The numbers of specimens where methylation was either (failed, or 0%) in <CIN1 for each of the genes were as follows: EPB41L3 (1, 68); EDNRB (1, 131); DPYS (0, 12); LMX1 (16, 21); MAL (1, 43); CADM1 (7, 211), respectively. For CIN2/3 the values for failed or 0% methylation were EPB41L3 (0, 12); EDNRB (0, 48); DPYS (0, 0); LMX1 (15, 9); MAL (2, 21); CADM1 (3, 101), respectively.
n indicates the number of specimens of each histological category tested for each gene.
The methylation of 26 genes was measured in the pilot set comprising 20 CIN3 and 20 normal controls, all were HR-HPV positive and randomly selected from the P1 study. No assays failed in the pilot set. Overall the methylation of genes was low with 14 genes—JAM3, GSTP1, CDKN2A, APC, HIN1, RARB, RASSF1A, TWIST1, ESR1, CDH13, SPARC, PTGS2, CTNNB1 and CCND2 showing median methylation ≤ 1% in both groups (Supplemental table 3), these were excluded from further consideration. Nine genes: EPB41L3, EDNRB, DPYS, CADM1, LMX1, JAM3, TERT, PAX1 and MDR1 showed elevated methylation in CIN3 and were individually significant at α = 0.05 (Supplemental table 3). However, after correction for multiple comparisons, only EPB41L3 (p < 0.0001) and EDNRB (p = 0.0004) remained significant and consequently they were tested in the entire P1 study. In addition, LMX1 and DPYS showed relatively high methylation (3–6%) with good ability to separate normal from CIN3 with AUC > 0.7 (Supplemental Table 3). Thus, these genes were also investigated in the entire P1 cohort. Additionally, MAL and CADM1 were tested in the P1 study because these two genes were extensively evaluated in earlier reports and shown as particularly promising methylation biomarkers. In total, 6 genes were tested in all remaining (759) samples available from the P1 cohort.
Methylation of EPB41L3, EDNRB, DPYS, LMX1, CADM1 and MAL in the P1 study
The methylation of the 6 selected genes was measured in all available P1 samples and was analyzed separately for 572 HR-HPV positive (Table 1) and 217 negative samples. The success rates for all the methylation assays was 97.0–99.9%. In HR-HPV positive patients, Mann–Whitney U test indicated that methylation was elevated in CIN2/3 for all 6 genes (p < 0.001). Particularly, EPB41L3 showed the best separation of high grade lesions from those with ≤ CIN1 with AUC 0.73 (95%CI 0.68–0.77). Although EDNRB was second best at separating the diagnostic groups with AUC 0.67 (95%CI 0.62–0.71), its methylation was also most highly correlated to methylation of EPB41L3 (Table 2). The AUC for DPYS, LMX1, MAL and CADM1 were 0.65 (95%CI 0.6–0.69), 0.63 (95%CI 0.58–0.68), 0.61 (95%CI 0.56–0.66) and 0.58 (95%CI 0.53–0.63) respectively. Except for DPYS and MAL, methylation of all genes showed significant correlation to methylation of the other genes (p < 0.001) (Table 2). A logistic regression model was used to explore the pairwise predictive power of the methylation of the 6 genes (Supplemental table S3). In a multivariate model with EPB41L3, other genes did not add significant information for identifying high grade disease; however DPYS was the strongest gene among the remaining 5 (Supplemental table S4). Therefore, EPB41L3 and DPYS were selected for investigation in all HR-HPV positive samples (884) in the P2 cohort. Although pre-selected due to its high profile in publications, CADM1 performed the poorest amongst the six genes, had the lowest AUC, median methylation (Supplemental Fig. S1), and had the lowest univariate LR-χ2 (Supplemental table S4). CADM1 was therefore excluded from further testing. MAL was however, taken forward for further evaluation.
Table 2.
Spearman correlation coefficient showing the level of correlation of gene methylation in P1 specimens.
| CADM1 | MAL | DPYS | EDNRB | EPB41L3 | LMX1 | |
|---|---|---|---|---|---|---|
| CADM1 | 1.00 | |||||
| MAL | 0.19 | 1.00 | ||||
| DPYS | 0.27 | − 0.02 | 1.00 | |||
| EDNRB | 0.26 | 0.55 | 0.19 | 1.00 | ||
| EPB41L3 | 0.27 | 0.45 | 0.20 | 0.58 | 1.00 | |
| LMX1 | 0.26 | 0.38 | 0.28 | 0.42 | 0.40 | 1.00 |
The methylation of the 6 selected genes was measured in all available P1 samples and was analyzed separately for 572 HR-HPV positive (Table 1) and 217 negative samples. The success rates for all the methylation assays was 97.0–99.9%. In HR-HPV positive patients, Mann–Whitney U test indicated that methylation was elevated in CIN2/3 for all 6 genes (p < 0.001). Particularly, EPB41L3 showed the best separation of high grade lesions from those with ≤CIN1 with AUC 0.73 (95%CI 0.68–0.77). Although EDNRB was second best at separating the diagnostic groups with AUC 0.67 (95%CI 0.62–0.72), its methylation was also most highly correlated to methylation of EPB41L3 (Table 2). The AUC for DPYS, LMX1, MAL and CADM1 were 0.65 (95%CI 0.60–0.69), 0.63 (95%CI 0.58–0.68), 0.62 (95%CI 0.57–0.67) and 0.58 (95%CI 0.53–0.62) respectively. Except for DPYS and MAL, methylation of all genes showed significant correlation to methylation of the other genes (p < 0.001) (Table 2). A logistic regression model was used to explore the pairwise predictive power of the methylation of the 6 genes (Supplemental table S3). In a multivariate model with EPB41L3, other genes did not add substantial information for identifying high grade disease; however DPYS was the strongest gene among the remaining 5 (Supplemental table S4). Therefore, EPB41L3 and DPYS were selected for investigation in all HR-HPV positive samples (884) in the P2 cohort. Although pre-selected due to its high profile in publications, CADM1 performed the poorest amongst the six genes, had the lowest AUC, median methylation (Supplemental Fig. S1), and had the lowest univariate LR-χ2 (Supplemental table S4). CADM1 was therefore excluded from further testing. MAL was however, taken forward for further evaluation.
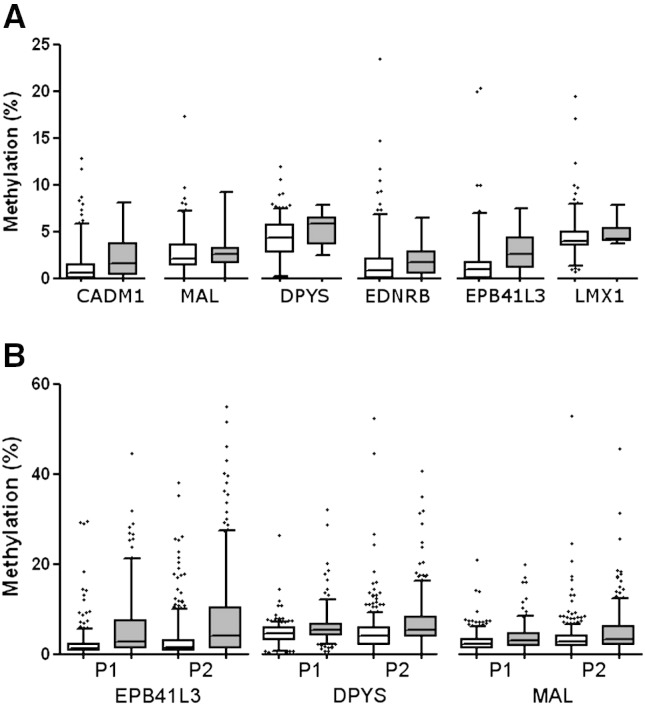
In HR-HPV negative patients, EPB41L3 (p = 0.02) and DPYS (p = 0.04) showed significantly elevated methylation in the 10 CIN2/3 cases while the other genes did not (Fig. 1A).
Fig. 1.
The distribution of methylation in HR-HPV negative and positive samples. A) Comparison of methylation distribution of 6 genes in 207 HR-HPV negative controls (white) and 10 CIN2/3 (grey) in the P1 study. EPB41L3 (p = 0.023) and DPYS (p = 0.043) showed elevated methylation in CIN2/3 while CADM1 (p = 0.051), MAL (p = 0.810) and LMX1 (p = 0.177) did not. B) Comparison of EPB41L3, DPYS and MAL methylation distribution in HR-HPV positive <CIN1 controls (white) and CIN2/3 (grey) from P1 and P2 cohort. Significantly elevated methylation was observed in CIN2/3 group for all three genes in both studies (p < 0.0001). Numbers of patients in each group are available in Supplemental table S1. For better visualisation of the low methylation results, 2 outliers are not shown, one at 70% EPB41L3 methylation in a normal tissue from P2 and one at 91% DPYS methylation in a normal tissue from P1. Whiskers of the boxplot mark the 5th and 95th percentiles, the box 25th percentile, median and 75 percentile, while extreme values are shown by (•).
Internal validation of EPB41L3 and DPYS genes in P2 HR-HPV positive samples
EPB41L3, DPYS and MAL showed a similar distribution of methylation in ≤ CIN1 and CIN2/3 in both studies (Fig. 1b). The combined methylation data from all HR-HPV positive P1 and P2 samples stratified by histology groups: normal, CIN1, CIN2, CIN3 and cancer is shown in Supplemental Fig. S1. The methylation in cancer tissues was highest, followed by CIN3.
EPB41L3, DPYS and MAL showed a similar distribution of methylation in ≤CIN1 and CIN2/3 in both studies (Fig. 1B). The combined methylation data from all HR-HPV positive P1 and P2 samples stratified by histology groups: normal, CIN1, CIN2, CIN3 and cancer is shown in Supplemental Fig. S1. The methylation in cancer tissues was highest, followed by CIN3.
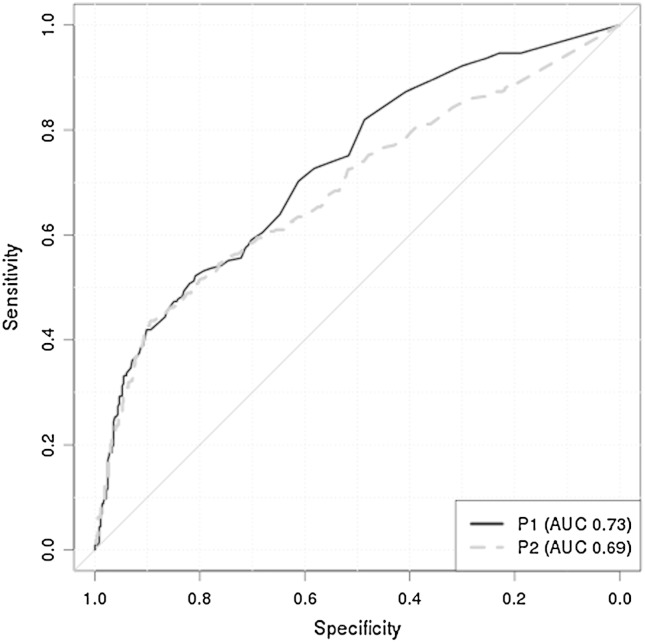
Based on the methylation data from the P1 specimens, a cut-off to reach a minimum of 90% sensitivity was selected for the 3 genes: EBP41L3 = 0.9, DPYS = 2.7 and MAL = 0.5 and then applied to data collected in P2 (Table 3). The AUC for all three genes were reproducible in the two studies and specificity and sensitivity at the selected cut-off indicated that all three genes preformed similarly in P1 and P2 (Table 3). EPB41L3 was the best at separating CIN2/3 while MAL was the poorest classifier. Furthermore EPB41L3 showed the best ability to identify CIN2/3 in both the P1 and P2 cohort with similar AUC of 0.73 (95% CI 0.68–0.77) and 0.69 (0.65–0.73) respectively (Fig. 2).
Table 3.
Univariate analysis of the ability of three human genes to separate ≤CIN1 from CIN2/3 in HR-HPV positive samples. The number of patients with successful methylation measurement in each group and study is presented. The AUCs with 95%CI, sensitivity and specificity in P1 and P2 are compared at a cut-off selected in the P1 study to reach minimum 90% sensitivity.
|
EPB41L3 |
DPYS |
MAL |
||||
|---|---|---|---|---|---|---|
| P1 | P2 | P1 | P2 | P1 | P2 | |
| No.a of ≤CIN1 | 363 | 535 | 364 | 541 | 363 | 543 |
| No.b of CIN2/3 | 208 | 323 | 208 | 330 | 206 | 326 |
| AUC | 0.73 | 0.69 | 0.65 | 0.64 | 0.62 | 0.59 |
| 95%CI | 0.68–0.77 | 0.65–0.73 | 0.61–0.70 | 0.61–0.68 | 0.57–0.67 | 0.55–0.63 |
| Cut-off | 0.9 | 2.7 | 0.5 | |||
| Sensitivity | 90 | 85 | 90 | 82 | 90 | 94 |
| Specificity | 31 | 31 | 22 | 31 | 12 | 6 |
Total number of ≤CIN1 in P1 and P2 was 364 and 545 respectively.
Total number of CIN2/3 in P1 and P2 was 208 and 330 respectively.
Fig. 2.
The comparison of ROC curves in P1 and P2 for EPB41L3.
Discussion
The absolute methylation of 26 genes in 20 normal cervical scrapes compared to 20 scrapes from women diagnosed with CIN3 showed elevated methylation of EBP41L3, EDNRB, DPYS, CADM1, LMX1, TERT, PAX1 and MDR1 (p < 0.05) (Supplemental table 3). However, after adjustment for multiple testing in the pilot, only EPB41L3 and EDNRB remained significant. To further investigate the biomarker potential of these genes, their methylation as well as DPYS, LMX1, CADM1 and MAL was measured in specimens from the P1 study. Although methylation of MAL did not appear interesting in the initial set of 40, it was investigated in the entire P1 cohort because in combination with CADM1, a QMSP assay based on these two genes has been repeatedly shown as promising by a team in the Netherlands [7–9]. Similarly, a different group from Taiwan suggested [12] a four gene model including LMX1, SOX1, PAX1, NKX6-1 for a QMSP assay. Although we were not able to obtain an assay for NKX6-1, out of the remaining 3 genes, LMX1 showed the best separation of CIN3 from normal tissues (Supplemental table 3) and therefore was also investigated in the P1 specimens. Overall, the methylation of the 26 genes was low, with median values less than 10%. For 14 genes median methylation of less than 1% was observed (Supplemental table 3); with such low methylation values there is an increased possibility of measurement errors and these genes were not further investigated, regardless of whether or not they were previously shown to have interesting potential [13,14]. A possible explanation for the disagreement in results between our study and those in earlier reports is that the small absolute differences we saw by pyrosequencing may be increased by primer-biased QMSP assays which give data in relative terms. Another possibility is that some of the CpG sites investigated were different from those previously reported due to assay design requirements. Although in close proximity, methylation can vary in different CpGs that are part of the same CpG island [23]. DAPK1 is an example where our pyrosequencing assay was approximately 200 bp downstream of the CpG sites most commonly investigated by other methods. DAPK1 has repeatedly been suggested to have significantly altered methylation in high grade lesions in cancers [14]. Recently, this gene was proposed as a complimentary test to HPV methylation assays to provide a comprehensive classifier for identification of CIN2 + by use of the bisulfite sequencing method [24]. Although our assay did not show any differences in DAPK1 methylation (Table 1), pyrosequencing or QMSP are more likely tests than bisulfite sequencing for use in a clinical setting and it is possible that pyrosequencing assays for EPB41L3 and HPV [25] can provide a similar combination test as DAPK1 and HPV.
The absolute methylation of 26 genes in 20 normal cervical scrapes compared to 20 scrapes from women diagnosed with CIN3 showed elevated methylation of EBP41L3, EDNRB, DPYS, CADM1, LMX1, JAM3, TERT, PAX1 and MDR1 (p < 0.05) (Supplemental table 3). However, after adjustment for multiple testing in the pilot, only EPB41L3 and EDNRB remained significant. To further investigate the biomarker potential of these genes, their methylation as well as DPYS, LMX1, CADM1 and MAL was measured in specimens from the P1 study. Although methylation of MAL did not appear interesting in the initial set of 40, it was investigated in the entire P1 cohort because in combination with CADM1, a QMSP assay based on these two genes has been repeatedly shown as promising by a team in the Netherlands [7–9]. Similarly, a different group from Taiwan suggested [12] a four gene model including LMX1, SOX1, PAX1, NKX6-1 for a QMSP assay. Although we were not able to obtain an assay for NKX6-1, out of the remaining 3 genes, LMX1 showed the best separation of CIN3 from normal tissues (Supplemental table 3) and therefore was also investigated in the P1 specimens. Overall, the methylation of the 26 genes was low, with median values less than 10%. For 14 genes median methylation of less than 1% was observed in CIN3 (Supplemental table 3); with such low methylation values there is an increased possibility of measurement errors and these genes were not further investigated, regardless of whether or not they were previously shown to have interesting potential [13,14]. A possible explanation for the disagreement in results between our study and those in earlier reports is that the small absolute differences we saw by pyrosequencing may be increased by primer-biased QMSP assays which give data in relative terms. Another possibility is that some of the CpG sites investigated were different from those previously reported due to assay design requirements. Although in close proximity, methylation can vary in different CpGs that are part of the same CpG island [23]. DAPK1 is an example where our pyrosequencing assay was approximately 200 bp downstream of the CpG sites most commonly investigated by other methods. DAPK1 has repeatedly been suggested to have significantly altered methylation in high grade lesions in cancers [14]. Recently, this gene was proposed as a complimentary test to HPV methylation assays to provide a comprehensive classifier for identification of CIN2 + by use of the bisulfite sequencing method [24]. Although our assay did not show any differences in DAPK1 methylation (Supplemental Table 3), pyrosequencing or QMSP are more likely tests than bisulfite sequencing for use in a clinical setting and it is possible that pyrosequencing assays for EPB41L3 and HPV [25] can provide a similar combination test as DAPK1 and HPV.
We included testing of MAL in our study as a pre-selected gene because of the strong data by QMSP that it is a useful biomarker for CIN2/3. Although we investigated the same CpGs as in the earlier QMSP studies MAL did not show any promise as a classifier in our pyrosequencing-based methylation assay indicating important differences between the methods used and possibly also samples used. The current main weakness of the proposed CADM1 and MAL QMSP assay is that it has not been validated by different groups and in different study populations.
Our primary goal was to investigate the biomarker potential of a relatively large set of candidate human genes for pyrosequencing methylation assays to triage HR-HPV positive women. However, in P1, we also assessed the biomarker utility of the genes in HR-HPV negative women. While, all 6 investigated genes showed significantly elevated methylation in 208 CIN2/3 HR-HPV positive samples, elevated methylation was observed only for EPB41L3 and DPYS in the 10 CIN2/3 HR-HPV negative women (Fig. 1A). This suggests that measurement of methylation of some genes might be useful to identify women who are at risk of developing cervical cancer irrespective of HR-HPV status.
A combination of the collected methylation data from both P1 and P2 HR-HPV positive women showed that methylation was several fold higher in cancers for all investigated genes (Supplemental fig. S1). Although the optimal cut-off for cancer was not established for EPB41L3, this assay may be useful for cancer detection with an observed 100% sensitivity and 91% specificity versus the group of CIN2/3 plus < CIN1in the P1 specimens (data not shown). It is noteworthy that the cut-offs for classification of cancers were higher than those of CIN2/3. This demonstrates that when searching for diagnostic biomarkers able to distinguish low from high grade precursor lesions, it is reasonable to exclude the cancers from the analyses to avoid potential skewing of cut-offs, it appears that virtually all the cancers may be detected regardless of the best final cut-offs chosen to detect the CIN2/3.
A combination of the collected methylation data from both P1 and P2 HR-HPV positive women showed that methylation was several fold higher in cancers for all investigated genes (Supplemental fig. S1). Although the optimal cut-off for cancer was not established for EPB41L3, this assay may be useful for cancer detection with an observed 100% sensitivity and 91% specificity versus the group of CIN2/3 plus <CIN1 in the P1 specimens (data not shown). It is noteworthy that the cut-offs for classification of cancers were higher than those of CIN2/3. This demonstrates that when searching for diagnostic biomarkers able to distinguish low from high grade precursor lesions, it is reasonable to exclude the cancers from the analyses to avoid potential skewing of cut-offs, it appears that virtually all the cancers may be detected regardless of the best final cut-offs chosen to detect the CIN2/3.
EPB41L3, encoding the DAL-1 protein, was originally identified as a down-regulated tumor suppressor in lung adenocarcinoma [26] with later evidence emerging that its down-regulations may be attributed to promoter hypermethylation [27]. Loss of DAL-1 and its associated tumor suppressor activity have also been seen in breast [28], prostate cancer [29] and meningiomas [30]. To the best of our knowledge, there are no reports illustrating the expression of DAL-1 in cervical cancer and taking into consideration its role in other cancers as well as the aberrant methylation seen in our study and reported by others, further studies are warranted.
Elevated methylation in HPV16 L1 in CIN2/3 lesions is becoming increasingly evident [31–33]. Recently, we showed that a classifier based on methylation in HPV16 L1 and L2 may be useful for identifying women with CIN2/3 [25]. Other HPV types such as HPV18, HPV31 and HPV45 have also shown similar characteristics [24,34,35]. The feasibility of including EPB41L3 in a QMSP assay with other genes has already been shown in cervical scrapes [10] and in cervico-vaginal lavages obtained by a self-sampling [11]. The potential for success of methylation classifiers in self-collected specimens increases their value and could lead to important improvements in preventing cervical cancer deaths, particularly in less developed regions of the world.
Some limitations of our study are that we used a different assay than most prior investigators and also that we used archived specimens from women referred to colposcopy. Conversely the limitations also provide some advantages in that our results are a stronger validation of the intrinsic value of the identified biomarkers because the biomarker diagnostic potential is preferably not limited to just certain assays. Furthermore, we were able to investigate methylation in a large group of CIN2 +. Another limitation is that our study did not include an assessment of all possible CpG sites in the gene promoters which requires more powerful methods such as directed deep sequencing. With respect to other potential triage methods for HR-HPV positive women such as reflex cytology, p16 staining and HPV genotyping, our results are quite modest and the best gene EPB41L3 provided an AUC of 0.69 (Table 3) in the validation set. Therefore we anticipate the need to improve the performance of our current gene assay or perhaps to use EPB41L3 in combination with other biomarkers such as methylation of HPV16 and other HR-HPV [24,25].
In summary, the measurement of methylation of several human genes may be considered for assays to separate CIN2/3 from normal women. Methylation of EPB41L3 was low in the ≤CIN1 group, was significantly elevated in CIN2/3 and was highest in cancers; this gene was the best classifier in the set of genes evaluated here. Our pyrosequencing assay using EPB41L3 accurately detected CIN2/3 and cervical cancers in two different sets of specimens and looks suitable for further evaluation both alone and in combination with other viral and/or human genes as a potential molecular approach to triage of HR-HPV positive women.
The following are the supplementary data related to this article.
Summary of worst reviewed histology (WRH) in patients analyzed from the P1 and P2 study.
PCR assay1 and primer details; forward (F), reverse (R) and sequencing (s) primers for additionally created assays are shown giving the size and position of the amplicons as well as the number of investigated CGs for each gene.
The median methylation %, interquartile range (IQR) and area under the curve (AUC) in the pilot set of 20 normal and 20 CIN3 for each of the 26 investigated genes. Mann–Whitney U test identified 9 genes (bold) with p-values < 0.05.
Univariate LR-χ2 and multivariate LR-χ2 demonstrating the value of information added by the next gene to each logistic regression model.
Distribution of methylation stratified by histological groups: normal, CIN1, CIN2, CIN3 and cancer. A) The methylation of EPB41L3, DPYS and MAL in all analyzed HR-HPV positive samples from P1 and P2 showed highest methylation of cancers. For better visualisation of the low methylation results, 2 outliers are not shown `here, one at 70% EPB41L3 methylation in a normal tissue from P2 and one at 91% DPYS methylation in a normal tissue from P1. B) The methylation of EDNRB, LMX1 and CADM1 in HR-HPV positive samples from P1 cohort also showed highest methylation in cancers. Whiskers of the boxplot mark the 5th and 95th percentiles, the box 25th percentile, median and 75 percentile, while extreme values are shown by (•).
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ygyno.2014.02.001.
Funding
This work was funded by Cancer Research UK [grant number C569/A10404].
Conflict of interest
None to declare.
Acknowledgments
The authors thank all the participating patients. We thank Louise Cadman, Janet Austin and Anne Szarewski for providing us with the samples and collecting, updating and maintaining the P1 and P2 cohorts. Dr Anne Szarewski passed away during the final stages of this study. We are very grateful to her for the wonderful enthusiasm and the great efforts she made to ensure that studies were done to the highest level.
References
- 1.Arbyn M., Castellsague X., de Sanjose S., Bruni L., Saraiya M., Bray F. Worldwide burden of cervical cancer in 2008. Ann Oncol. 2011;22:2675–2686. doi: 10.1093/annonc/mdr015. [DOI] [PubMed] [Google Scholar]
- 2.zur Hausen H. Papillomaviruses in human cancers. Proc Assoc Am Physicians. 1999;111:581–587. doi: 10.1046/j.1525-1381.1999.99723.x. [DOI] [PubMed] [Google Scholar]
- 3.Ho G.Y., Bierman R., Beardsley L., Chang C.J., Burk R.D. Natural history of cervicovaginal papillomavirus infection in young women. N Engl J Med. 1998;338:423–428. doi: 10.1056/NEJM199802123380703. [DOI] [PubMed] [Google Scholar]
- 4.Robertson K.D. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 5.Portela A., Esteller M. Epigenetic modifications and human disease. Nat Biotechnol. 2010;28:1057–1068. doi: 10.1038/nbt.1685. [DOI] [PubMed] [Google Scholar]
- 6.Steenbergen R.D., Kramer D., Braakhuis B.J., Stern P.L., Verheijen R.H., Meijer C.J. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst. 2004;96:294–305. doi: 10.1093/jnci/djh031. [DOI] [PubMed] [Google Scholar]
- 7.Overmeer R.M., Louwers J.A., Meijer C.J., van Kemenade F.J., Hesselink A.T., Daalmeijer N.F. Combined CADM1 and MAL promoter methylation analysis to detect (pre-)malignant cervical lesions in high-risk HPV-positive women. Int J Cancer. 2010;129:2218–2225. doi: 10.1002/ijc.25890. [DOI] [PubMed] [Google Scholar]
- 8.Hesselink A.T., Heideman D.A., Steenbergen R.D., Coupe V.M., Overmeer R.M., Rijkaart D. Combined promoter methylation analysis of CADM1 and MAL: an objective triage tool for high-risk human papillomavirus DNA-positive women. Clin Cancer Res. 2011;17:2459–2465. doi: 10.1158/1078-0432.CCR-10-2548. [DOI] [PubMed] [Google Scholar]
- 9.Bierkens M., Hesselink A.T., Meijer C.J., Heideman D.A., Wisman G.B., van der Zee A.G. CADM1 and MAL promoter methylation levels in hrHPV-positive cervical scrapes increase proportional to degree and duration of underlying cervical disease. Int J Cancer. 2013;133:1293–1299. doi: 10.1002/ijc.28138. [DOI] [PubMed] [Google Scholar]
- 10.Eijsink J.J., Lendvai A., Deregowski V., Klip H.G., Verpooten G., Dehaspe L. A four-gene methylation marker panel as triage test in high-risk human papillomavirus positive patients. Int J Cancer. 2012;130:1861–1869. doi: 10.1002/ijc.26326. [DOI] [PubMed] [Google Scholar]
- 11.Eijsink J.J., Yang N., Lendvai A., Klip H.G., Volders H.H., Buikema H.J. Detection of cervical neoplasia by DNA methylation analysis in cervico-vaginal lavages, a feasibility study. Gynecol Oncol. 2011;120:280–283. doi: 10.1016/j.ygyno.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 12.Lai H.C., Lin Y.W., Huang R.L., Chung M.T., Wang H.C., Liao Y.P. Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer. 2010;116:4266–4274. doi: 10.1002/cncr.25252. [DOI] [PubMed] [Google Scholar]
- 13.Kim J.H., Choi Y.D., Lee J.S., Lee J.H., Nam J.H., Choi C. Quantitative assessment of DNA methylation for the detection of cervical neoplasia in liquid-based cytology specimens. Virchows Arch. 2010;457:35–42. doi: 10.1007/s00428-010-0936-2. [DOI] [PubMed] [Google Scholar]
- 14.Wentzensen N., Sherman M.E., Schiffman M., Wang S.S. Utility of methylation markers in cervical cancer early detection: appraisal of the state-of-the-science. Gynecol Oncol. 2009;112:293–299. doi: 10.1016/j.ygyno.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schussel J., Zhou X.C., Zhang Z., Pattani K., Bermudez F., Jean-Charles G. EDNRB and DCC salivary rinse hypermethylation has a similar performance as expert clinical examination in discrimination of oral cancer/dysplasia versus benign lesions. Clin Cancer Res. 2013;19:3268–3275. doi: 10.1158/1078-0432.CCR-12-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tahara T., Maegawa S., Chung W., Garriga J., Jelinek J., Estecio M.R., Shibata T., Hirata I., Arisawa T., Issa J.P. Examination of whole blood DNA methylation as a potential risk marker for gastric cancer. Cancer Prev Res (Phila) 2013;6:1093–1100. doi: 10.1158/1940-6207.CAPR-13-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bebek G., Bennett K.L., Funchain P., Campbell R., Seth R., Scharpf J. Microbiomic subprofiles and MDR1 promoter methylation in head and neck squamous cell carcinoma. Hum Mol Genet. 2012;21:1557–1565. doi: 10.1093/hmg/ddr593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007;373:15–24. doi: 10.1385/1-59745-377-3:15. [DOI] [PubMed] [Google Scholar]
- 19.Szarewski A., Ambroisine L., Cadman L., Austin J., Ho L., Terry G. Comparison of predictors for high-grade cervical intraepithelial neoplasia in women with abnormal smears. Cancer Epidemiol Biomarkers Prev. 2008;17:3033–3042. doi: 10.1158/1055-9965.EPI-08-0508. [DOI] [PubMed] [Google Scholar]
- 20.Szarewski A., Mesher D., Cadman L., Austin J., Ashdown-Barr L., Ho L. Comparison of seven tests for high-grade cervical intraepithelial neoplasia in women with abnormal smears: the Predictors 2 study. J Clin Microbiol. 2012;50:1867–1873. doi: 10.1128/JCM.00181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vasiljevic N., Wu K., Brentnall A.R., Kim D.C., Thorat M., Kudahetti S.C. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Dis Markers. 2011;30:151–161. doi: 10.3233/DMA-2011-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 23.Vasiljevic N., Ahmad A.S., Beesley C., Thorat M.A., Fisher G., Berney D.M. Association between DNA methylation of HSPB1 and death in low Gleason score prostate cancer. Prostate Cancer Prostatic Dis. 2012;16:35–40. doi: 10.1038/pcan.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalantari M., Osann K., Calleja-Macias I.E., Kim S., Yan B., Jordan S. Methylation of human papillomavirus 16, 18, 31, and 45L2 and L1 genes and the cellular DAPK gene: considerations for use as biomarkers of the progression of cervical neoplasia. Virology. 2014;448:314–321. doi: 10.1016/j.virol.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorincz A.T., Brentnall A.R., Vasiljevic N., Scibior-Bentkowska D., Castanon A., Fiander A. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int J Cancer. 2013;133:637–644. doi: 10.1002/ijc.28050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran Y.K., Bogler O., Gorse K.M., Wieland I., Green M.R., Newsham I.F. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res. 1999;59:35–43. [PubMed] [Google Scholar]
- 27.Kikuchi S., Yamada D., Fukami T., Masuda M., Sakurai-Yageta M., Williams Y.N. Promoter methylation of DAL-1/4.1B predicts poor prognosis in non-small cell lung cancer. Clin Cancer Res. 2005;11:2954–2961. doi: 10.1158/1078-0432.CCR-04-2206. [DOI] [PubMed] [Google Scholar]
- 28.Takahashi Y., Iwai M., Kawai T., Arakawa A., Ito T., Sakurai-Yageta M. Aberrant expression of tumor suppressors CADM1 and 4.1B in invasive lesions of primary breast cancer. Breast Cancer. 2012;19:242–252. doi: 10.1007/s12282-011-0272-7. [DOI] [PubMed] [Google Scholar]
- 29.Bernkopf D.B., Williams E.D. Potential role of EPB41L3 (protein 4.1B/Dal-1) as a target for treatment of advanced prostate cancer. Expert Opin Ther Targets. 2008;12:845–853. doi: 10.1517/14728222.12.7.845. [DOI] [PubMed] [Google Scholar]
- 30.Gutmann D.H., Donahoe J., Perry A., Lemke N., Gorse K., Kittiniyom K. Loss of DAL-1, a protein 4.1-related tumor suppressor, is an important early event in the pathogenesis of meningiomas. Hum Mol Genet. 2000;9:1495–1500. doi: 10.1093/hmg/9.10.1495. [DOI] [PubMed] [Google Scholar]
- 31.Mirabello L., Schiffman M., Ghosh A., Rodriguez A.C., Vasiljevic N., Wentzensen N. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J Cancer. 2012;132:1412–1422. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sun C., Reimers L.L., Burk R.D. Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer. Gynecol Oncol. 2011;121:59–63. doi: 10.1016/j.ygyno.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalantari M., Calleja-Macias I.E., Tewari D., Hagmar B., Lie K., Barrera-Saldana H.A. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78:12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wentzensen N., Sun C., Ghosh A., Kinney W.K., Mirabello L., Wacholder S. Methylation of HPV18, HPV31, and HPV45 genomes is associated with cervical intraepithelial neoplasia grade 3. J Natl Cancer Inst. 2012;104:1738–1749. doi: 10.1093/jnci/djs425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turan T., Kalantari M., Calleja-Macias I.E., Cubie H.A., Cuschieri K., Villa L.L. Methylation of the human papillomavirus-18L1 gene: a biomarker of neoplastic progression? Virology. 2006;349:175–183. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of worst reviewed histology (WRH) in patients analyzed from the P1 and P2 study.
PCR assay1 and primer details; forward (F), reverse (R) and sequencing (s) primers for additionally created assays are shown giving the size and position of the amplicons as well as the number of investigated CGs for each gene.
The median methylation %, interquartile range (IQR) and area under the curve (AUC) in the pilot set of 20 normal and 20 CIN3 for each of the 26 investigated genes. Mann–Whitney U test identified 9 genes (bold) with p-values < 0.05.
Univariate LR-χ2 and multivariate LR-χ2 demonstrating the value of information added by the next gene to each logistic regression model.
Distribution of methylation stratified by histological groups: normal, CIN1, CIN2, CIN3 and cancer. A) The methylation of EPB41L3, DPYS and MAL in all analyzed HR-HPV positive samples from P1 and P2 showed highest methylation of cancers. For better visualisation of the low methylation results, 2 outliers are not shown `here, one at 70% EPB41L3 methylation in a normal tissue from P2 and one at 91% DPYS methylation in a normal tissue from P1. B) The methylation of EDNRB, LMX1 and CADM1 in HR-HPV positive samples from P1 cohort also showed highest methylation in cancers. Whiskers of the boxplot mark the 5th and 95th percentiles, the box 25th percentile, median and 75 percentile, while extreme values are shown by (•).