Abstract
Mycobacterium chelonae is a rapidly growing mycobacterial opportunistic pathogen closely related to Mycobacterium abscessus that causes cornea, skin and soft tissue infections in humans. Although M. chelonae and the emerging mycobacterial pathogen M. abscessus have long been considered to belong to the same species, these two microorganisms considerably differ in terms of optimum growth temperature, drug susceptibility, pathogenicity and the types of infection they cause. The whole genome sequencing of clinical isolates of M. chelonae and M. abscessus is opening the way to comparative studies aimed at understanding the biology of these pathogens and elucidating the molecular bases of their pathogenicity and biocide resistance. Key to the validation of the numerous hypotheses that this approach will raise, however, is the availability of genetic tools allowing for the expression and targeted mutagenesis of genes in these species. While homologous recombination systems have recently been described for M. abscessus, genetic tools are lacking for M. chelonae. We here show that two different allelic replacement methods, one based on mycobacteriophage-encoded recombinases and the other on a temperature-sensitive plasmid harboring the counterselectable marker sacB, can be used to efficiently disrupt genes in this species. Knock-out mutants for each of the three porin genes of M. chelonae ATCC 35752 were constructed using both methodologies, one of which displays a significantly reduced glucose uptake rate consistent with decreased porin expression.
Introduction
Rapidly growing non-tuberculous mycobacteria (RGM) are opportunistic pathogens widespread in the environment that can cause a wide spectrum of infections [1]. Among RGM, the Mycobacterium chelonae-abscessus group is considered to be the most pathogenic for humans [1], [2]. On the basis of their almost identical biochemical features, M. chelonae and M. abscessus were originally considered to belong to the same species (‘M. chelonei’ or ‘M. chelonae’). Advances in molecular biology techniques then led to their separation into two distinct species in 1992 [1], [3]. M. abscessus has since been shown to consist of three subspecies, M. abscessus subsp. abscessus, M. abscessus subsp. massiliense and M. abscessus subsp. bolletii [4], [5], even though massiliense and bolletii have recently been proposed to represent one single subspecies [6]. Distinguishing between M. chelonae and M. abscessus (sensu lato) is important because these organisms are typically not associated with the same types of infections in humans, differ in their pathogenicity and show very different antimicrobial susceptibility patterns [2], [7]. M. abscessus (sensu lato) is the most pathogenic and antibiotic-resistant RGM. It is responsible for more than 80% of all pulmonary infections caused by RGM in the United States [1], [8] and it is also associated with contaminated traumatic skin wounds, post-surgical soft tissue infections, central nervous system infections and disseminated infections in immunodeficient patients [9]–[11]. M. chelonae in contrast is only rarely a cause of chronic lung disease [8] and is better known as a causative agent of cornea, skin and soft tissue infections [1]. Pulmonary infections with M. abscessus (sensu lato) have been increasing in prevalence throughout the world, particularly in patients with structural lung disease such as chronic obstructive pulmonary disease, bronchiectasis, and cystic fibrosis [2], [12], [13]. Patient-to-patient transmission involving M. abscessus subsp. massiliense has recently been proposed [13], [14]. Another important distinction between M. chelonae and M. abscessus is their optimum growth temperature. The temperature optimum of M. chelonae is 28–30°C whereas M. abscessus (sensu lato) grows best at 35–37°C.
The first genome sequence of M. abscessus subsp abscessus (strain ATCC 19977) was completed and released in 2009 [15], followed by that of several other M. abscessus subsp. abscessus, massiliense and bolletii isolates. The genome sequencing of the two first M. chelonae isolates (strains ATCC 35752 and 1518) is in progress. Comparative genomics of these two closely related RGM is expected to provide significant insights into the molecular bases underlying the important differences associated with the physiology, pathogenicity and drug susceptibility of RGM. A prerequisite to the validation of the numerous hypotheses that comparative genomics will raise, however, is the availability of expression and mutagenesis systems allowing for the knock-in and knock-out of genes in these microorganisms.
Progress was made recently toward the development of conditional expression and homologous recombination systems for M. abscessus. Although orders of magnitude less efficient than in M. smegmatis, these methodologies were successfully used to construct a null and a conditional mutant [16], [17]. Foremost among the obstacles that genetic strategies have to overcome in RGM pathogens in general is their low electrotransformation efficiency, intrinsic high level of resistance to antibiotics and propensity to develop spontaneous resistance to commonly used antibiotic resistance markers, and relatively narrow temperature growth range limiting the effectiveness of counterselectable temperature-sensitive plasmids and phages [16], [17]. To the best of our knowledge, no mutagenesis system has yet been optimized for M. chelonae. With the goal of facilitating future studies on the biology of RGM pathogens, we here undertook to address the current lack of suitable genetic systems in M. chelonae. Various selectable and counterselectable markers and allelic replacement methods, namely the temperature sensitive – SacB (Ts-SacB) system [18], the thermosensitive mycobacteriophage system [19] and the recombinase-based system [20] were tested and compared. Both the Ts-sacB and the recombinase-based systems were found to be functional in M. chelonae as long as appropriate selectable markers are used.
Materials and Methods
Bacterial strains and culture media
Escherichia coli DH5α, the strain used for cloning, was grown in LB Lennox (BD, Difco) medium at 37°C. M. chelonae strains ATCC 35752 and 9917 [21], and M. abscessus ATCC 19977 were grown in LB or Middlebrook 7H9-OADC broth (BD, Difco) supplemented with 0.05% Tween 80, 7H11-OADC agar (BD, Difco) or minimal Sauton's medium supplemented with 0.05% tyloxapol. Kanamycin (Kan) and Gentamicin (Gen) were added to final concentrations of 200 µg/ml. Zeocin (Zeo) was added to a final concentration of 100 µg/ml.
Electrotransformation
Electrocompetent M. chelonae and M. abscessus cells were prepared by washing bacterial pellets two times with double distilled water containing 0.05% Tween 80 and one time with 10% glycerol–0.05% Tween 80 at 4°C. One hundred microliter aliquots of freshly prepared competent cells in 10% glycerol–0.05% Tween 80 were electroporated in the presence of 1 µg of plasmid DNA or 300 ng of linear DNA in 0.2-cm cuvettes with a single pulse (2.5 kV; 25 µF; 1,000 ohms). One milliliter of fresh 7H9-OADC-Tween 80 medium was then added, and the culture was incubated at 30°C (M. chelonae) or 37°C (M. abscessus) for 4 h before plating on 7H11-OADC agar plates.
Allelic replacement using the Ts-SacB system
The MCH_4689c gene and flanking regions was PCR-amplified from M. chelonae ATCC 35752 genomic DNA with primers MCH4689Fw2 (5′-TTTTTCTAGAGTGTATCGGCTGCGAGTTAGC-3′) and MCH4689Rv2 (5′-TTTTTCTAGAGCGGGTGGTAATGGTCGCAT-3′) and the disrupted alleles, MCH_4689c::kan and MCH_4689c::zeo, were obtained by inserting either the Tn903 kanamycin resistance cassette from pUC4K (GE Healthcare) or the zeocin resistance cassette from pEM7/Zeo (Invitrogen) at the unique SgfI restriction site of MCH_4689c. Following similar strategies, the MCH_4690c and MCH_4691c genes and their flanking regions were PCR-amplified individually from M. chelonae ATCC 35752 genomic DNA using the following primer combinations: MCH4690Fw (5′-TTTTTCTAGATTACGTAGGTCGAGGCGCCG-3′) and MCH4690Rv (5′-TTTTTCTAGACGTCCCGATCACTGCCAGCC-3′) (for MCH_4690c); and MCH4691Fw (5′-TTTTTCTAGAGTCTTGGGTTCGGCTAACTTC-3′) and MCH4691Rv (5′-TTTTTCTAGATGTACGAAGCCTCAGGACCA-3′) (for MCH_4691c). Disrupted alleles of MCH_4690c and MCH_4691c were obtained by insertion of the kan or zeo resistance cassettes at the unique SgfI site of these genes. All six disrupted alleles were then cloned in the XbaI-cut pPR27-xylE [18], [22], yielding pPR27-4689-KX, pPR27-4689-ZX, pPR27-4690-KX, pPR27-4690-ZX, pPR27-4691-KX and pPR27-4691-ZX, the plasmids used to achieve allelic replacement at the MCH_4689c, MCH_4690c and MCH_4691c loci of M. chelonae (Table 1).
Table 1. Strains and plasmids used in this study.
| Plasmid or strain | Features | Source or reference |
| Strains | ||
| M. chelonae ATCC 35752 | M. chelonae reference strain | ATCC |
| M. chelonae 9917 | Glutaraldehyde-resistant isolate of M. chelonae | [21] |
| M. abscessus ATCC 19977 | M. abscessus reference strain | ATCC |
| Plasmids | ||
| pOMK | pBluescript KS- derivative carrying a mycobacterial origin of replication and a Kan resistance gene | [43] |
| pOMK-zeo | pOMK derivative carrying the zeo resistance gene from pEM7/Zeo | This study |
| pOMK_4691 | pOMK derivative expressing MCH_4691c under control of its own promoter | This study |
| pOMK_[4691_4689] | pOMK derivative expressing MCH_4691c, MCH_4690c and MCH_4689c under control of their own promoter | This study |
| pJV53 | Recombineering plasmid carrying the Che9c gp60 and gp61 genes under control of the acetamidase promoter | [20] |
| pJV53-xylE | pJV53 derivative carrying the xylE gene | This study |
| pEM7/Zeo | Plasmid carrying the Zeo resistance cassette | Invitrogen |
| pUC4K | Plasmid carrying the Kan resistance cassette | GE Healthcare |
| pPR27-xylE | Mycobacterial shuttle plasmid carrying the sacB counterselectable marker, a Gen resistance cassette and xylE | [22] |
| pPR27-4689-KX | pPR27-xylE derivative carrying MCH4689c::kan | This study |
| pPR27-4690-KX | pPR27-xylE derivative carrying MCH4690c::kan | This study |
| pPR27-4691-KX | pPR27-xylE derivative carrying MCH4691c::kan | This study |
| pPR27-4689-ZX | pPR27-xylE derivative carrying MCH4689c::zeo | This study |
| pPR27-4690-ZX | pPR27-xylE derivative carrying MCH4690c::zeo | This study |
| pPR27-4691-ZX | pPR27-xylE derivative carrying MCH4691c::zeo | This study |
The pPR27-xylE constructs carrying the disrupted porin genes were electroporated in M. chelonae ATCC 35752 and transformants were selected on Kan or Zeo-containing medium at 30°C. One to three XylE positive colonies (i.e., turning yellow upon exposure to catechol) [22] were inoculated in 7H9-OADC-Tween 80 broth at 30°C for 5–7 days and finally plated onto 7H11-OADC containing Kan or Zeo and 10% sucrose at 37°C. Colony forming units (CFU) displaying the expected phenotype for allelic exchange mutants (i.e., resistance to sucrose and Kan or Zeo, XylE−) were picked after 14 to 21 days and analyzed by PCR using primers MCHDCO1 (5′-ATCTGGCAGGTCGCGAAGTC-3′) and MCHDCO2 (5′-ACCGGAGATATCGTCGACATC-3′) to amplify the entire porin gene cluster (Fig. S1). That allelic replacement occurred at the right porin locus was confirmed by sequencing the regions immediately flanking the antibiotic resistance cassette.
pOMK_4691 and pOMK_[4691_4689], the constructs used for complementation of the MCH_4691c knock-out mutant, were constructed by cloning the MCH_4691c gene or the entire porin gene cluster encompassing MCH_4691c, MCH_4690c and MCH_4689c plus 169 bp of upstream and 522 bp (pOMK_[4691_4689]) to 525 bp (pOMK_4691) of downstream DNA sequence into the XbaI restriction site of pOMK (Table 1).
Allelic replacement using the recombineering system
This system consists of performing allelic exchange in strains expressing the gp60 and gp61 recombineering proteins from mycobacteriophage Che9c that effectively promote homologous recombination in mycobacteria [20]. The gp60 and gp61genes are expressed from the replicative plasmid pJV53 under control of an acetamide-inducible promoter [20]. Acetamide-induced strains harboring pJV53 are electro-transformed with allelic exchange substrates under the form of linear DNA and double crossover mutants are isolated on selective medium.
To facilitate the detection of pJV53 transformants, a derivative of pJV53 carrying the colored marker xylE was first constructed by cloning the xylE gene from pPR27-xylE in the unique SpeI restriction site of pJV53 (Table 1). pJV53-xylE was introduced in M. chelonae ATCC 35752 by electroporation and a kanamycin-resistant-XylE+ transformant was then cultured at 30°C in LB broth containing Kan, 0.05% tyloxapol and 0.2% succinate to mid-log phase (A600nm = 0.4) at which point 0.2% acetamide was added. After 3 h of induction, electrocompetent cells were prepared as described above. The competent cells were transformed with 300 ng of linear allelic exchange substrates. The linear zeo cassette-disrupted alleles used to achieve gene replacement at the MCH_4689c, MCH_4690c and MCH_4691c loci of M. chelonae ATCC 35752 were identical to those used in the Ts-sacB system. Candidate allelic exchange mutants were analyzed by PCR and sequencing as described above (Fig. S1).
Drug susceptibility testing
MIC values were determined in 7H9-OADC-Tween 80 and Muller-Hinton II broth (BD, Difco) in a total volume of 200 µl in 96-well microtiter plates. M. chelonae cultures grown to early log phase (A600nm = 0.2) were diluted to a final concentration of 5×10e5 CFU/ml and incubated in the presence of serial dilutions of the drugs for 5 to 7 days at 30°C. MICs were determined using the colorimetric resazurin assay and confirmed by visually scanning for growth.
Glucose uptake experiment
[U-14C]glucose uptake experiments were essentially carried out as described by Stahl et al. [23]. Briefly, 100 ml M. chelonae cultures grown to an OD600 of 0.5 were harvested by centrifugation, washed twice in uptake buffer (50 mM Tris-HCl pH 7.1, 15 mM KCl, 10 mM NH4SO4, 1 mM MgSO4, 0.1% Tween 80), and resuspended in 25 ml of the same buffer. [U-14C]glucose (specific activity, 5 mCi/mmol, American Radiolabeled Chemicals) was mixed with cold glucose and added to the cell suspension to a final concentration of 20 µM. The mixtures were incubated at 30°C and 200 µl samples were removed at times ranging from 1 to 128 min. The cells were then added 200 µl of killing buffer (10% formalin: 0.1 M LiCl; 2∶1 by vol.) before being filtered through a 0.45 µm pore size membrane filter, washed twice with killing buffer and counted in a liquid scintillation counter. The uptake of glucose was expressed as pmol/mg (dry weight) cells. Glucose uptake experiments were performed twice using independent culture batches.
Results and Discussion
Porin genes as candidates for allelic replacement experiments
The particular structure and composition of the mycobacterial cell envelope is generally thought to be one of the major determinant of the intrinsic resistance of mycobacteria to most common antibiotics and other biocides [24], [25]. The presence of a highly impermeable outer membrane [26], [27] implies the existence of pore proteins spanning the outer membrane for the uptake of nutrients and efflux of waste products [28]. The Msp porins of the rapidly growing Mycobacterium species, M. smegmatis, represent the main general diffusion pathway for small hydrophilic molecules across the outer membrane of this bacterium [28], [29]. Msp porins have been extensively studied and showed to not only be major determinants of the susceptibility of M. smegmatis to multiple antibiotics and biocides [28], [30]–[33], but to also impact its vulnerability to killing by reactive nitrogen intermediates inside host phagocytic cells [34], [35]. As in other bacteria [36], porin expression in M. smegmatis appears to be tightly regulated [23], [37]. Whereas Msp-type porins are apparently not found in slow growing mycobacteria, the availability of a growing number of genome sequences from various Mycobacterium species and earlier biochemical and DNA hybridization studies indicate that they are likely to be widespread among RGM [28], [38]. Incidentally, the first Gram positive porin ever reported was that of an M. chelonae isolate [39], [40]. Evidence to date therefore points to an important role of porins in the physiology, biocide resistance and intracellular survival of RGM and the availability of isogenic knock-out mutants of M. chelonae deficient in their expression will prove useful for subsequent studies on the biology of this opportunistic pathogen.
Our previous work identified three porin genes (MCH_4689c, MCH_4690c and MCH_4691c) clustered on the chromosomes of the M. chelonae strain ATCC 35752 (Fig. 1A) and a glutaraldehyde-resistant M. chelonae isolate, 9917 [21]. 115- and 222-bp of intergenic space separate MCH_4691c from MCH_4690c and MCH_4690c from MCH_4689c, respectively, suggesting that these genes are expressed as single transcriptional units. MCH_4689c, MCH_4690c and MCH_4691c from M. chelonae ATCC 35752 differ from one another at one to four positions of the mature proteins (Fig. S2). Allelic exchange substrates were generated by inserting an antibiotic (kanamycin or zeocin) resistance cassette at the unique SgfI restriction site of each of the three porin genes of M. chelonae ATCC 35752 as described under Materials and Methods.
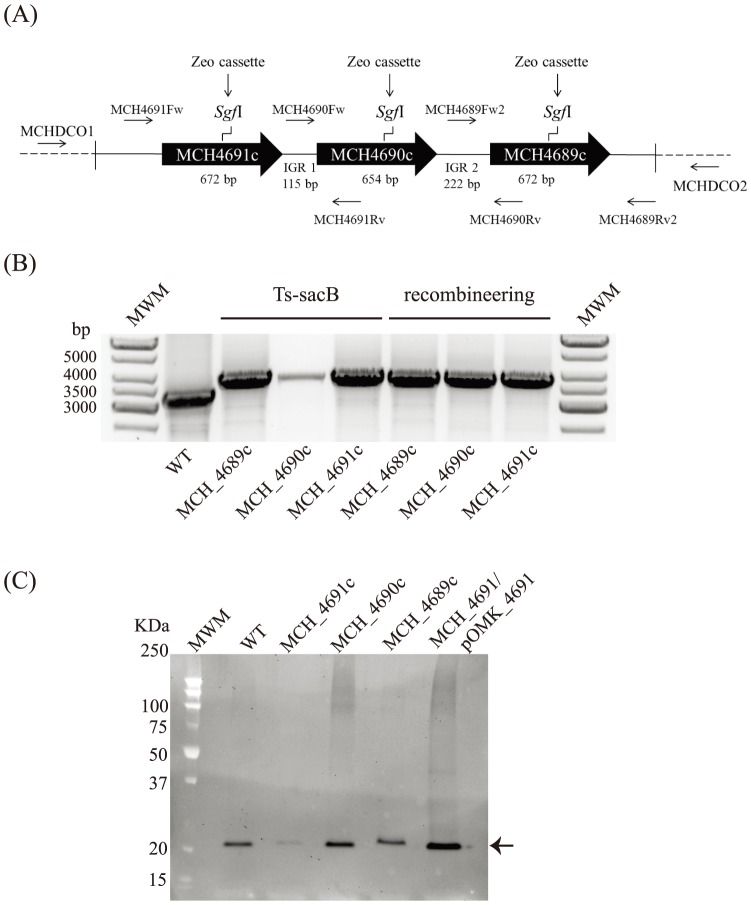
Figure 1. Gene replacement at the MCH_4689c, MCH_4690c and MCH_4691c porin loci of M. chelonae ATCC 35752 using the Ts-sacB and recombineering systems.
(A) Porin gene cluster of M. chelonae ATCC 35752. The positions of the primers used to generate the allelic exchange substrates and analyze the candidate mutants are indicated. IGR1 and IGR2 represent the intergenic regions. (B) Candidate mutants obtained for each of the porin genes using the Ts-sacB or the recombineering systems were analyzed by PCR as described under Materials and Methods and confirmed by sequencing the regions flanking the resistance cassette. The expected size of the PCR fragments is 3.3 kb for the wild-type parent strain and 3.8 kb for the knock-out mutants. MWM, molecular weight marker. WT, wild-type. (C) Immunoblot analysis of porin production in the wild-type, mutant and complemented mutant strains. Strains were grown in 7H9-OADC-Tween 80 broth at 30°C to mid-log phase (OD600 = 1) and porins were selectively extracted from whole cells at 100°C using 0.5% n-octylpolyoxyethylene as a detergent as described [44]. Protein samples prepared from the same amount of cells for each strain were denatured by boiling in 80% DMSO followed by acetone precipitation [23]. Denatured proteins were loaded volume to volume, separated by SDS-PAGE, blotted onto a nitrocellulose membrane, and porins were detected using rabbit antiserum to purified MspA [23]. Immune complexes were detected by chemiluminescence (Pierce, ELC) and semi-quantified using the Image Lab software (Biorad).
Choice of allelic exchange methodologies
Mainly three homologous recombination systems have been used thus far in mycobacteria: (i) The Ts-sacB system, based on the use of a temperature-sensitive plasmid carrying the counterselectable marker sacB [18]; (ii) the temperature-sensitive mycobacteriophage system [19]; and the recombineering system [20]. Ts-SacB allows for allelic exchange substrates to be delivered on a replicative plasmid harboring a mycobacterial temperature-sensitive origin of replication and therefore to persist inside the cells until grown at a non-permissive temperature. Growth at non-permissive temperature in the presence of sucrose selects for allelic exchange mutants [18]. The recombineering system takes advantage of the transient expression of highly active mycobacteriophage-encoded recombinases to promote homologous recombination between the chromosomal copy of a target gene and a disrupted copy of this gene delivered to the cells as a linear DNA substrate [20]. The mycobacteriophage system compensates for the low transformation efficiency of mycobacteria by allowing allelic exchange substrates to be delivered to the cells with high efficiency on conditionally replicating mycobacteriophages. Thus far, the only two conditionally (temperature-sensitive) replicating phage systems available for this purpose are based on the mycobacteriophages TM4 and D29 [19]. Earlier reports indicated that none of these two mycobacteriophages efficiently transfect M. chelonae isolates [19], [41]. Our own assays confirmed these findings in the reference M. chelonae strain ATCC 35752 in that no plaques were obtained upon transfection of this isolate with high titers of TM4 or D29. Therefore, our efforts focused on the construction of porin knock-out mutants of M. chelonae using the recombineering and Ts-sacB systems.
Transformation efficiency and spontaneous resistance to antibiotics
Key to the success of any allelic replacement methodology is the availability of selection markers allowing for the efficient selection of transformants and recombinant strains resulting from single or double crossover events. Given the high frequencies of spontaneous antibiotic-resistance mutations reported in M. abscessus subsp. abscessus [16], we first set out to test and compare different antibiotic selection markers (gen, zeo and kan) in M. chelonae. M. chelonae ATCC 35752 being intrinsically highly resistant to hygromycin (MIC>500 µg/ml), ampicillin (MIC>500 µg/ml), and streptomycin (MIC>200 µg/ml), the corresponding resistance cassettes were not tested here. M. chelonae ATCC 35752 was transformed with plasmids pOMK-zeo, conferring resistance both to Kan and Zeo, and pJV53-xylE conferring resistance to Kan (Table 1). In addition, the pPR27-based plasmids harboring zeo or kan-disrupted alleles of the porin genes were used. For comparison purposes, some plasmids were also transformed in M. chelonae 9917, although, in this case, the high intrinsic level of resistance of this particular isolate to zeocin (MIC>200 µg/ml compared to an MIC of 50 µg/ml for ATCC 35752) precluded selection on Zeo-containing media.
Transformation efficiencies in M. abscessus ATCC 19977 were in line with those reported previously [16]. Transformation efficiencies in M. chelonae ATCC 35752 were two orders of magnitude greater than that measured for M. abscessus ATCC 19977 with pOMK-zeo, and considerably greater (>400-fold) with pOMK-zeo and pJV53-xylE than with the pPR27-derived constructs (Table 2). Compared to the ATCC 35752 strain, transformation efficiencies were 70 to a 500-fold less in the 9917 isolate, suggestive of important variations in the transformation efficiency of different M. chelonae isolates (Table 2). All M. chelonae transformants harboring xylE-containing plasmids (pJV53-xylE; pPR27-derived constructs) stained yellow upon spraying with catechol [22] indicating that XylE is a functional colored marker in this species. Interestingly, these transformations also revealed highly variable selection efficiencies for the kan cassette depending on the plasmid used (Table 2). Whereas Kan selection was very efficient with pOMK-zeo and pJV53-xylE in M. abscessus ATCC 19977, M. chelonae ATCC 35752 and M. chelonae 9917 (56 to a 100% of the tested KanR colonies contained the plasmids), less than 1% of the selected KanR M. chelonae ATCC 35752 CFUs actually contained plasmids when pPR27-based constructs were used. The selection efficacy of Gen upon transformation of pPR27-based constructs was 5 to 20-fold greater than that of Kan (Table 2). Overall, and similar to the situation in M. abscessus ATCC 19977 [16], selection with zeo was consistently the most efficient independent of the type of plasmid used (Table 2). That the variable selection efficacy of kan was due to the origin of replication and copy number of the plasmids is unlikely given that all of them harbor the mycobacterial origin of replication from pAL5000 (albeit, a temperature-sensitive version of it in the pPR27 plasmids). Most noticeable is the different sizes of the plasmids, which are greater than 12 kb for the pPR27 constructs compared to 7.7 kb for pOMK-zeo and 9.8 kb for pJV53-xylE. This difference in size may account, at least in part, for the lower transformation efficiencies observed with the larger pPR27 plasmids (Table 2).
Table 2. Comparative electrotransformation efficiency and spontaneous resistance to different antibiotics in M. chelonae strains ATCC 35752 and 9917, and M. abscessus ATCC 19977.
| Transformant | M. chelonae | M. chelonae | M. abscessus |
| ATCC 35752 | 9917 | ATCC 19977 | |
| pOMK-zeo | Kan: 2.3×105 | Kan: 394 | Kan: 3.0×103 |
| [100%] | [100%] | [100%] | |
| Zeo: 2.8×105 | Zeo: 3.6×103 | ||
| [100%] | [100%] | ||
| pPR27-4689-ZX | Zeo: 9 [100%] | nd | nd |
| Gen: 32 [20.3%] | |||
| pPR27-4690-ZX | Zeo: 4 [100%] | nd | nd |
| Gen: 27 [27.1%] | |||
| pPR27-4691-ZX | Zeo: 5 [100%] | nd | nd |
| Gen: 3 [17.2%] | |||
| pPR27-4689-KX | Kan: 335 [0.9%] | nd | nd |
| Gen: 18 [16.6%] | |||
| pPR27-4690-KX | Kan: 263 [0.8%] | nd | nd |
| Gen: 18 [5.6%] | |||
| pPR27-4691-KX | Kan: 311 [1.0%] | nd | nd |
| Gen: 23 [13.0%] | |||
| pJV53-xylE | Kan: 2.1×103 | Kan: 30 | Kan: 2.5×103 |
| [56.8%] | [100%] | [99.2%] |
Transformation efficiencies upon selection with the indicated antibiotics on 7H11-OADC agar are expressed as numbers of drug-resistant CFUs per µg of DNA electroporated. The percentage below each transformation efficiency value represents the percentage of Kan, Zeo or Gen-resistant CFUs confirmed to be actual transformants either by PCR (pOMK-zeo) or determination of their XylE phenotype (all other plasmids).
Taken together, the results of the experiments presented in Table 2 indicated that: (i) transformation efficiencies of M. chelonae vary with the isolate and the plasmid transformed but are up to 100 times greater than that of M. abscessus ATCC 19977; (ii) Gen, Kan and Zeo can all be used as selection markers in M. chelonae ATCC 35752; however, Zeo seems to be the least prone to spontaneous resistance; (iii) XylE is a functional colored marker in M. chelonae.
Construction of allelic exchange mutants using the Ts-sacB system
Both kan and zeo-disrupted alleles of the porin genes were constructed for use in allelic exchange experiments with the Ts-sacB system (Table 1). Attempts to grow M. chelonae ATCC 35752 at increasing temperatures set the maximum at which the temperature-sensitive pPR27 plasmids could be counterselected at about 37°C since no colonies formed on 7H11-OADC plates beyond this temperature (39°C). M. chelonae ATCC 35752 colonies appeared after 6 days at 37°C instead of 3 days at 30°C.
Transformation of M. chelonae ATCC 35752 with pPR27-4691-ZX followed by plating on 7H11-OADC-Zeo at 30°C or 37°C in the presence of absence of sucrose in the culture medium allowed the counterselection efficacy of the Ts-sacB system to be determined. Shifting the temperature from 30°C to 37°C resulted in a two-fold reduction in CFU counts while sucrose alone reduced CFU counts by about two orders of magnitude (Table 3). The combination of both counterselections showed a clear additive effect reducing CFU counts by 3 to 4 orders of magnitude. Thus, although about 10 times less efficient than the counterselection efficiency measured in M. smegmatis [16], [42], results were encouraging in that they indicated that, in spite of the relatively narrow temperature growth range of M. chelonae, the Ts-sacB plasmid could successfully be used to deliver allelic exchange substrates to the cells. This is in contrast to the situation in M. abscessus ATCC 19977 where the Ts-sacB system was shown not to be functional due to the lack of counterselective efficacy of sacB [16]. We thus next proceeded to the transformation of all six (Zeo and Kan) porin knock-out constructs in M. chelonae ATCC 35752 and selected for transformants at 30°C on Zeo or Kan plates. One to three XylE+ transformants of each were propagated in liquid medium at 30°C in the presence of Zeo or Kan and then serially diluted and plated onto agar plates containing 10% sucrose and Kan or Zeo at 37°C. The outcome of these experiments is summarized in Table 4. Results clearly illustrated the superiority of zeo over kan in the selection of double crossover mutants. Whereas, 50 to 100% of the sucrose resistant, ZeoR and XylE− clones isolated at the last selection step corresponded to knock-out mutants (Fig. 1B), in none but one case (MCH_4689; transformant T2) did the plating of KanR transformants yield mutants. The sucrose resistant/KanR/XylE− clones instead most likely corresponded to Kan spontaneous resistant clones that arose during the multiple culturing steps in the presence of Kan. The fact that MCH_4689 knock-out mutants were isolated upon plating of the pPR27-4689-KX transformant T2 suggests that double crossover events probably occurred earlier in this particular culture relative to the development of spontaneous resistance facilitating the detection of knock-out mutants at the final selection step.
Table 3. Counterselection efficiency of the Ts-sacB system in M. chelonae ATCC 35752.
| Recovered colonies | |||||
| Transformant | 7H11-OADC | 7H11-OADC | 7H11-OADC | 7H11-OADC | Counterselection |
| Zeo (30°C) | Zeo (37°C) | Zeo/Suc (30°C) | Zeo/Suc (37°C) | efficiency (a) | |
| MCH (ATCC) | 9.7×108 | 4.1×108 | 1.6×107 | 4.9×105 | 3.38×10−4 |
| pPR27-4691-ZX | (+/−1.5×10−4) | ||||
The experiment was conducted on three independent transformants and mean counterselection efficiencies +/− standard deviations are indicated.
Table 4. Comparative efficiency of the Ts-sacB system using zeo and kan disrupted allelic exchange substrates in M. chelonae ATCC 35752.
| Transformant | % of XylE− SucR | Number of XylE− | Number of |
| ZeoR or KanR CFUs | CFUs analyzed by | confirmed double | |
| PCR | crossover mutants | ||
| pPR27-4689-ZX | |||
| T1 | 100% | 4 | 3 |
| pPR27-4690-ZX | |||
| T1 | 100% | 4 | 3 |
| T2 | 11% | 4 | 2 |
| pPR27-4691-ZX | |||
| T1 | 49% | 4 | 4 |
| T2 | 29% | 4 | 3 |
| pPR27-4689-KX | |||
| T1 | 100% | 5 | 0 |
| T2 | 74% | 8 | 8 |
| T3 | 100% | 5 | 0 |
| pPR27-4690-KX | |||
| T1 | 100% | 8 | 0 |
| T2 | 99% | 10 | 0 |
| T3 | 100% | 8 | 0 |
| pPR27-4691-KX | |||
| T1 | 100% | 10 | 0 |
| T2 | 100% | 8 | 0 |
One to three transformants (T1, T2 and T3) were selected on plates upon transformation with the pPR27-derived plasmids, grown in 7H9-OADC broth at 30°C for 5 to 7 days, and finally plated onto 7H11-OADC containing Kan or Zeo and 10% sucrose at 37°C. The percentage of CFUs presenting the expected phenotype for allelic exchange mutants at the last selection step of the Ts-SacB procedure (sucrose resistant; KanR or ZeoR and XylE−) is indicated for each construct. Four to ten candidate mutants were analyzed by PCR in each case and the number of double crossover mutants identified is indicated in the last column.
Construction of allelic exchange mutants using the recombineering system
The same MCH_4689c, MCH_4690c and MCH_4691c zeo cassette-disrupted alleles as the ones used in the Ts-SacB system were used in the recombineering system. Care was taken in preparing the linear DNA substrates to generate fragments with incompatible ends that would not recircularize in the bacteria, in order to avoid the selection of single crossover events. All three allelic exchange substrates were electroporated in M. chelonae ATCC 35752 harboring the pJV53-xylE plasmid (Table 1) and transformants selected on Zeo plates at 30°C. Twenty-nine (MCH_4689c), 65 (MCH_4690c) and 57 (MCH_4691c) ZeoR colonies were obtained in a typical transformation experiment with 300 ng of linear DNA. The number of transformants, however, increased three-fold when the quantity of linear DNA substrate was increased from 300 ng to 1 µg indicating that saturation conditions had not been reached. Ten to fourteen clones were picked for each gene and propagated in liquid broth prior to genomic DNA isolation. PCR analysis followed by sequencing confirmed that allelic replacement had occurred in 36% of the MCH_4691c mutant candidates, 80% of the MCH_4690c candidates and 30% of the MCH_4689c candidates (Fig. 1B). Therefore, more than 30% of the ZeoR transformants corresponded to allelic exchange mutants, a selection efficiency lower than that reported for M. smegmatis (>90%) [16], [20] but greater than that reported for M. abscessus ATCC 19977 (7%) [16].
Because of concerns that genetic rearrangements may occur in the knock-out mutants as a result of the retention of the recombinase expression plasmid pJV53-xylE, attempts were then made to cure this plasmid from the porin mutants by culturing on medium devoid of Kan. Interestingly, the direct selection of recombineering mutants on Zeo-containing plates in the absence of Kan yielded ZeoR colonies, 15 to 26% of which were XylE− and Kan-susceptible, indicative of the loss of pJV53-xylE. In the case of knock-out mutants that had retained XylE positivity and Kan resistance, two passages in liquid broth devoid of Kan followed by plating on Zeo-containing agar were sufficient to yield cultures entirely cured of the plasmid. PCR amplification of the kan cassette further confirmed that the pJV53-xylE plasmid had been lost from the selected porin mutants. Thus, in the absence of Kan selective pressure, the pJV53-xylE plasmid can efficiently be cured either during the mutant selection step or upon one or two passages of the selected knock-out clones in medium devoid of Kan.
Porin production in each of the mutant strains was analyzed by immunoblotting using polyclonal antibodies raised against the purified MspA protein of M. smegmatis [23]. The mature MCH_4691c, MCH_4690c and MCH_4689c porin products have an expected molecular weight of about 19.6 KDa and display about 73% amino acid identity with the mature MspA protein. Of all mutants, the MCH_4691c knock-out strain was the one with the lowest porin expression which was also significantly (approximately 50%) less than in the wild-type parent strain (Fig. 1C). In contrast, relative to the wild-type parent, porin production was similar in the MCH_4689c mutant (94.4% of the wild-type porin content) and slightly increased in the MCH_4690c mutant (130% of the wild-type porin content) (Fig. 1C). Thus, M. chelonae appears to compensate for the disruption of MCH_4690c and MCH_4689c by overexpressing one or two of the remaining porin genes. Complementation of the MCH_4691c knock-out mutant with MCH_4691c expressed from the replicative multicopy plasmid pOMK restored porin production beyond wild-type levels (Fig. 1C).
Growth rate, glucose uptake and biocide susceptibility of the M. chelonae porin knock-out mutants
The growth rate of the MCH_4691c mutant in 7H9-OADC-Tween 80 broth was slightly but reproducibly decreased compared to the wild-type strain and other knock-out mutants (Fig. 2). Wild-type growth was restored in the MCH_4691c mutant upon complementation with MCH_4691c expressed from pOMK (Fig. 2).
Figure 2. Growth rates of wild-type M. chelonae ATCC 35752, its isogenic porin knock-out mutants and complemented MCH_4691 mutant strains in 7H9-OADC-Tween 80 broth at 30°C.
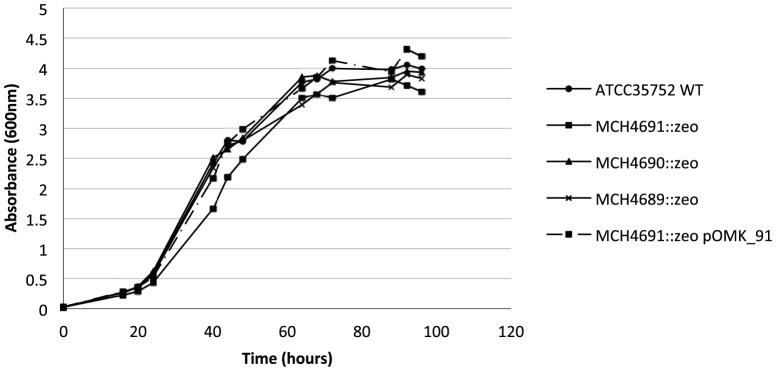
Shown are representative results of two to three independent experiments using different culture batches.
Consistent with decreased porin expression, the MCH_4691c mutant was also 2.3-fold less proficient at taking up [14C]-glucose than wild-type M. chelonae ATCC 35752 (Fig. 3). Glucose uptake rates were restored beyond wild-type levels in the MCH_4691c mutant complemented with MCH_4691c or with the three-porin gene cluster (Fig. 3). The MCH_4690c and MCH_4689c mutants that produce slightly more or equivalent amounts of porins as the M. chelonae ATCC 35752 strain (Fig. 1C) consistently showed 1.6 and 1.2-fold increased [14C]-glucose uptake rates compared to their wild-type parent.
Figure 3. Glucose uptake by M. chelonae ATCC 35752 and its isogenic porin knock-out mutants.
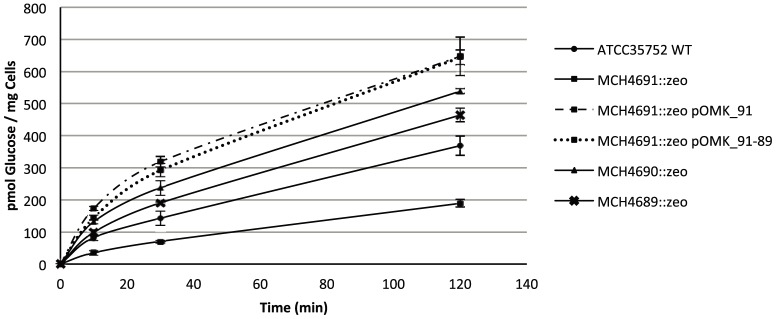
The accumulation of [U-14C]glucose by the strains over time was measured as described under Materials and Methods. Glucose uptake rates were calculated on the first 10 min of the reactions. Uptake experiments were performed in triplicates and are shown with their standard deviations.
The wild-type and mutant strains, however, did not differ in terms of their susceptibility to tetracycline (MIC = 16–32 µg/ml), ethambutol (MIC = 8–16 µg/ml), chloramphenicol (MIC = 8–16 µg/ml), erythromycin (MIC = 4–8 µg/ml), linezolid (MIC = 8–16 µg/ml), and rifampicin (MIC = 256 µg/ml).
Altogether, the results suggest that MCH_4691c is the main porin of M. chelonae ATCC 35752 under the culture conditions used in this study. That polar effects of MCH_4691c disruption on the expression of downstream porin genes account for this result is unlikely given the 115-bp of intergenic space separating MCH_4691c from MCH_4690c and the 222-bp separating MCH_4690c from MCH_4689c. The presence of an A/T-rich region 60 bp upstream from the start codon of MCH_4690c further supports the existence of a promoter region upstream this gene.
Conclusions
In conclusion, both the Ts-sacB and the recombineering homologous recombination systems can be used to inactivate genes in M. chelonae as long as efficient antibiotic resistance selection markers are used. The genetic methodologies described here open the way to the genetic dissection of key aspects of the physiology, biocide resistance and virulence of M. chelonae. The detailed characterization of the roles of each of the three porins of M. chelonae ATCC 35752 in the physiology and virulence of this bacterium is out of the scope of the present work but further experiments have begun in our laboratory to study their expression and regulation, and the effects that their combined inactivation might have on bacterial growth, biocide susceptibility and virulence.
Supporting Information
Details of the gene replacement protocols used in M. chelonae ATCC 35752. See text for further details.
(PDF)
Sequence alignment of the three porins from M. chelonae ATCC 35752.
(PDF)
Acknowledgments
We thank Dr. William Jacobs Jr. (Albert Einstein College of Medicine, Bronx, New York) for kindly providing us with phages TM4 and D29, Dr. Graham Hatfull (University of Pittsburgh, Pennsylvania) for the generous gift of plasmid pJV53, and Dr. Michael Niederweis (University of Alabama at Birmingham) for the anti-MspA antibodies.
Funding Statement
This work was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases grant AI089718. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Brown-Elliott BA, Wallace RJ Jr (2002) Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin Microbiol Rev 15: 716–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Petrini B (2006) Mycobacterium abscessus: an emerging rapid-growing potential pathogen. APMIS 114: 319–328. [DOI] [PubMed] [Google Scholar]
- 3. Kusunoki S, Ezaki T (1992) Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica, et al.) to species status: Mycobacterium abscessus comb. nov. Int J Syst Bacteriol 42: 240–245. [DOI] [PubMed] [Google Scholar]
- 4. Adékambi T, Berger P, Raoult D, Drancourt M (2006) rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int J Syst Evol Microbiol 56: 133–143. [DOI] [PubMed] [Google Scholar]
- 5. Adékambi T, Reynaud-Gaubert M, Greub G, Gevaudan M-J, La Scola B, et al. (2004) Amoebal coculture of “Mycobacterium massiliense” sp. nov. from the sputum of a patient with hemoptoic pneumonia. J Clin Microbiol 42: 5493–5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Leao SC, Tortoli E, Euzeby JP, Garcia MJ (2011) Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus . Int J Syst Evol Microbiol 61: 2311–2313. [DOI] [PubMed] [Google Scholar]
- 7. De Groote MA, Huitt G (2006) Infections due to rapidly growing mycobacteria. Clin Infect Dis 42: 1756–1763. [DOI] [PubMed] [Google Scholar]
- 8. Griffith DE, Girard WM, Wallace Jr RJ (1993) Clinical features of pulmonary disease caused by rapidly growing mycobacteria. An analysis of 154 patients. Am Rev Respir Dis 147: 1271–1278. [DOI] [PubMed] [Google Scholar]
- 9. Tortoli E, Gabini R, Galanti I, Mariottini A (2008) Lethal Mycobacterium massiliense sepsis, Italy. Emerg Infect Dis 14: 984–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Talati NJ, Rouphael N, Kuppalli K, Franco-Paredes (2008) Spectrum of CNS disease caused by rapidly growing mycobacteria. Lancet Infect Dis 8: 390–398. [DOI] [PubMed] [Google Scholar]
- 11. Duarte RS, Lourenco MCS, de Souza Fonseca LS, Leao SC, Amorim ELT, et al. (2009) An epidemic of postsurgical infections caused by Mycobacterium massiliense . J Clin Microbiol 47: 2149–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prevots DR, Shaw PA, Strickland D, Jackson LA, Raebel MA, et al. (2010) Nontuberculous mycobacterial lung disease prevalence at four integrated health care delivery systems. Am J Respir Crit Care Med 182: 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bryant JM, Grogono DM, Greaves D, Foweraker J, Roddick I, et al. (2013) Whole-genome sequencing to identify transmission of Mycobacterium abscessus between patients with cystic fibrosis: a retrospective cohort study. Lancet 381: 1551–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Aitken ML, Limaye A, Pottinger P, Whimbey E, Goss CH, et al. (2012) Respiratory outbreak of Mycobacterium abscessus subspecies massiliense in a lung transplant and cystic fibrosis center. Am J Respir Crit Care Med 185: 231–232. [DOI] [PubMed] [Google Scholar]
- 15. Ripoll F, Pasek S, Schenowitz C, Dossat C, Barbe V, et al. (2009) Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus . PLoS ONE 4: e5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Medjahed H, Reyrat J-M (2009) Construction of Mycobacterium abscessus defined glycopeptidolipid mutants: Comparison of genetic tools. Appl Environ Microbiol 75: 1331–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cortes M, Singh AK, Reyrat JM, Gaillard JL, Nassif X, et al. (2011) Conditional gene expression in Mycobacterium abscessus . PLoS One 6: e29306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pelicic V, Jackson M, Reyrat JM, Jacobs Jr WR, Gicquel B, et al. (1997) Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis . Proc Natl Acad Sci USA 94: 10955–10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, et al. (1997) Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis . Proc Natl Acad Sci USA 94: 10961–10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Kessel JC, Hatfull GF (2007) Recombineering in Mycobacterium tuberculosis . Nat Methods 4: 147–152. [DOI] [PubMed] [Google Scholar]
- 21. Svetlíková Z, Škovierová H, Niederweis M, Gaillard J-L, McDonnell G, et al. (2009) The role of porins in the susceptibility of Mycobacterium smegmatis and Mycobacterium chelonae to aldehyde-based disinfectants and drugs. Antimicrob Agents Chemother 53: 4015–4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson M, Camacho LR, Gicquel B, Guilhot C (2001) Gene replacement and transposon delivery using the negative selection marker sacB In: Parish T, Stocker NG, editors. Mycobacterium tuberculosis protocols. vol. 54. Humana Press, Totowa N J. pp. 59–75. [DOI] [PubMed]
- 23. Stahl C, Kubetzko S, Kaps I, Seeber S, Engelhardt H, et al. (2001) MspA provides the main hydrophilic pathway through the cell wall of Mycobacterium smegmatis . Mol Microbiol 40: 451–464. [DOI] [PubMed] [Google Scholar]
- 24. Jarlier V, Nikaido H (1994) Mycobacterial cell wall: Structure and role in natural resistance to antibiotics. FEMS Microbiol Lett 123: 11–18. [DOI] [PubMed] [Google Scholar]
- 25. Brennan PJ, Nikaido H (1995) The envelope of mycobacteria. Annu Rev Biochem 64: 29–63. [DOI] [PubMed] [Google Scholar]
- 26. Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, et al. (2008) Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 190: 5672–5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H (2008) Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA 105: 3963–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Niederweis M (2003) Mycobacterial porins - new channel proteins in unique outer membranes. Mol Microbiol 49: 1167–1177. [DOI] [PubMed] [Google Scholar]
- 29. Stephan J, Bender J, Wolschendorf F, Hoffmann C, Roth E, et al. (2005) The growth rate of Mycobacterium smegmatis depends on sufficient porin-mediated influx of nutrients. Mol Microbiol 58: 714–730. [DOI] [PubMed] [Google Scholar]
- 30. Stephan J, Mailaender C, Etienne G, Daffé M, Niederweis M (2004) Multidrug resistance of a porin deletion mutant of Mycobacterium smegmatis . Antimicrob Agents Chemother 48: 4163–4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Danilchanka O, Pavlenok M, Niederweis M (2008) Role of porins for uptake of antibiotics by Mycobacterium smegmatis . Antimicrob Agents Chemother 52: 3127–3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frenzel E, Schmidt S, Niederweis M, Steinhauer K (2011) Importance of porins for biocide efficacy against Mycobacterium smegmatis . Appl Environ Microbiol 77: 3068–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rodrigues L, Ramos J, Couto I, Amaral L, Viveiros M (2011) Ethidium bromide transport across Mycobacterium smegmatis cell-wall: correlation with antibiotic resistance. BMC Microbiol 11: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharbati-Tehrani S, Stephan J, Holland G, Appel B, Niederweis M, et al. (2005) Porins limit the intracellular persistence of Mycobacterium smegmatis . Microbiology 151: 2403–2410. [DOI] [PubMed] [Google Scholar]
- 35. Fabrino DL, Bleck CKE, Anes E, Hasilik A, Melo RCN, et al. (2009) Porins facilitate nitric oxide-mediated killing of mycobacteria. Microb Infect 11: 868–875. [DOI] [PubMed] [Google Scholar]
- 36. Achouak W, Heulin T, Pages J-M (2001) Multiple facets of bacterial porins. FEMS Microbiol Lett 199: 1–7. [DOI] [PubMed] [Google Scholar]
- 37. Hillmann D, Eschenbacher I, Thiel A, Niederweis M (2007) Expression of the major porin gene mspA is regulated in Mycobacterium smegmatis . J Bacteriol 189: 958–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sharbati S, Schramm K, Rempel S, Wang H, Andrich R, et al. (2009) Characterisation of porin genes from Mycobacterium fortuitum and their impact on growth. BMC Microbiol 9: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trias J, Benz R (1993) Characterization of the channel formed by the mycobacterial porin in lipid bilayer membranes. Demonstration of voltage gating and of negative point charges at the channel mouth. J Biol Chem 268: 6234–6240. [PubMed] [Google Scholar]
- 40. Trias J, Jarlier V, Benz R (1992) Porins in the cell wall of mycobacteria. Science 258: 1479–1481. [DOI] [PubMed] [Google Scholar]
- 41. Rybniker J, Kramme S, Small PL (2006) Host range of 14 mycobacteriophages in Mycobacterium ulcerans and seven other mycobacteria including Mycobacterium tuberculosis - application for identification and susceptibility testing. J Med Microbiol 55: 37–42. [DOI] [PubMed] [Google Scholar]
- 42. Pelicic V, Reyrat JM, Gicquel B (1996) Expression of the Bacillus subtilis sacB gene confers sucrose sensitivity on mycobacteria. J Bacteriol 178: 1197–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jackson M, Berthet FX, Otal I, Rauzier J, Martin C, et al. (1996) The Mycobacterium tuberculosis purine biosynthetic pathway: isolation and characterization of the purC and purL genes. Microbiology 142: 2439–2447. [DOI] [PubMed] [Google Scholar]
- 44.Heinz C, Roth E, Niederweis M (2003) Purification of porins from Mycobacterium smegmatis. In: Selinsky BS, editor. Methods in Molecular Biology - Membrane Protein Protocols: Expression, Purification and Characterization. Totowa, NJ: Humana Press Inc. pp. 139–150. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details of the gene replacement protocols used in M. chelonae ATCC 35752. See text for further details.
(PDF)
Sequence alignment of the three porins from M. chelonae ATCC 35752.
(PDF)