Abstract
A novel carbonyl reductase (AcCR) catalyzing the asymmetric reduction of ketones to enantiopure alcohols with anti-Prelog stereoselectivity was found in Acetobacter sp. CCTCC M209061 and enriched 27.5-fold with an overall yield of 0.4% by purification. The enzyme showed a homotetrameric structure with an apparent molecular mass of 104 kDa and each subunit of 27 kDa. The gene sequence of AcCR was cloned and sequenced, and a 762 bp gene fragment was obtained. Either NAD(H) or NADP(H) can be used as coenzyme. For the reduction of 4′-chloroacetophenone, the Km value for NADH was around 25-fold greater than that for NADPH (0.66 mM vs 0.026 mM), showing that AcCR preferred NADPH over NADH. However, when NADH was used as cofactor, the response of AcCR activity to increasing concentration of 4′-chloroacetophenone was clearly sigmoidal with a Hill coefficient of 3.1, suggesting that the enzyme might possess four substrate-binding sites cooperating with each other The Vmax value for NADH-linked reduction was higher than that for NADPH-linked reduction (0.21 mM/min vs 0.17 mM/min). For the oxidation of isopropanol, the similar enzymological properties of AcCR were found using NAD+ or NADP+ as cofactor. Furthermore, a broad range of ketones such as aryl ketones, α-ketoesters and aliphatic ketones could be enantioselectively reduced into the corresponding chiral alcohols by this enzyme with high activity.
Introduction
Enantiopure alcohols are widely used as building blocks in pharmaceutical, agrochemical, flavor, and functional material industries. The “green” synthesis catalyzed by enzymes, microorganisms and plant cells for the production of various enantiopure alcohols has attracted considerable attention due to its high efficiency, high enantioselectivity, mild reaction condition, and low environmental pollution [1], [2]. Carbonyl reductase is one of the attractive biocatalysts for synthesis of enantiopure alcohols [3], and widely expressed in various organisms from microorganisms to mammals such as Bacillus [4], Neurospora crassa [5] and human brain [6]. Carbonyl reductases belong to the oxidoreductase family that often need cofactors such as NAD(H) or NADP(H) to be functionally active, which could be divided into three subtypes based on the coenzyme requirements: (1) NAD(H)-specific, like carbonyl reductase originated from Candida viswanathii [7]; (2) NADP(H)-specific, such as carbonyl reductase from Neurospora crassa [5]; (3) dual coenzyme specific, like carbonyl reductase from Candida parapsilosis [8].
The substrate specificity of an enzyme is largely determined by the structure of substrate-binding site and the general plasticity of this region. Most carbonyl reductases often have more than one type of substrates. For example, the carbonyl reductase from Neurospora crassa could catalyze asymmetric reduction of ketones, diketones, α-keto esters, and β-keto esters [5] and the carbonyl reductase from Streptomyces coelicolor could be used for synthesis of chiral alcohols from aryl ketones, α-keto esters, and β-keto esters with high enantioselectivity [9].
Obviously, carbonyl reductases with good catalytic efficiency and excellent stereoselectivity can improve the product yield and simplify the purification processes. Some carbonyl reductases with high enantioselectivity have been reported. For example, the carbonyl reductases from Kluyveromyces thermotolerans [10] and Candida glabrata [11] were able to catalyze the asymmetric reduction of methyl o-chlorobenzoylformate to methyl (R)-o-chloromandelate with high enantioselectivity (99% e.e.). Combined with the effective cofactor regeneration systems, the carbonyl reductases showed great potential for the industrial application.
In the present study, the discovery, purification, and characterization of a novel carbonyl reductase from Acetobacter sp. CCTCC M209061 (AcCR) have been described. It was found that AcCR was capable of catalyzing the anti-Prelog asymmetric reduction of various types of ketones with excellent enantioselectivity. Furthermore, the potential of AcCR acted as a robust industrial biocatalyst was examined for efficient synthesis of several pharmaceutically important chiral alcohol intermediates.
Materials and Methods
Chemicals
2′-Methoxyacetophenone (99% purity), 3′-methoxyacetophenone (97% purity), 4′-methoxyacetophenone (99% purity), (R)-1-(4-fluorophenyl)ethanol (97% purity), (R)-1-(4-chlorophenyl)ethanol (95% purity), 4′-bromoacetophenone (98% purity), (R)-1-(4-bromophenyl)ethanol (95% purity), (S)-1-(4-bromophenyl)ethanol (95% purity), 4′-methylacetophenone(96% purity), methyl acetoacetate (99% purity), methyl (R)-3-hydroxybutyrate (99% purity), methyl (S)-3-hydroxybutyrate (99% purity), ethyl 3-hydroxybutyrate (98% purity), ethyl (R)-3-hydroxybutyrate (98% purity), 3,3-dimethyl-2-butanone (97% purity), 4-(trimethylsilyl)-3-butyn-2-one (97% purity), 4-(trimethylsilyl)-3-butyn-2-ol (97% purity), 1-(trimethylsilyl)ethanone (97% purity) and 1-(trimethylsilyl)ethanol (98% purity) were purchased from Sigma–Aldrich (St Louis, MO, USA). 1-(2-Methoxylphenyl) ethanol (99% purity), 1-(3-methoxylphenyl) ethanol (97% purity) and 1-(4-nitrophenyl))ethanol (95% purity), were purchased from J&K Scientific Ltd (Beijing, China). 1-(4-Methoxylphenyl)ethanol (95% purity), 4′-fluoroacetophenone (99% purity), 1-(4-fluorophenyl)ethanol (98% purity), 4′-chloroacetophenone (98% purity), 1-(4-chlorophenyl)ethanol (97% purity), 4′-nitroacetophenone (98% purity), 1-(4-methylphenyl)ethanol (97% purity), 3,3-dimethyl-2-butanol (99% purity), n-decane (99% purity), 2-pentanone (99% purity), 2-pentanol (99% purity), (S)-2-pentanol (97% purity), 2-octanone (98% purity), 2-octanol (98% purity) and (R)-2-octanol (99% purity) were purchased from Alfa Aesar (Ward Hill, MA, USA). Coenzymes, glucose-6-phosphate (G-6-P) and glucose-6-phosphate dehydrogenase (GPDH) were purchased from Qiyun Biotechnology Company (Guangzhou, China). All other reagents and solvents were of analytical grade and used without further purification.
Microorganism and Growth
Acetobacter sp. CCTCC M209061 was previously isolated from Chinese kefir grains [12] and kept in our laboratory at −80°C. It was cultivated and harvested as described previously [13].
Purification of Carbonyl Reductase
After harvesting, 10 g of wet cells were re-suspended in 30 ml buffer A (20 mM sodium phosphate, pH 6.5, 0.5 mM phenylmethanesulfonylfluoride, 1.0 mM β-mercaptoethanol, 0.5 mM MnSO4), and disintegrated by 40 min sonication at 4°C. Cell debris was removed by centrifugation (12,000×g) at 4°C for 15 min, and the supernatant (crude extract enzyme) was mixed with ammonium sulfate (30% saturation). After centrifugation (12,000×g at 4°C for 15 min), the concentration of ammonium sulfate in supernatant was increased to 80% saturation under which the crude enzyme was precipitated and collected by centrifugation (12,000×g) at 4°C for 15 min. After dialysis against buffer A overnight, the crude enzyme (around 70 mg protein) was loaded on a pre-equilibrated DEAE-Sepharose column (GE Healthcare, Uppsala, Sweden) and purified on an ÄKTA Purifier 100 system (GE Healthcare, Piscataway, NJ, USA) at 4°C. The elution was carried out with increasing concentration of NaCl (0, 0.1, 0.2, 0.5 and 1.0 M) in buffer B (20 mM sodium phosphate, pH 6.5, 0.5 mM MnSO4). The active fraction was eluted by buffer B with 0.2 M NaCl, pooled and concentrated by ultrafiltration with Centriprep YM-10 from Millipore (Bedford, MA, USA). Then Native-PAGE was performed according to the procedure reported by Chen et al [14]. The separating and stacking gels contained 9% and 5% acrylamide, respectively. The electrophoresis was performed at 4°C and the subsequent experiment was performed as described by Chen et al [14]. The target segment was chopped into small pieces, transferred into a dialysis bag with a cutoff molecular weight of 11 kDa and electrophoresed for 10 min to recollect the enzyme. The purified enzyme solution was subsequently desalted and concentrated by ultrafiltration using Centriprep YM-10.
Molecular Mass Determination
The purified enzyme was loaded on a Superdex G-200 column (1.0 cm ×30 cm, GE Healthcare, Piscataway, NJ, USA) and the apparent molecular mass was determined by gel permeation chromatography with the mobile phase of phosphate buffer (50 mM, pH 7.0) containing 0.15 M NaCl at a flow rate of 0.2 ml/min. The column was pre-calibrated with standard molecular mass markers including ovalbumin (43.0 kDa), conalbumin (75.0 kDa), aldolase (158.0 kDa), ferritin (440.0 kDa), and thyroglobulin (669.0 kDa).
SDS-PAGE was conducted according to the procedure reported by Laemmli [15]. The subunit molecular mass was determined by the RF values. The calibration curve was obtained using standard low molecular mass markers including lysozyme (14.3 kDa), trypsin inhibitor (20.1 kDa), carbonic anhydrase (29.0 kDa), ovalbumin (44.3 kDa), bovine serum albumin (66.4 kDa), and phosphorylase b (97.2 kDa).
Enzyme Activity Assays
The initial rate of AcCR-catalyzed reduction of ketone was determined by monitoring the change in absorbance at 340 nm over 1 min on a Shimadzu UV-3010 spectrophotometer (Shimadzu, Japan). The NADH- and NADPH-based reductions were carried out at 25°C in 0.25 ml phosphate-citrate buffer (50 mM, pH 5.0 or 7.5). One unit (U) of AcCR was defined as the amount of enzyme that catalyzed the conversion of 1 µmol NADH or NADPH per minute.
Since there were other NADH- and NADPH-oxidases in crude enzyme preparations (crude extract and ammonium sulfate precipitate), the change in the absorbance of NADH or NADPH at 340 nm might not be only caused by the AcCR-mediated reduction reaction. Therefore, the conversion of 4′-chloroacetophenone was determined by gas chromatography (GC). Reactions were carried out with 30 mM NADH or NADPH and 5 mM 4′-chloroacetophenone at 25°C in 0.25 ml phosphate-citrate buffer (50 mM, pH 5.0 or 7.5). One unit (U) of AcCR was defined as the amount of enzyme that catalyzed the conversion of 1 µmol 4′-chloroacetophenone per min.
The NAD+- and NADP+-linked oxidations were conducted with NAD+ or NADP+ and isopropanol at 35°C in 0.25 ml of sodium phosphate buffer (50 mM, pH 8.0). One unit (U) of AcCR was defined as the amount of enzyme that catalyzed the production of 1 µmol NADH or NADPH per min.
Protein contents were determined by the method of Bradford [16].
Effects of pH and Temperature
The pH dependence of enzyme activity was studied at various pHs (pH 4.0–9.5) using 50 mM citrate–phosphate (pH 4.0–8.0), Tris-HCl (pH 7.5–8.5) and glycine–NaOH (pH 8.6–9.5) as the buffer, respectively. The effect of temperature on enzyme activity was investigated at varied temperatures from 10 to 45°C. The pH stability of the purified enzyme was assessed by determining the enzyme activity after 5 days incubation at 4°C in buffers with various pHs. The thermal stability of the enzyme was assessed after 5 h incubation at varied temperatures from 15 to 40°C. The enzyme activity was measured as described above.
Effects of Various Additives
The effects of metal ions, metal ion chelators, and thiol reagents on the catalytic activity of AcCR were determined by pre-incubating the enzyme with various additives in phosphate-citrate buffer (50 mM, pH 5.0) at 25°C for 15 min. The enzyme activity was then measured as described above. The relative activity was calculated as the percentage of the activity of the enzyme observed without the examined additives.
Substrate Specificity
The activity of AcCR for each specific substrate was determined by the method described above using NADH as the cofactor. The relative activity towards 4′-chloroacetophenone was defined as 100%.
Bioconversion of Various Ketones
The reductions of ketones were performed in a 10-ml Erlenmeyer flask with 2 ml phosphate-citrate buffer (50 mM, pH 5.0) containing 30 mM NADH and 5 mM ketones. The flask was capped with a septum and pre-incubated in an incubator with shaking at 25°C for 10 min. The purified AcCR was added to initiate the reaction. Samples (50 µl) were withdrawn at specified time intervals. The product and the residual substrate were extracted twice with 100 µl isopropyl ether (aryl ketones) or n-hexane (other ketones) containing 5 mM n-decane (as an internal standard) prior to GC analysis.
Kinetic Parameters Assays
The initial reaction rates were determined under the optimum conditions. For the reduction of 4′-chloroacetophenone, NADH or NADPH concentrations varied from 0.04 to 1.50 mM or 0.02 to 0.40 mM, and 4′-chloroacetophenone concentrations varied from 1.5 to 20.0 mM. For the oxidation of isopropanol, NAD+ and NADP+ concentrations varied from 0.2 to 20.0 mM, and isopropanol concentrations varied from 13 to 208 mM. All measurements were performed in triplicate. Michaelis-Menten or Hill equation was used to fit the data (initial reaction rate vs. substrate concentration), and the kinetic parameters of AcCR-catalyzed reduction and oxidation reactions, Km and Vmax values, were obtained from the fit.
Supply of Cofactors
The reactions were carried out in 0.25 ml of 50 mM phosphate-citrate buffer at pH 5.0 (NADH acted as cofactor) or pH 7.5 (NADPH acted as cofactor) containing 5 mM 4′-chloroacetophenone, 10 mM 2-octanone, or 15 mM ethyl acetoacetate. The cofactor was supplied by three methods: (1) direct addition of 30 mM NADH or NADPH; (2) addition of 30 mM G-6-P, 0.2 U/ml GPDH and 0.2 mM NADPH; (3) 130 mM isopropanol and 0.2 mM or 1 mM NADH/NADPH. Samples (50 µl) were withdrawn at specified time intervals. The product and the residual substrate were extracted twice for GC analysis.
In addition, using G-6-P/GPDH system to regenerate NADPH, the reactions were carried out in 25 ml of sodium phosphate buffer (50 mM, pH 7.5). When the reaction was completed with no further product formation (2–4 h), the aqueous layer was extracted three times with 10 ml of isopropyl ether for 4′-chloroacetophenone or ethyl ester for 2-octanone and ethyl acetoacetate. The organic layers were combined and dried by Na2SO4, and analyzed by GC and Polarimeter SGW-2.
Analytical Methods
The organic extracts of reaction mixtures were analyzed with a GC 2010 g from Shimadzu Corp. (Kyoto, Japan) equipped with chiral columns {HP Chiral 20% and CP-ChiraSil-DEX CB from J&W Scientific (Folsom, CA, USA) and HP Chiral 10% from Hewlett Packard (USA)} and a flame ionization detector. Nitrogen was used as the carrier gas. The split ratio was 1∶100 (v/v). The injector and the detector temperatures were both set at 250°C. The detailed methods were summarized in Table S1, and the R- or S-configuration of the formed product (chiral alcohol) was determined by comparison with the reference compound.
The [α]D of the product was measured by Polarimeter SGW-2. The [α]D of (R)-1-(4-chlorophenyl)ethanol, ethyl (R)-3-hydroxybutyrate, and (R)-2-octanol were [α]24.3 D +49.4° (C = 0.32, isopropyl ether), [α]24.3 D - 45.4° (C = 0.17, ethyl ester), and [α]24.3 D - 11.5° (C = 0.23, ethyl ester), respectively.
Hunting for AcCR gene sequence from genome of Acetobacter sp. CCTCC M209061
Genomic DNA of Acetobacter sp. CCTCC M209061 was extracted and purified using a bacterial genomic DNA Kit (Generay, Shanghai, China). Oligonucleotide primers were designed according to the published gene sequence of oxidoreductase from Acetobacter pasteurianus 386B (Sequence ID: ref|YP_008391729.1). Primer 1: 5′-ATGGCACGTGTAGCAGGCAAGGTT-3′; primer 2: 5′-TTATTGTGCAGTGTACCCACCATCAAT-3′. The DNA fragment of AcCR was amplified by polymerase chain reaction (PCR) using the KOD DNA polymerase (TOYOBO. Osaka, Japan). The PCR amplification was performed with 2 min of pre-denaturation at 94°C, followed by 30 cycles of 15 s at 98°C, 30 s annealing at 64°C, 40 s extension at 68°C, and a final extension of 7 min at 68°C. The PCR products were analyzed and purified using 1.2% agarose gel. After electrophoresis, the amplified AcCR DNA band was collected and sequenced. The obtained sequences were classified by comparison with those in public DNA databases.
Results and Discussion
Activity of Carbonyl Reductase over Cultivation Time
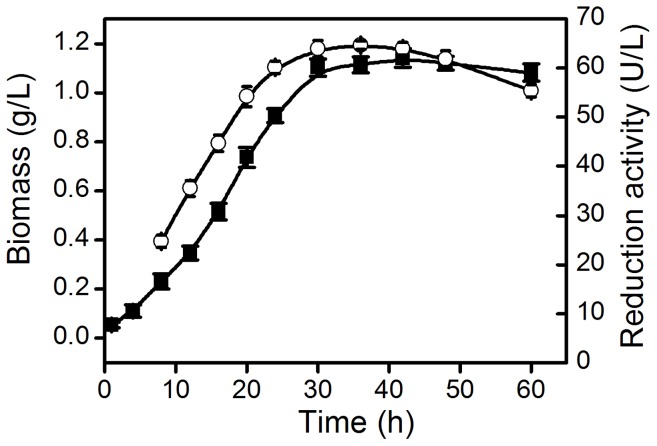
Acetobacter sp. CCTCC M209061 was cultivated and harvested under conditions reported previously [13]. The growth curve of Acetobacter sp. CCTCC M209061, and the dependence of the reduction activity of AcCR from Acetobacter sp. CCTCC M209061 cells on cultivation time were illustrated in Figure 1. As can be seen, the biomass and the total activity of AcCR remarkably increased with increasing cultivation time up to 30 h (growth phase). A further increase in cultivation time (beyond 30 h) resulted in a moderate fall in the total activity of AcCR, but led to no significant increase in the biomass. Additionally, the specific activity of AcCR (relative to the biomass) reached the highest value at about 15 h cultivation (mid-log phase) but leveled off afterwards. The total activity of AcCR stayed at a high level (above 60 U/L) between 24 and 42 h of cultivation time. Interestingly, it was found that the storage stability of AcCR from Acetobacter sp. CCTCC M209061 was dependent on the cultivation time of the cell. The activities of AcCRs from the cells collected at different cultivation times were assessed after the storage for seven days at 4°C, and the enzyme AcCR from the cells harvested between 24 and 36 h of cultivation time remained around 70–80% of its original activity, and the enzyme from the cells collected at less than 24 h or more than 36 h of cultivation time maintained approximately 50–60% of its initial activity. Obviously, the optimal cultivation time is 30 h for the reduction activity of AcCR isolated and purified from Acetobacter sp. CCTCC M209061.
Figure 1. The biomass of Acetobacter sp. CCTCC M209061 and the reduction activity of AcCR during its growing process.
Biomass (▪); Reduction activity (○).
Purification of Carbonyl Reductase
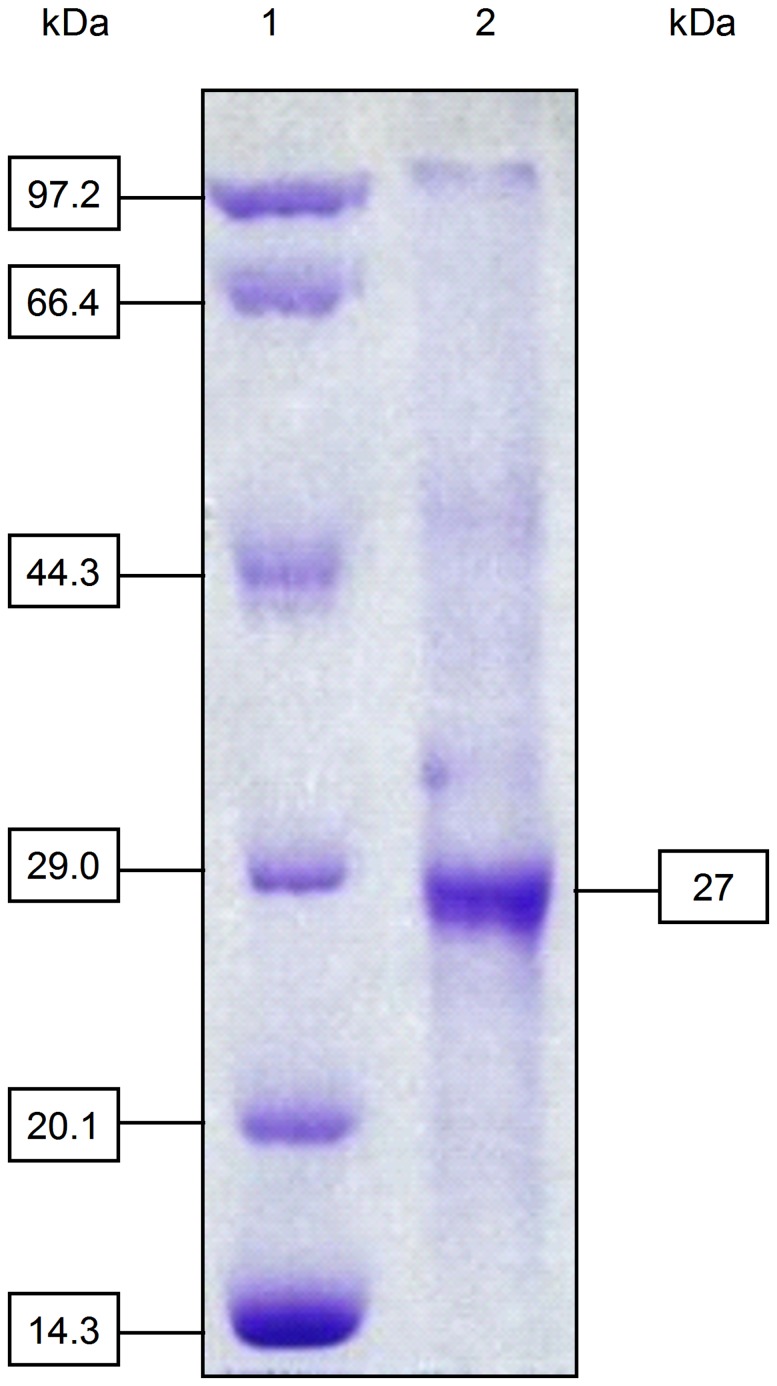
In our early attempts to purify the AcCR from Acetobacter sp. CCTCC M209061 cells, the enzyme quickly lost its activity during ion exchange chromatography or Native-PAGE, and only about 40% of activity was obtained after the precipitation with ammonium sulphate. After many purification conditions were examined, surprisingly, the addition of 0.5 mM Mn2+ to the buffer was found to effectively prevent the enzyme AcCR deactivation during the purification process, which was similar to that observed during the purification of (R)-phenylethanol dehydrogenase [17]. From the data for each purification step summarized in Table 1, the cell-free extract was precipitated with ammonium sulfate fractionation and the active enzyme protein was collected in the range of 30–80% saturation. This crude enzyme was then purified by the DEAE-Sepharose column. The active enzyme was detected in the eluent of 0.2 M NaCl with a 5.0-fold enrichment. The enzyme was concentrated and subsequently purified by Native-PAGE (Figure S1). After the final purification step, the AcCR was purified by 27.5-fold to a specific activity of 3.85 U/mg-protein with a total yield of 0.4%. The apparent molecular mass of the native enzyme was estimated to be 104 kDa by gel filtration and the molecular mass of the subunit was estimated to be 27 kDa by SDS-PAGE (Figure 2), indicating that the enzyme AcCR has a homotetrameric structure.
Table 1. Purification of AcCR from Acetobacter sp. CCTCC M209061.
| Purification step | Total protein (mg) | Total activity (U) | Specific activity (U/mg) | Yield (%) | Enrichment (fold) |
| Cell-free extract | 757.8 | 105.9 | 0.14 | 100.0 | 1.0 |
| (NH4)2SO4 precipitation | 407.6 | 76.1 | 0.19 | 71.9 | 1.3 |
| DEAE Sepharose | 51.0 | 35.6 | 0.70 | 33.6 | 5.0 |
| Native-PAGE | 0.1 | 0.4 | 3.85 | 0.4 | 27.5 |
All operations were carried out at 4°C.
Cell-free extract and (NH4)2SO4 precipitation: 1 U = reduction of 1 µmol 4′-chloroacetophenone per min.
DEAE Sepharose and Native-PAGE: 1 U = oxidation of 1 µmol NADPH per min.
Figure 2. SDS-PAGE analysis of the purified AcCR.
Gel was stained with 0.05% Coomassie Blue R-250: lane 1, molecular weight markers; lane 2, the purified enzyme with a molecular mass of 27 kDa.
Effect of Temperature on the Activity and Stability of AcCR
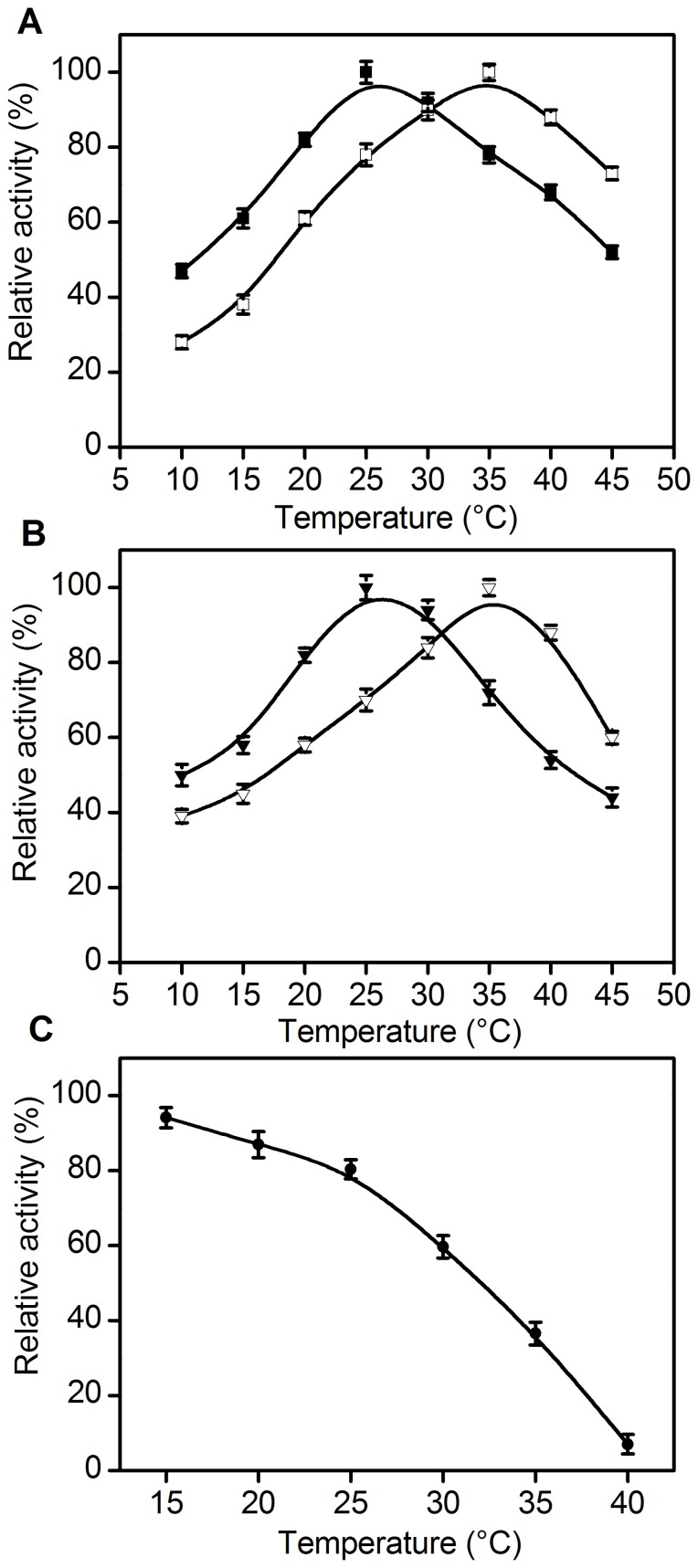
AcCR was found to be a dual coenzyme specific enzyme and showed reduction and oxidation activities in the presence of either NAD(H) or NADP(H). This might be due to its unique structure that allows dual coenzyme binding. Initially, the effects of various temperatures on the activity of AcCR was investigated for both reduction and oxidation reactions. The optimal temperature was found to be 25°C, for reduction of 4′-chloroacetophenone when NADPH (Figure 3A) or NADH (Figure 3B) was used as cofactor, which was similar to that observed with carbonyl reductase from G. capitatum JCM 3908 [18], but was much lower than that with carbonyl reductases from Thermus thermophilus HB27 (85°C) [19] and Geotrichum candidum (65°C) [20]. The maximum activity for oxidation of isopropanol was observed at 35°C using NADP+ (Figure 3A) or NAD+ (Figure 3B) as cofactor.
Figure 3. Relative activity of AcCR at different temperatures.
(A) Reduction of 4′-chloroacetophenone using NADPH as cofactor (▪); Oxidation of isopropanol using NADP+ as cofactor (□); (B) Reduction of 4′-chloroacetophenone using NADH as cofactor (▾); Oxidation of isopropanol using NAD+ as cofactor (□); The activity was measured in the temperature range of 10–45°C. The pH value was 5.0 (NADH as cofactor) or 7.5 (NADPH as cofactor) for reduction of 4′-chloroacetophenone or 8.0 for oxidation of isopropanol, respectively. The relative activity of AcCR at each optimal temperature for reduction and oxidation reactions was defined as 100%. (C) Thermal stability (•). The residual activity of AcCR after 5 h incubation (buffer pH 6.5) at varying temperatures were measured at 25°C.
The thermal stability of AcCR was determined at varying temperatures from 15 to 40°C (Figure 3C). The enzyme was very sensitive to reaction temperature and lost its activity quickly when temperature was higher than 35°C. The enzyme retained more than 80% of its original activity after 5 h incubation at a temperature range of 15–25°C. However, a significant drop in AcCR's activity was observed when reaction temperature was raised to 30°C, and AcCR remained only about 7% of its initial activity after 5 h incubation at 40°C. Obviously, the thermal stability of AcCR was not very satisfactory. Protein engineering strategy might be promising approach to further improve the activity and stability of the enzyme AcCR [21].
Effect of pH on the Activity and Stability of AcCR
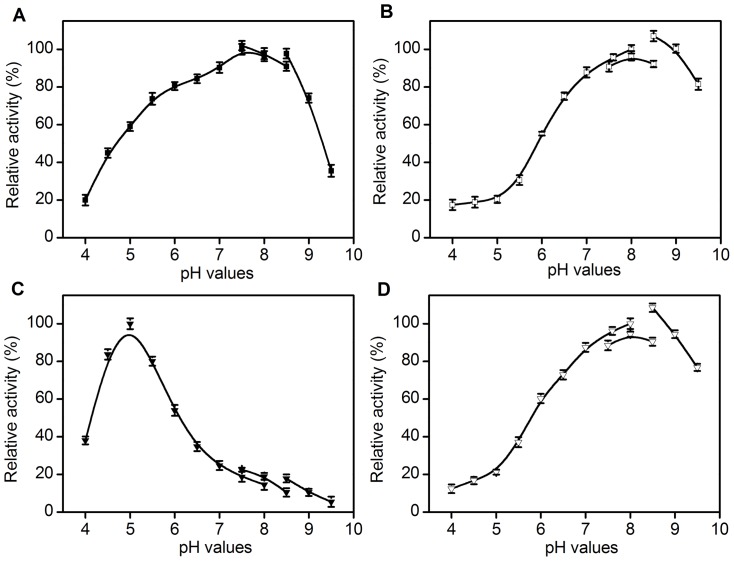
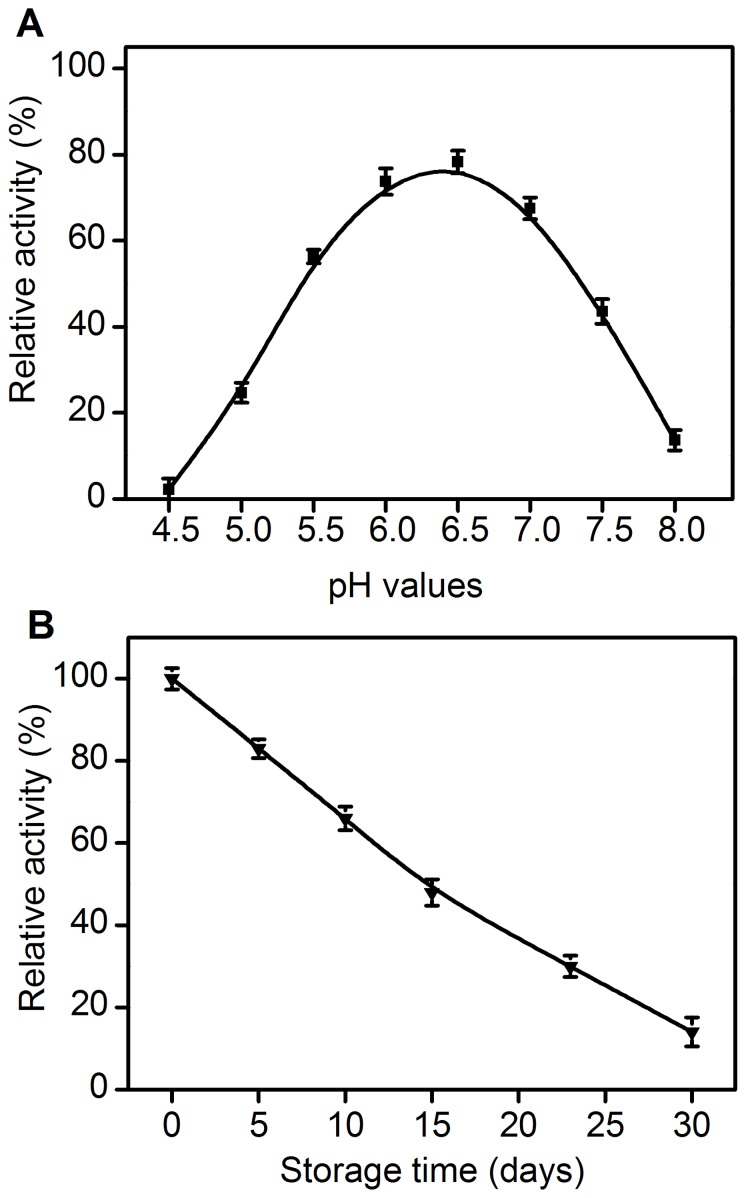
The effect of pH on the activity of AcCR was examined for both reduction and oxidation at a range of pH 4.0–9.5 (Figure 4). AcCR manifested the maximum reduction activity towards of 4′-chloroacetophenone using NADPH (Figure 4A) and NADH (Figure 4C) as cofactors at pH 7.5 and pH 5.0, respectively. The optimal pH for oxidation of isopropanol was found to be pH 8.0 when either NADP+ (Figures 4B) or NAD+ (Figures 4D) was acted as cofactor. Interestingly, it was noted that AcCR showed significantly different activities in the used three buffers at the same pH value (pH 7.5, 8.0 or 8.5) (Figure 4), which might be mainly attributable to the influences of the ionic strengths and types of these buffers. The detailed reasons for this unexpected observation need further investigations and are underway in our laboratory. When NADPH and NADP+ were used as cofactors, the optimal pH values for reduction and oxidation reactions with AcCR were very close (7.5 and 8.0), which was similar to that with alcohol dehydrogenase from Lactobacillus kefir [22]. When NADH and NAD+ were used as cofactors, on the contrary, the optimal pH values for reduction and oxidation indicated great difference (5.0 and 8.0), which was in good accordance with the observation with alcohol dehydrogenase from Candida maris IFO10003 [22]. Additionally, in the case of AcCR-catalyzed oxidation of isopropanol, both NADP+ and NAD+ as cofactors gave the same optimal pH value (pH 8.0), which was similar to that with isocitrate dehydrogenase from Rhodomicrobium vannielii [23].
Figure 4. Relative activity of AcCR at different pH values.
(A) Reduction of 4′-chloroacetophenone using NADPH as cofactor (▪); (B) Oxidation of isopropanol using NADP+ as cofactor (□); (C) Reduction of 4′-chloroacetophenone using NADH as cofactor (▾); (D) Oxidation of isopropanol using NAD+ as cofactor (□). The buffers used were 50 mM citrate-phosphate (pH 4.0–8.0), Tris-HCl (pH 7.5–8.5) and glycine–NaOH (pH 8.6–9.5) buffers. The enzyme's activity was measured in the pH range of 4.0–9.5 at 25°C for reduction of 4′-chloroacetophenone and at 35°C for oxidation of isopropanol. The relative activity of AcCR at each optimal pH for reduction and oxidation reactions was defined as 100%.
The pH stability of AcCR was investigated by incubation in different buffers at varied pH values between 4.5 and 8.0 at 4°C (Figure 5A). The enzyme still retained around 80% of its initial activity after incubation for five days at pH 6.5. However, a considerable loss in activity was observed over a five-day period at pH values lower than 5.5 or higher than 7.0. Furthermore, the enzyme lost its total activity after being incubated in buffers with pH ≤4.5 or pH ≥8.0 for a same period.
Figure 5. The stability of AcCR.
(A) pH stability (▪); The residual activities after incubation at various pH values (pH 4.5–8.0) for 5 days at 4°C were measured at buffer pH 5.0 and 25°C (NADH as cofactor). (B) Storage stability (▾). The residual activities after incubation at pH 6.5 for 30 days at 4°C were measured at buffer pH 5.0 and 25°C (NADH as cofactor). The relative activity of AcCR without incubation was defined as 100%.
Storage Stability of AcCR
The storage stability of the enzyme was investigated in sodium phosphate buffer (50 mM, pH 6.5) at 4°C. The purified AcCR showed the unsatisfactory storage stability, which was supported by the observation that the enzyme lost about 17% and 50% of activity after storage for five days and two weeks, respectively (Figure 5B). Clearly, the AcCR purified from Acetobacter sp. CCTCC M209061 cells was less stable than immobilized Acetobacter sp. CCTCC M209061 cells. Therefore, it is of great interest to enhance the stability (including storage stability) of AcCR by immobilization in our further studies.
Effects of Metal ions, Metal ion Chelators, and Thiol Reagents
It is well known that additives including metal ions have remarkable effects on the activity of carbonyl reductases from different sources. For example, Ca2+, Mn2+ and Mg2+ activated carbonyl reductase from Leifsonia xyli [24] and Zn2+ strongly inhibited carbonyl reductases from Candida viswanathii MTCC 5158 [7] and Geotrichum candidum [20]. So it is of great significance to explore the effects of various additives (metal ions, metal ion chelators, and thiol reagents) on the activity of AcCR using reduction of 4′-chloroacetophenone as a model reaction. As evident from the data summarized in Table 2, Cu2+ (2 mM) showed inhibitory effect on AcCR with 74% loss of activity, and Hg2+ and Ag+ inhibited completely the activity of AcCR. Other metal ions tested exhibited positive effects on the enzyme's activity. Zn2+ and Ca2+ slightly increased the activity, and Mn2+, Ni2+ and Fe2+ greatly activated the enzyme, which was consistent with our previous observation that the activity of Acetobaceter sp. CCTCC M209061 cell was remarkably improved in the presence of Mn2+ during its cultivation [13]. Also, Co2+ and Mg2+ manifested significant activation to the enzyme. Thiol-binding reagent iodoacetamide had only weak effect on the enzyme's activity, suggesting that there were no essential -SH groups at the catalytic sites of AcCR. β-Mercaptoethanol with a high concentration (40 mM) exhibited no significant effect on the enzyme's activity, indicating there were also no significant disulfide linkages present in the catalytic groups of the enzyme. Moreover, chelating agent EDTA had a strong inhibitory effect on the enzyme, indicating that metal ions were required for the enzyme's activity. Various surfactants such as SDS, Tween 80, and Triton X-100 were also examined for their effects on AcCR's activity. All these detergents inhibited the enzyme's activity, which might be attributable to the disruption of hydrophobic interactions and enhancement in internal repulsive forces [25].
Table 2. Effects of various additives on the activity of AcCR.
| Additives | Concentration (mM) | Relative activity (%) |
| Control | 0 mM | 100 |
| Mn2+ | 2 mM | 319.9±3.6 |
| 5 mM | 334.2±4.2 | |
| Mg2+ | 2 mM | 172.4±2.8 |
| 5 mM | 233.4±3.4 | |
| Ni2+ | 2 mM | 286.9±4.6 |
| 5 mM | 315.3±5.9 | |
| Co2+ | 2 mM | 201.4±5.3 |
| 5 mM | 232.9±1.9 | |
| Fe2+ | 2 mM | 271.4±5.2 |
| 5 mM | 253.0±4.5 | |
| Ca2+ | 2 mM | 115.8±4.7 |
| 5 mM | 135.7±3.9 | |
| Cu2+ | 2 mM | 36.0±1.5 |
| 5 mM | 23.5±0.6 | |
| Zn2+ | 2 mM | 141.5±1.9 |
| 5 mM | 131.3±1.0 | |
| Hg2+ | 2 mM | 0 |
| 5 mM | 0 | |
| Ag+ | 2 mM | 0 |
| 5 mM | 0 | |
| EDTA | 2 mM | 14.0±0.3 |
| 5 mM | 0 | |
| 2-Mercaptoethanol | 20 mM | 99.5±1.5 |
| 40 mM | 97.1±1.7 | |
| Iodoacetamide | 20 mM | 93.0±2.5 |
| 40 mM | 70.3±2.7 | |
| SDS | 2 mM | 91.1±2.5 |
| 5 mM | 69.2±1.3 | |
| Tween 80 | 15 mM | 49.3±1.1 |
| 30 mM | 28.8±0.7 | |
| Triton X-100 | 30 mM | 30.5±1.1 |
The reduction activity of AcCR was measured after being incubated at 25°C (NADH as cofactor) for 5 min with various additives in 50 mM phosphate-citrate buffer (pH 5.0). The relative activity was calculated by setting the activity without additive as 100%.
Obviously, the activity of AcCR was strongly inhibited by some heavy metal ions, metal ion chelating reagents and protein detergents, and also effectively activated by specific metal ions such as Mn2+. Moreover, the above-described results clearly showed that there were no essential thiol groups or disulfide linkages at or near the catalytic sites of AcCR.
Substrate Specificity and Enantioselectivity of AcCR
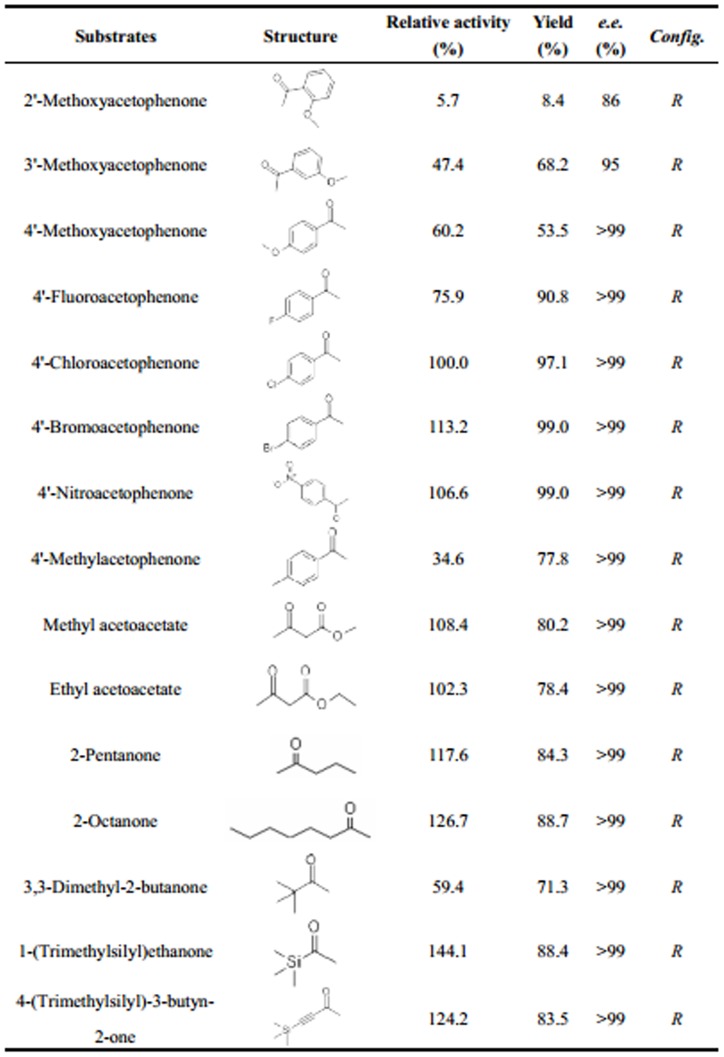
Substrate specificity and enantioselectivity are important characteristics of enzyme. Therefore, it was of great interest to investigate the substrate specificity and enantioselectivity of the novel AcCR purified from Acetobacter sp. CCTCC M209061 cells. A broad range of carbonyl compounds (including 4′-chloroacetophenone) such as aryl ketones, α-ketoesters and aliphatic ketones were examined as the substrates for AcCR. Initially, both NADH and NADPH were used as cofactors for AcCR-catalyzed reductions of carbonyl compounds. The reaction rates and yields were measured at buffer pH 5.0 for NADH and buffer pH 7.5 for NADPH, respectively. The obtained yield and reaction rate with NADH as cofactor were clearly superior to those with NADPH. The higher Vmax value was observed in the presence of 30 mM NADH. Thus, NADH was more suitable cofactor for AcCR-catalyzed reduction than NADPH.
As shown in Figure 6, NADH was used as cofactor and the relative activity of AcCR for reduction of model substrate 4′-chloroacetophenone was defined as 100%. High product yield and excellent enantioselectivity (>99%) were achieved when the 4′-position of acetophenone was substituted by electron-withdrawing groups such as -Br, whereas the electron-releasing groups like -methyl had negative effects. Moreover, the enzyme did not perform well with 2′- and 3′- substituted acetophenone such as 2′-methoxyacetophenone (5.7%) and 3′-methoxyacetophenone (47.4%), which might be affected by steric hindrance [7]. Specifically, α-ketoesters, e.g. methyl acetoacetate and ethyl acetoacetate, whose chiral alcohol derivatives were potential building blocks for the synthesis of (+)-decarestrictine [26], were successfully converted with high yields (80.2% and 78.4%) and excellent enantioselectivity (>99%). Among the aliphatic ketones, 2-pentanone and 2-octanone were converted with a high yield and enantioselectivity. Interestingly, the enzyme seemed more active towards silicon-containing ketones than the natural ketones. Relative activities above 100% were achieved even though the reaction conditions were not optimized for each specific substrate. These results could be attributable to specific properties of the silicon atom, such as its larger atomic radius and smaller electronegativity compared with the carbon atom. Indeed, the replacement of certain specific carbon atoms by silicon makes chemical and physical characteristics of silicon-contained molecules different from those of conventional organic compounds [27].
Figure 6. Effects of different substrates on AcCR-catalyzed asymmetric reduction of prochiral ketones.
AcCR was capable of catalyzing the anti-Prelog [28] enantioselective reductions of all the tested substrates to the corresponding (R)-alcohols. Except for 2′-methoxyacetophenone, the enzyme exhibited high enantioselectivity towards these carbonyl compounds. Compared with other carbonyl reductases, the substrate specificity of AcCR was similar to that of carbonyl reductase from Leifsonia xyli HS0904 [24] and very different from that of the enzyme from Pichia stipitis [29].
Kinetic Study
The kinetic parameters of AcCR for 4′-chloroacetophenone reduction and isopropanol oxidation were determined by measuring initial reaction rates with varying concentrations of substrates, and the obtained results were listed in Table 3. By analyzing of the kinetic data with Michaelis-Menten model and Hill model, it could be clearly seen that there was no cooperativity of the enzyme for these substrates except that NADH was used as the coenzyme for reduction of 4′-chloroacetophenone. The dependence of AcCR's activity on concentrations of substrates used and the corresponding Michaelis-Menten curve or Hill curve were illustrated in Figures S2–5.
Table 3. Kinetic parameters of AcCR for reduction of 4′-chloroacetophenone and oxidation of isopropanol.
| Substrates | Km c (mM) | Vmax (mM/min) |
| NADH | 0.66±0.021 | 0.21±0.008 |
| 4′-Chloroacetophenonea | 4.13±0.157c | -- |
| NADPH | 0.026±0.001 | 0.17±0.005 |
| 4′-Chloroacetophenoneb | 2.96±0.087 | -- |
| NAD+ | 1.33±0.045 | 0.069±0.003 |
| Isopropanola | 48.32±0.986 | -- |
| NADP+ | 1.12±0.035 | 0.074±0.004 |
| Isopropanolb | 67.82±2.321 | -- |
Kinetic constants were spectrophotometrically analyzed by determining the initial reaction rate at 340 nm at different substrate concentrations. Reactions were carried out in the optimal conditions. The dependence of AcCR's activity on substrate concentrations and the corresponding Michaelis–Menten curve or Hill curve were illustrated in Figures S2–5.
Determination for the NAD(H)-linked reaction.
Determination for the NADP(H)-linked reaction.
The apparent Km value was considered as S0.5.
For the reduction of 4′-chloroacetophenone, the Km value for NADH (0.66 mM) was over 25-fold greater than that for NADPH (0.026 mM), showing that the enzyme had a much higher affinity for NADPH than for NADH when the cofactors were at relatively low concentration levels, which was similar with the carbonyl reductase from Candida parapsilosis with dual cofactor specificity and strong preference for NADPH [30]. In contrast, some reductases preferred NADH such as the carbonyl reductase from Pichia stipitis [29]. The cofactor preferences exhibited by carbonyl reductases might be attributed to the electrostatic environment surrounding the 2′-hydroxyl (or phosphate) group of the adenosine ribose moiety of NADH (or NADPH) [31]. When NADH was used as cofactor, the response of AcCR's activity to increasing concentration of 4′-chloroacetophenone was clearly sigmoidal with a Hill coefficient of 3.1, suggesting that the enzyme might possess four substrate-binding sites cooperated with each other when NADH was present. Similar substrate binding cooperativity was observed with the matrix isocitrate dehydrogenase from potato mitochondria when the response of enzyme activity to isocitrate concentration was studied, while the dependence of initial velocity on the NAD+ concentration indicated that there was no cooperativity between subunits [32]. The apparent Km value (S0.5) for 4′-chloroacetophenone was 4.13 mM, which was also higher than 4′-chloroacetophenone Km (2.96 mM) using NADPH as cofactor. These results showed that the enzyme preferred NADPH when catalyzing reduction of 4′-chloroacetophenone. However, it is interesting that the Vmax value for NADH-based reduction of 4′-chloroacetophenone was slightly higher than that for NADPH-based reduction (0.21 mM/min vs 0.17 mM/min). The reasons for this result are not clear and are the subject of ongoing investigations in our research group.
For the oxidation of isopropanol, the Km value for NAD+ (1.33 mM) and NADP+ (1.12 mM) were similar and higher than that for NADH and NADPH (0.66 mM and 0.026 mM). The Km values for isopropanol were 48.32 mM (NAD+ acted as cofactor) and 67.82 mM (NADP+ acted as cofactor), which were also higher than that for 4′-chloroacetophenone (4.13 mM and 2.96 mM). The Vmax values for oxidation of isopropanol (0.069 mM/min and 0.074 mM/min) were lower than those for reduction of ketones (0.21 mM/min and 0.17 mM/min). All of these results indicated that the enzyme had a clear preference for reduction over oxidation (Table 3).
Coenzyme Supply for the Reduction of Ketones
For AcCR, expensive coenzymes (NADH or NADPH) were required for asymmetric reduction of prochiral ketones. Three different methods for coenzyme supply were tested and compared: (1) direct addition of NADH or NADPH; (2) regeneration of NADPH by adding G-6-P and GPDH; (3) addition of isopropanol. In the case of the second method, about 0.2 U/ml GPDH was used to keep 1/1 ratio of the two enzymes.
As can be seen in Table 4, AcCR gave different yields using the three cofactor supply methods. Although direct addition of NADH/NADPH could effectively prompt the reduction of ketones with high yields of 75–97%, it increased the cost for large-scale industrial applications because of the very expensive price of NADH or NADPH [33].
Table 4. Effects of coenzyme supply methods on AcCR-mediated asymmetric reduction of prochiral ketones.
| Coenzyme supply | Yield (%) (R)-1-(4-chlorophenyl)ethanol | Yield (%) (R)-2-octanol | Yield (%) (R)-3-hydroxybutyrate | ||||||
| 30 min | 60 min | 120 min | 30 min | 60 min | 120 min | 30 min | 60 min | 120 min | |
| 30 mM NADH | 84.1±2.8 | 95.5±2.5 | 97.1±3.3 | 66.8±2.5 | 82.0±3.1 | 88.7±2.6 | 29.2±1.1 | 49.7±1.7 | 78.4±1.8 |
| 30 mM NADPH | 78.4±2.4 | 87.3±3.2 | 96.2±2.6 | 61.2±1.4 | 74.9±1.9 | 86.6±2.8 | 27.8±1.0 | 45.3±1.4 | 75.5±2.5 |
| G-6-P/GPDH +0.2 mM NADPH | 80.6±3.1 | 98.3±2.4 | 97.6±2.8 | 43.4±1.2 | 60.6±2.2 | 96.0±2.4 | 29.6±0.9 | 58.3±2.1 | 84.2±2.7 |
| Isopropanol +0.2 mM NADH | 7.5±0.2 | 11.3±0.3 | 19.7±0.7 | 4.5±0.1 | 7.6±0.2 | 13.4±0.2 | 3.4±0.1 | 4.5±0.1 | 7.2±0.2 |
| Isopropanol +0.2 mM NADPH | 8.9±0.3 | 14.5±0.5 | 24.6±0.8 | 5.8±0.2 | 9.2±0.2 | 16.3±0.3 | 4.6±0.2 | 6.1±0.2 | 9.8±0.3 |
| Isopropanol +1 mM NADH | 32.6±1.2 | 47.2±1.5 | 81.0±2.1 | 21.7±0.7 | 32.2±0.9 | 53.1±1.7 | 10.4±0.3 | 20.2±0.6 | 29.8±1.1 |
| Isopropanol +1 mM NADPH | 33.7±1.1 | 48.3±1.6 | 84.2±3.2 | 22.6±0.5 | 31.5±1.2 | 55.6±1.5 | 12.8±0.4 | 22.5±0.9 | 32.6±0.9 |
The reactions were performed in phosphate-citrate buffer (50 mM, pH 5.0 or 7.5) containing substrates (5 mM 4′-chloroacetophenone, 10 mM 2-octanone or 15 mM ethyl acetoacetate) and different cofactor supply systems at 25°C.
G-6-P: Glucose-6- phosphate.
GPDH: Glucose-6- phosphate dehydrogenase.
The highest yields (84–98%) were obtained by using the cost-efficient NADPH-regenerating system (G-6-P/GPDH), which has been widely used in enzymatic synthesis of chiral alcohols [34]. Clearly, this NADPH-regenerating system surpassed the direct addition of coenzymes.
The in situ regeneration of cofactors can be also achieved using isopropanol without addition of GPDH. The AcCR-catalyzed oxidation of isopropanol can produce the reduced form of coenzymes, which in turn is used as cofactor for reduction of ketones. This method might be less expensive for cofactor supply and has been effectively used in many organic synthesis [17]. For the in situ regeneration of cofactors with addition of isopropanol, the achieved yields (30–85%) by together adding 1 mM NADH/NADPH to the reaction system were much higher than those with 0.2 mM NADH/NADPH.
In comparison with carbonyl reductases from other microorganisms such as Leifsonia xyli HS0904 [24], Candida viswanathii MTCC 5158 [7], Geotrichum candidum [20] and Pichia stipitis [29], AcCR from Acetobacter sp. CCTCC M209061 showed unique biochemical properties (dual coenzyme, cooperativity) and excellent anti-Prelog enantioslectivity in asymmetric reduction of a broad range of ketones, and had tremendous potential for synthesis of enantiopure alcohols.
Cloning of AcCR gene
A genome mining approach was used for hunting AcCR gene sequence according to the similar reports [35], [36]. The apparent molecular mass of the native AcCR was estimated to be 104 kDa by gel filtration and the molecular mass of the subunit was estimated to be 27 kDa by SDS-PAGE (Figure 2), suggesting that the enzyme has a homotetrameric structure and similar catalytic properties to that of alcohol dehydrogenase from Lactobacillus kefir and Lactobacillus brevis [37]. Thus, by searching alcohol dehydrogenase from Lactobacillus brevis with GENBANK, it was found that an oxidoreductase from Acetobacter pasteurianus 386B (Sequence ID: ref|YP_008391729.1) had 51% sequence similarity with alcohol dehydrogenase from Lactobacillus brevis. The corresponding gene sequence of oxidoreductase from Acetobacter pasteurianus 386B (NC_021991.1) was found according to its amino acid sequence. Consequently, the primers were designed for hunting AcCR gene sequence from Acetobacter sp. CCTCC M209061. A DNA fragment of 762 bp was obtained (Figure S6), and had 98% sequence similarity with the gene sequence of oxidoreductase from Acetobacter pasteurianus 386B (Figure S7). Thus, we successfully got the gene sequence of the novel AcCR from Acetobacter sp. CCTCC M209061 (Figure 7).
Figure 7. The gene sequences of AcCR from Acetobacter sp. CCTCC M209061.
Conclusion
The novel AcCR was purified from cytoplasm of Acetobacter sp. CCTCC M209061 and found to be a dual coenzyme specific enzyme, showing unique characteristics in reduction/oxidation reactions. Kinetic study indicated that the Km and Vmax values with NADH were higher than those with NADPH. The high anti-Prelog enantioselectivity of AcCR in catalyzing reduction of various ketones highlighted its application potential for synthesis of chiral alcohols. The efficiency of the biocatalytic process could be further improved by co-expression of AcCR and GPDH in a heterologous host.
Supporting Information
Clear Native-PAGE for purification of Ac CR. (A) Crude enzyme after purification with DEAE-Sepharose; (B) AcCR. The separating and stacking gels contained 9% and 5% acrylamide, respectively. The electrophoresis was performed at 4°C. The target segment (A) was chopped into small pieces, transferred into a dialysis bag with a cutoff molecular weight of 11 kDa and electrophoresed for 10 min to recollect the enzyme. The purified AcCR was subsequently desalted and concentrated by ultrafiltration using Centriprep YM-10, which showed a single band by Native-PAGE (B). Gel was stained with 0.05% Coomassie Blue R-250.
(TIF)
Dependence of Ac CR's activity on concentrations of NADH (A) and 4′-chloroacetophenone (B). Substrate saturation curves were shown with Lineweaver-Burk plot (A) and Hill plot (B) inset. The enzyme's activity was estimated under standard assay conditions, and NADH concentrations varied from 0.04 to 1.50 mM (4′-chloroacetophenone concentration was fixed at 5 mM) and 4′-chloroacetophenone concentrations varied from 1.5 to 20.0 mM (NADH concentration was fixed at 1 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk and Hill plotting.
(TIF)
Dependence of Ac CR's activity on concentrations of NADPH (A) and 4′-chloroacetophenone (B). Michaelis–Menten curves were shown with Lineweaver-Burk plots inset. The enzyme's activity was estimated under standard assay conditions, and NADPH concentrations varied from 0.02 to 0.40 mM (4′-chloroacetophenone concentration was fixed at 5 mM) and 4′-chloroacetophenone concentrations varied from 1.5 to 20.0 mM (NADPH concentration was fixed at 0.1 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk plotting.
(TIF)
Dependence of Ac CR's activity on concentrations of NAD+ (A) and isopropanol (B). Michaelis–Menten curves were shown with Lineweaver-Burk plots inset. The enzyme's activity was estimated under standard assay conditions, and NAD+ concentrations varied from 0.2 to 20.0 mM (isopropanol concentration was fixed at 110 mM) and isopropanol concentrations varied from 13 to 208 mM (NAD+ concentration was fixed at 5 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk plotting.
(TIF)
Dependence of Ac CR's activity on concentrations of NADP+ (A) and isopropanol (B). Michaelis–Menten curves were shown with Lineweaver-Burk plots inset. The enzyme's activity was estimated under standard assay conditions, and NADP+ concentrations varied from 0.2 to 20.0 mM (isopropanol concentration was fixed at 110 mM) and isopropanol concentrations varied from 13 to 208 mM (NADP+ concentration was fixed at 5 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk plotting.
(TIF)
PCR amplification of Ac CR gene sequence. Lane A: 10 µl sample, lane M: marker.
(TIF)
Comparative analysis gene sequences of oxidoreductase from Acetobacter pasteurianus 386B and Ac CR from Acetobacter sp. CCTCC M209061. A: gene sequence of oxidoreductase from Acetobacter pasteurianus 386B; B: gene sequence of AcCR from Acetobacter sp. CCTCC M209061.
(TIF)
GC analytic methods for various carbonyl compounds and their corresponding chiral alcohols.
(DOC)
Acknowledgments
We are grateful to Dr. Xiao-Feng Li for critical reading and commenting the manuscript.
Funding Statement
This study was partially supported by the National Science Fund for Excellent Young Scholars (21222606), the NSFC (21376096), the Key Program of Guangdong Natural Science Foundation (S2013020013049), the Foundation for the Author of National Excellent Doctoral Dissertation of China (201504) and the Fundamental Research Funds for SCUT (2013ZG0003). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sheldon RA (2012) Fundamentals of green chemistry: efficiency in reaction design. Chem Soc Rev 41: 1437–1451. [DOI] [PubMed] [Google Scholar]
- 2. Ni Y, Xu JH (2012) Biocatalytic ketone reduction: A green and efficient access to enantiopure alcohols. Biotechnol Adv 30: 1279–1288. [DOI] [PubMed] [Google Scholar]
- 3. Gao C, Zhang LJ, Xie YJ, Hu CH, Zhang Y, et al. (2013) Production of (3S)-acetoin from diacetyl by using stereoselective NADPH-dependent carbonyl reductase and glucose dehydrogenase. Bioresource Technol 137: 111–115. [DOI] [PubMed] [Google Scholar]
- 4. Xie Y, Xu JH, Xu Y (2010) Isolation of a Bacillus strain producing ketone reductase with high substrate tolerance. Bioresource Technol 101: 1054–1059. [DOI] [PubMed] [Google Scholar]
- 5. Richter N, Hummel W (2011) Biochemical characterisation of a NADPH-dependent carbonyl reductase from Neurospora crassa reducing α-and β-keto esters. Enzyme Microb Tech 48: 472–479. [DOI] [PubMed] [Google Scholar]
- 6. Wermuth B (1981) Purification and properties of an NADPH-dependent carbonyl reductase from human brain. Relationship to prostaglandin 9-ketoreductase and xenobiotic ketone reductase. J Biol Chem 256: 1206–1213. [PubMed] [Google Scholar]
- 7. Soni P, Kansal H, Banerjee UC (2007) Purification and characterization of an enantioselective carbonyl reductase from Candida viswanathii MTCC 5158. Process Biochem 42: 1632–1640. [Google Scholar]
- 8. Zhang RZ, Geng YW, Xu Y, Zhang WC, Wang SS, et al. (2011) Carbonyl reductase SCRII from Candida parapsilosis catalyzes anti-Prelog reaction to (S)-1-phenyl-1, 2-ethanediol with absolute stereochemical selectivity. Bioresource Technol 102: 483–489. [DOI] [PubMed] [Google Scholar]
- 9. Wang LJ, Li CX, Ni Y, Zhang J, Liu X, et al. (2011) Highly efficient synthesis of chiral alcohols with a novel NADH-dependent reductase from Streptomyces coelicolor Bioresource Technol. 102: 7023–7028. [DOI] [PubMed] [Google Scholar]
- 10. Xu GC, Yu HL, Zhang XY, Xu JH (2012) Access to optically active aryl halohydrins using a substrate-tolerant carbonyl reductase discovered from Kluyveromyces thermotolerans . Acs Catal 2: 2566–2571. [Google Scholar]
- 11. Ma HM, Yang LL, Ni Y, Zhang J, Li CX, et al. (2012) Stereospecific reduction of methyl o-chlorobenzoylformate at 300 g L−1 without additional cofactor using a carbonyl reductase mined from Candida glabrata . Adv Synth Cata 354: 1765–1772. [Google Scholar]
- 12. Xiao ZJ, Zong MH, Lou WY (2009) Highly enantioselective reduction of 4-(trimethylsilyl)-3-butyn-2-one to enantiopure (R)-4-(trimethylsilyl)-3-butyn-2-ol using a novel strain Acetobacter sp. CCTCC M209061. Bioresource Technol 100: 5560–5565. [DOI] [PubMed] [Google Scholar]
- 13. Chen XH, Lou WY, Zong MH, Smith TJ (2011) Optimization of culture conditions to produce high yields of active Acetobacter sp. CCTCC M209061 cells for anti-Prelog reduction of prochiral ketones. BMC Biotechnol 11: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen AJ, Poulter CD (1993) Purification and characterization of farnesyl diphosphate/geranylgeranyl diphosphate synthase. A thermostable bifunctional enzyme from Methanobacterium thermoautotrophicum . J Biol Chem 268: 11002–11007. [PubMed] [Google Scholar]
- 15. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 16. Bradford MM (1976) Foreign patent documents. Biochemistry 72: 248–254. [Google Scholar]
- 17. Hummel W (1990) Reduction of acetophenone to R (+)-phenylethanol by a new alcohol dehydrogenase from Lactobacillus kefir . Appl Microbiol Biot 34: 15–19. [Google Scholar]
- 18. Yamada-Onodera K, Fukui M, Tani Y (2007) Purification and characterization of alcohol dehydrogenase reducing N-benzyl-3-pyrrolidinone from Geotrichum capitatum . J Biosci Bioeng 103: 174–178. [DOI] [PubMed] [Google Scholar]
- 19. Rocha-Martín J, Vega D, Bolivar JM, Hidalgo A, Berenguer J, et al. (2012) Characterization and further stabilization of a new anti-prelog specific alcohol dehydrogenase from Thermus thermophilus HB27 for asymmetric reduction of carbonyl compounds. Bioresource Technol 103: 343–350. [DOI] [PubMed] [Google Scholar]
- 20. Singh A, Bhattacharyya MS, Banerjee UC (2009) Purification and characterization of carbonyl reductase from Geotrichum candidum . Process Biochem 44: 986–991. [Google Scholar]
- 21. Jakoblinnert A, van den Wittenboer A, Shivange AV, Bocola M, Heffele L, et al. (2013) Design of an activity and stability improved carbonyl reductase from Candida parapsilosis . J Biotechnol 165: 52–62. [DOI] [PubMed] [Google Scholar]
- 22. Kawano S, Yano M, Hasegawa J, Yasohara Y (2011) Purification and characterization of an NADH-dependent alcohol dehydrogenase from Candida maris for the synthesis of optically active 1-(Pyridyl) ethanol derivatives. Biosci Biotech Bioch 75: 1055–1060. [DOI] [PubMed] [Google Scholar]
- 23. Leyland ML, Hamblin MJ, Kelly DJ (1989) Eubacterial isocitrate dehydrogenase with dual specificity for NAD and NADP from Rhodomicrobium vannielii . FEMS Microbiol Lett 58: 165–169. [Google Scholar]
- 24. Wang NQ, Huang J, Luo HD, Wang P, Li J (2013) Purification and characterization of a new carbonyl reductase from Leifsonia xyli HS0904 involved in stereoselective reduction of 3, 5-bis (trifluoromethyl) acetophenone. J Mol Catal B: Enzym 92: 1–6. [Google Scholar]
- 25. Adler JJ, Singh PK, Patist A, Rabinovich YI, Shah DO, et al. (2000) Correlation of particulate dispersion stability with the strength of self-assembled surfactant films. Langmuir 16: 7255–7262. [Google Scholar]
- 26. Clark JS, Fessard TC, Whitlock GA (2006) A concise enantioselective synthesis of the fungal metabolite (+)-decarestrictine L. Tetrahedron 62: 73–78. [Google Scholar]
- 27. Zhang BB, Lou WY, Zong MH, Wu H (2008) Efficient synthesis of enantiopure (S)-4-(trimethylsilyl)-3-butyn-2-ol via asymmetric reduction of 4-(trimethylsilyl)-3-butyn-2-one with immobilized Candida parapsilosis CCTCC M203011 cells. J Mol Catal B: Enzym 54: 122–129. [Google Scholar]
- 28. Prelog V (1964) Specification of the stereospecificity of some oxidoreductases by diamond lattice sections. Pure Appl Chem 9: 119–130. [Google Scholar]
- 29. Cao H, Mi L, Ye Q, Zang GL, Yan M, et al. (2011) Purification and characterization of a novel NADH-dependent carbonyl reductase from Pichia stipitis involved in biosynthesis of optically pure ethyl (S)-4-chloro-3-hydroxybutanoate. Bioresource Technol 102: 1733–1739. [DOI] [PubMed] [Google Scholar]
- 30. Nie Y, Xiao R, Xu Y, Montelione GT (2011) Novel anti-Prelog stereospecific carbonyl reductases from Candida parapsilosis for asymmetric reduction of prochiral ketones. Org Biomol Chem 9: 4070–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tanaka N, Nonaka T, Nakanishi M, Deyashiki Y, Hara A, et al. (1996) Crystal structure of the ternary complex of mouse lung carbonyl reductase at 1.8 Å resolution: the structural origin of coenzyme specificity in the short-chain dehydrogenase/reductase family. Structure 4: 33–45. [DOI] [PubMed] [Google Scholar]
- 32. Tezuka T, Laties GG (1983) Isolation and characterization of inner membrane-associated and matrix NAD-specific isocitrate dehydrogenase in potato mitochondria. Plant Physiol 72: 959–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura K, Yamanaka R, Matsuda T, Harada T (2003) Recent developments in asymmetric reduction of ketones with biocatalysts. Tetrahedron: Asymmetr 14: 2659–2681. [Google Scholar]
- 34. Weckbecker A, Gröger H, Hummel W (2010) Regeneration of nicotinamide coenzymes: principles and applications for the synthesis of chiral compounds. Biosyst Eng I: 195–242. [DOI] [PubMed] [Google Scholar]
- 35. Zhang Z-J, Xu J-H, He Y-C, Ouyang L-M, Liu Y-Y (2011) Cloning and biochemical properties of a highly thermostable and enantioselective nitrilase from Alcaligenes sp. ECU0401 and its potential for (R)-(−)-mandelic acid production. Bioprocess and biosystems engineering 34: 315–322. [DOI] [PubMed] [Google Scholar]
- 36. Xu G-C, Yu H-L, Zhang X-Y, Xu J-H (2012) Access to Optically Active Aryl Halohydrins Using a Substrate-Tolerant Carbonyl Reductase Discovered from Kluyveromyces thermotolerans. ACS Catalysis 2: 2566–2571. [Google Scholar]
- 37. Weckbecker A, Hummel W (2006) Cloning, expression, and characterization of an (R)-specific alcohol dehydrogenase from Lactobacillus kefir. Biocatalysis and Biotransformation 24: 380–389. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clear Native-PAGE for purification of Ac CR. (A) Crude enzyme after purification with DEAE-Sepharose; (B) AcCR. The separating and stacking gels contained 9% and 5% acrylamide, respectively. The electrophoresis was performed at 4°C. The target segment (A) was chopped into small pieces, transferred into a dialysis bag with a cutoff molecular weight of 11 kDa and electrophoresed for 10 min to recollect the enzyme. The purified AcCR was subsequently desalted and concentrated by ultrafiltration using Centriprep YM-10, which showed a single band by Native-PAGE (B). Gel was stained with 0.05% Coomassie Blue R-250.
(TIF)
Dependence of Ac CR's activity on concentrations of NADH (A) and 4′-chloroacetophenone (B). Substrate saturation curves were shown with Lineweaver-Burk plot (A) and Hill plot (B) inset. The enzyme's activity was estimated under standard assay conditions, and NADH concentrations varied from 0.04 to 1.50 mM (4′-chloroacetophenone concentration was fixed at 5 mM) and 4′-chloroacetophenone concentrations varied from 1.5 to 20.0 mM (NADH concentration was fixed at 1 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk and Hill plotting.
(TIF)
Dependence of Ac CR's activity on concentrations of NADPH (A) and 4′-chloroacetophenone (B). Michaelis–Menten curves were shown with Lineweaver-Burk plots inset. The enzyme's activity was estimated under standard assay conditions, and NADPH concentrations varied from 0.02 to 0.40 mM (4′-chloroacetophenone concentration was fixed at 5 mM) and 4′-chloroacetophenone concentrations varied from 1.5 to 20.0 mM (NADPH concentration was fixed at 0.1 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk plotting.
(TIF)
Dependence of Ac CR's activity on concentrations of NAD+ (A) and isopropanol (B). Michaelis–Menten curves were shown with Lineweaver-Burk plots inset. The enzyme's activity was estimated under standard assay conditions, and NAD+ concentrations varied from 0.2 to 20.0 mM (isopropanol concentration was fixed at 110 mM) and isopropanol concentrations varied from 13 to 208 mM (NAD+ concentration was fixed at 5 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk plotting.
(TIF)
Dependence of Ac CR's activity on concentrations of NADP+ (A) and isopropanol (B). Michaelis–Menten curves were shown with Lineweaver-Burk plots inset. The enzyme's activity was estimated under standard assay conditions, and NADP+ concentrations varied from 0.2 to 20.0 mM (isopropanol concentration was fixed at 110 mM) and isopropanol concentrations varied from 13 to 208 mM (NADP+ concentration was fixed at 5 mM). The kinetic parameters, Km and Vmax values, were graphically determined from the Lineweaver-Burk plotting.
(TIF)
PCR amplification of Ac CR gene sequence. Lane A: 10 µl sample, lane M: marker.
(TIF)
Comparative analysis gene sequences of oxidoreductase from Acetobacter pasteurianus 386B and Ac CR from Acetobacter sp. CCTCC M209061. A: gene sequence of oxidoreductase from Acetobacter pasteurianus 386B; B: gene sequence of AcCR from Acetobacter sp. CCTCC M209061.
(TIF)
GC analytic methods for various carbonyl compounds and their corresponding chiral alcohols.
(DOC)