Abstract
The nature of the gain-of-function toxicity found in polyglutamine (polyQ) diseases has been the subject of considerable debate. Duvick et al. (2010) and Nedelsky et al. (2010) now show that in two of these diseases, pathology is mediated by normal protein activity.
While many neurodegenerative diseases have been linked to causative proteins, controversy remains. When does a protein contribute to disease through gain of function (GOF), and when is it via loss of function (LOF)? While the interpretation (if not the molecular details) of a pathological LOF is generally straight-forward, the etiology behind a GOF disease can be more obscure. A GOF can be a novel property that emerges in a disease-associated protein, or alternatively, it might result from an enhancement of one or more of the protein’s normal functions to toxic levels.
Monogenic diseases offer tractable models to examine the nature of a GOF. Two papers in this issue of Neuron seek to do just this by exploring in vivo models of two poly-glutamine (polyQ) diseases. PolyQ diseases include at least nine neurodegenerative diseases caused by a pathological expansion of CAG repeats in the coding regions of different genes (Orr and Zoghbi, 2007). Despite widespread expression, these diseases vary remarkably in the susceptibility of specific brain and spinal cord regions. Together, they represent the most common cause of inherited neurodegenerative disease.
Since all are caused by a glutamine expansion and result in similar protein aggregates, direct toxicity of the polyQ stretch has been hypothesized as a common mechanism of disease. In animal models, a polyQ stretch is sufficient for pathology and aggregation when inserted into a larger protein (Ordway et al., 1997) or as an independent peptide (Marsh et al., 2000). In this model, the rest of the protein influences only the details—perhaps the where and the when—of the death and dysfunction.
More recent studies, however, have shown that the protein context is critical. In fact, in some cases polyQ expansion by itself is not sufficient for disease. For instance, phosporylation at serine 421 (Humbert et al., 2002) or serines 13 and 16 (Gu et al., 2009) of the protein that causes Huntington’s disease (HD) abrogates its toxicity. This connection between host protein context and toxicity suggests that the normal function of the protein may be linked to disease. Work by Nedelsky and colleagues (2010) on spinal bulbar muscular atrophy (SBMA) in this issue of Neuron suggests just that.
SBMA, also known as Kennedy’s disease, is an X-linked neurodegenerative disease characterized by late-onset muscle weakness and wasting. Pathology is notable for degeneration of motor neurons in the brainstem and spinal cord (Orr and Zoghbi, 2007). The polyQ expansion in SBMA occurs in the androgen receptor (AR), a classic nuclear hormone receptor. Accordingly, the AR binds its ligand, testosterone or dihydroxytestosterone, in the cytosol and then translocates to the nucleus to affect gene expression. This makes SBMA one of the few polyQ diseases in which the function of the host protein is well understood.
Nedelsky et al. (2010) combine existing knowledge about the AR with the power of fly genetics to hone in on the source of polyQ toxicity. The authors expressed the human AR with wild-type (12Q) or pathogenic (52Q) glutamine stretches in the adult Drosophila eye or larval salivary glands and motor neurons. Specific point mutations then allowed a systematic exploration of the conditions necessary for toxicity. In agreement with previous findings, ligand binding and nuclear translocation were necessary to evoke the degenerative phenotype (Katsuno et al., 2002; Takeyama et al., 2002). However, nuclear translocation alone was insufficient; DNA binding was necessary to cause degeneration. Thus, the first steps of normal AR function are necessary for toxicity.
After DNA binding, the AR normally recruits various transcriptional coregulators via its AF-1 and AF-2 domains. Preventing normal AF-2 interaction with coregulators rescued the toxicity of mutant AR, suggesting that normal AR-coregulator interactions are necessary for disease. If this is true, then altering the levels of coregulators should also affect the phenotype. As expected, an RNAi screen of 61 orthologues of human AR coregulators identified 19 that modify degeneration. Finally, as predicted, overexpression or knock-down of a specific coregulator known to bind at the AF-2 domain significantly affected the degenerative phenotype. The authors conclude that the polyQ expansion in the AR mediates its toxicity via normal AR interactions. Which interactions are most important for toxicity and their pathogenic mechanism(s) remain unresolved.
How might normal interactions cause toxicity? Two observations in the report suggest that toxicity is caused by enhanced AR activity. First, high levels of expression of wild-type AR showed a similar, albeit more minor, degenerative phenotype as mutant AR. This implies that high levels of normal AR activity, even in the absence of the polyQ expansion, are sufficient to cause degeneration. Second, gene expression profiling showed that expression of the wild-type AR yielded changes similar to those evoked by the mutant AR but to a lesser degree. The result hints that high levels of wild-type activity can phenocopy the degeneration caused by the mutant protein. Furthermore, the similarity in gene expression changes argues that the mild degenerative phenotype seen in flies expressing wild-type AR at high levels is not due to a non-specific toxicity of overexpression. These data implicate polyQ-mediated enhanced activity of wild-type function as the source of toxicity in SBMA.
Independent work in this issue by Duvick et al. (2010) on the polyQ disease spinocerebellar ataxia type 1 (SCA1) arrives at a similar conclusion. SCA1 is caused by a polyQ-expanded version of the protein ataxin-1 and usually presents in middle age as progressive ataxia, tremor, and dysarthria. Pathology is characterized by marked cerebellar atrophy with Purkinje cell death (Orr and Zoghbi, 2007). Protein context is also important in SCA1: phosphorylation at serine 776 of ataxin-1 is necessary for toxicity (Emamian et al., 2003). Apparently, mutant ataxin-1 alters the balance of protein-protein interactions between itself and two normal binding partners, but abolishing phosphorylation at S776 restores the wild-type equilibrium (Lim et al., 2008). Intriguingly, mimicking tonic phosphorylation by mutating serine to aspartic acid (S776D) causes wild-type ataxin-1 to interact with these binding partners as if it were polyQ expanded.
Duvick and colleagues (2010) follow up on this finding that wild-type ataxin-1-S776D adopts a biochemical attribute of mutant ataxin-1 to determine if it can evoke dysfunction and pathology in vivo. They generated transgenic mice expressing wild-type (30Q) or mutant (82Q) ataxin-1 with the S776D mutation. For comparison, they had previously generated transgenic mice expressing wild-type or mutant ataxin-1 with serine 776 intact. All mice expressed ataxin-1 under the Pcp2 promoter, directing expression to Purkinje cells. All appropriate Q-length control S776 mice express the transgenic protein at levels at least as high as those found in the S776D mice. Thus, any toxicity enhanced by the S776D mutation cannot be explained by grossly higher expression levels.
Mice expressing ataxin-1-82Q demonstrated behavioral and histological pathology (Orr and Zoghbi, 2007). Furthermore, the ataxin-1-82Q-S776D mice displayed similar pathology despite significantly less transgene expression, reflecting the enhanced toxicity of the mimicked tonic phosphorylation. The surprise came with analysis of the respective wild-type ataxin-1-30Q mouse lines. The ataxin-1-30Q-S776D cohort showed atrophy of Pukinje cell dendritic branches and a retraction of climbing fiber-Purkinje cell synapses, similar to that seen in those mice expressing mutant ataxin-1. The rotarod confirmed a progressive functional deficit. Hence, pathology caused by an expansion of a polyQ stretch was evoked in mice by the same protein with a non-mutant polyQ stretch if the phosphorylation site was mutated. It will be interesting to determine in the future if polyQ expansion facilitates phosphorylation at S776 in vivo.
If polyQ expansion is not necessary for ataxin-1 toxicity, then protein context is clearly critical in polyQ disease, and the wild-type functions of ataxin-1 alone might be sufficient for disease. Along these lines, mice expressing ataxin-1-30Q with no alteration at S776—the line coincidentally with the highest transgene expression level—showed evidence of pathology by immunohistochemistry. This suggests that excessive levels of normal ataxin-1 activity represent the GOF caused by polyQ expansion and argues against a novel property as the source of the GOF.
Thus, two reports from two independent groups on two separate polyQ diseases conclude that normal function of the host protein is crucial to disease pathogenesis. Furthermore, they implicate excessive levels of normal function as the GOF that causes toxicity. Are there any reasons to worry that the specific features of the model systems used in these studies might not fully reflect the mechanisms at play in the human conditions?
If a model is to demonstrate that toxicity is due to wild-type functions, then the protein must be able to execute them. The AR has no direct orthologue in flies, so its interactions are occurring in a non-native setting. However, the AF-1 and AF-2 domains are conserved in other native nuclear hormone receptors in Drosophila, and 61 of the 73 known human nuclear hormone coregulators have orthologues in the fly. It is also comforting that conclusions from fly models of SBMA have broadly agreed with those from mouse models (Katsuno et al., 2002; Takeyama et al., 2002).
Another point worth mentioning is that the study of HD has led to increasing evidence of contributions to disease from cell types other than neurons expressing the polyQ expanded protein (Bradford et al., 2009). If similar contributions are made in other polyQ diseases, then they might not be detected in the Drosophila eye or in a mouse in which transgene expression is restricted to one cell type. Any developmental contribution to disease, as has been suggested in the case of SCA1 (Orr and Zoghbi, 2007), might similarly be difficult to study.
Evidence for other causes of toxicity in these diseases has been published, including LOF. In SBMA, for example, patients sometimes experience gynecomastia and hypogonadism, clear signs of mild androgen insensitivity. On the other hand, frank feminization is not observed, and loss of the AR does not cause neurodegeneration (Orr and Zoghbi, 2007), seemingly excluding complete LOF as the sole cause of toxicity. A mixed GOF and LOF picture for SBMA may ultimately best explain patient phenotypes, consistent with cell-culture analysis that shows mutant AR expression leading to a combination of up- and downregulation of its target genes (Lieberman et al., 2002). Further study of transcripts from human samples should be helpful in clarifying any GOF and LOF changes, respectively.
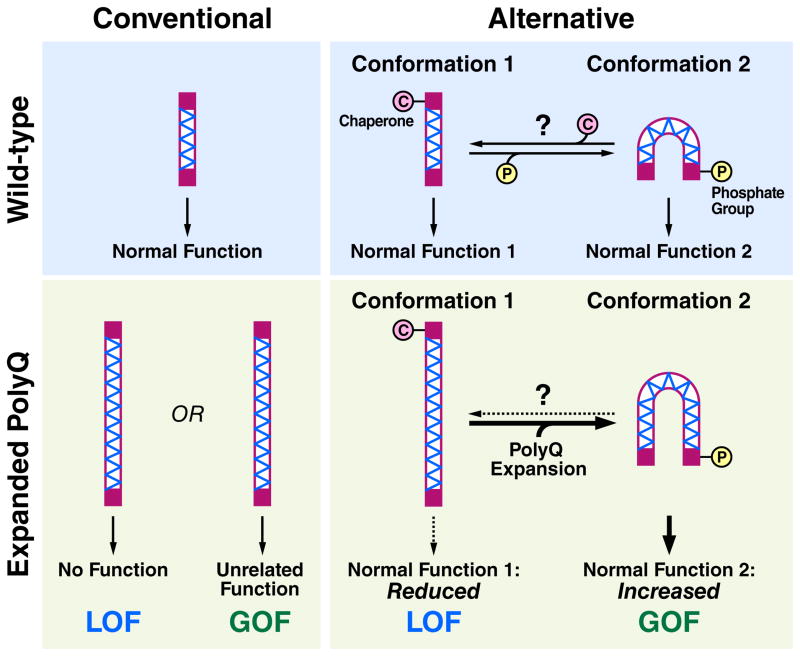
The studies discussed here provide clues as to how polyQ diseases could be explained by a dominant GOF together with a role for LOF (Fig. 1). In the wild-type setting, the host protein might adopt one of several conformations, depending on specific post-translational modifications, such as phosphorylation. Different conformations might, in turn, be associated with particular functions. In the disease setting, polyQ expansion could stabilize certain normal conformations of the host protein at the expense of others. That is, the wild-type equilibrium of protein conformations would be disrupted so that functions associated with preferred conformations are enhanced, whereas the remainder would be deficient.
Figure 1. Conventional and alternative hypotheses on the role of an expanded polyglutamine (polyQ) stretch in disease.
In the conventional view, a protein with a polyQ stretch (hatched region) has a specific normal function. Expansion of the polyQ stretch causes disease due to an unrelated toxic gain of function (GOF) or a loss of normal function (LOF). In an alternative view, the protein exists in an equilibrium of distinct conformations, some associated with particular and distinct normal functions. The equilibrium could be influenced in multiple ways, including chaperone binding (pink circle) and post-translational modifications, such as phosphorylation (yellow circle). With polyQ expansion, the equilibrium is disturbed, leading to GOF associated with enhanced function of the stabilized conformer. There is also LOF due to decreased function associated with the less preferred conformer. The polyQ expansion could affect the equilibrium by stabilizing one of the conformers directly or by promoting an interaction with a conformation-specific binding partner (not depicted), or indirectly by reducing chaperone availability through stress on the proteostasis machinery.
PolyQ expansions are also known to place an extra burden on a cell’s protein folding machinery (Powers et al., 2009). Disease might occur because the burden disrupts proper folding of other metastable proteins, contributing to their LOF. However, it is possible to imagine that the burden on the chaperone system could cause disease by enhancing a normal function of the instigating polyQ protein. If certain conformations of the polyQ protein require or are recognized by chaperones, then the reduced availability of chaperones could shift the distribution of its conformers towards those that do not require chaperones (Fig. 1). The increase in the function that corresponds to that conformer would represent a GOF linked to the protein’s normal function. Together, these changes might bring about cell dysfunction and eventual death.
The work discussed here also opens the door to new therapeutic opportunities. There are no effective treatments for the polyQ diseases, and the seemingly promising approach of chemical castration for SBMA failed a recent clinical trial (Katsuno et al., 2010). These studies suggest that targeting specific native protein functions may be required for effective treatment. Indeed, for the polyQ diseases and perhaps other neurodegenerative diseases, too much of even normal function may be a fundamental cause of toxicity.
References Cited
- Bradford J, Shin JY, Roberts M, Wang CE, Li XJ, Li S. Proc Natl Acad Sci U S A. 2009;106(52):22480–5. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvick L, Barnes J, Ebner B, Agrawal S, Andresen M, Lim J, Giesler GJ, Zoghbi HY, Orr HT. Neuron. 2010 doi: 10.1016/j.neuron.2010.08.022. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Kaytor MD, Duvick LA, Zu T, Tousey SK, Zoghbi HY, Clark HB, Orr HT. Neuron. 2003;38(3):375–87. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- Gu X, Greiner ER, Mishra R, Kodali R, Osmand A, Finkbeiner S, Steffan JS, Thompson LM, Wetzel R, Yang XW. Neuron. 2009;64(6):828–40. doi: 10.1016/j.neuron.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert S, Bryson EA, Cordelieres FP, Connors NC, Datta SR, Finkbeiner S, Greenberg ME, Saudou F. Dev Cell. 2002;2(6):831–7. doi: 10.1016/s1534-5807(02)00188-0. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Adachi H, Kume A, Li M, Nakagomi Y, Niwa H, Sang C, Kobayashi Y, Doyu M, Sobue G. Neuron. 2002;35(5):843–54. doi: 10.1016/s0896-6273(02)00834-6. [DOI] [PubMed] [Google Scholar]
- Katsuno M, Banno H, Suzuki K, Takeuchi Y, Kawashima M, Yabe I, Sasaki H, Aoki M, Morita M, Nakano I, Kanai K, Ito S, Ishikawa K, Mizusawa H, Yamamoto T, Tsuji S, Hasegawa K, Shimohata T, Nishizawa M, Miyajima H, Kanda F, Watanabe Y, Nakashima K, Tsujino A, Yamashita T, Uchino M, Fujimoto Y, Tanaka F, Sobue G. Lancet Neurol. 2010;9(9):875–84. doi: 10.1016/S1474-4422(10)70182-4. [DOI] [PubMed] [Google Scholar]
- Lieberman AP, Harmison G, Strand AD, Olson JM, Fischbeck KH. Hum Mol Genet. 2002;11(17):1967–76. doi: 10.1093/hmg/11.17.1967. [DOI] [PubMed] [Google Scholar]
- Lim J, Crespo-Barreto J, Jafar-Nejad P, Bowman AB, Richman R, Hill DE, Orr HT, Zoghbi HY. Nature. 2008;452(7188):713–8. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh JL, Walker H, Theisen H, Zhu YZ, Fielder T, Purcell J, Thompson LM. Hum Mol Genet. 2000;9(1):13–25. doi: 10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- Nedelsky NB, Pennuto M, Smith RB, Palazzolo I, Moore J, Nie Z, Neale G, Taylor JP. Neuron. 2010 doi: 10.1016/j.neuron.2010.08.034. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordway JM, Tallaksen-Greene S, Gutekunst CA, Bernstein EM, Cearley JA, Wiener HW, St Dure L, Lindsey R, Hersch SM, Jope RS, Albin RL, Detloff PJ. Cell. 1997;91(6):753–63. doi: 10.1016/s0092-8674(00)80464-x. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Powers ET, Morimoto RI, Dillin A, Kelly JW, Balch WE. Annu Rev Biochem. 2009;78:959–91. doi: 10.1146/annurev.biochem.052308.114844. [DOI] [PubMed] [Google Scholar]
- Takeyama K, Ito S, Yamamoto A, Tanimoto H, Furutani T, Kanuka H, Miura M, Tabata T, Kato S. Neuron. 2002;35(5):855–64. doi: 10.1016/s0896-6273(02)00875-9. [DOI] [PubMed] [Google Scholar]