Abstract
Differentiation therapy is a means to treat cancer and is induced by different agents with low toxicity and more specificity than traditional ones. Diosgenin, a plant steroid, is able to induce megakaryocytic differentiation or apoptosis in human HEL erythroleukemia cells in a dose-dependent manner. However, the exact mechanism by which diosgenin induces megakaryocytic differentiation has not been elucidated. In this study, we studied the involvement of Sonic Hedgehog in megakaryocytic differentiation induced by diosgenin in HEL cells. First, we showed that different elements of the Hedgehog pathway are expressed in our model by qRT-PCR. Then, we focused our interest on key elements in the Sonic Hedgehog pathway: Smoothened receptor, GLI transcription factor and the ligand Sonic Hedgehog. We showed that Smoothened and Sonic Hedgehog were overexpressed in disogenin-treated cells and that GLI transcription factors were activated. Then, we showed that SMO inhibition using siSMO or the GLI antagonist GANT-61, blocked megakaryocytic differentiation induced by diosgenin in HEL cells. Furthermore, we demonstrated that Sonic Hedgehog pathway inhibition led to inhibition of ERK1/2 activation, a major physiological pathway involved in megakaryocytic differentiation. In conclusion, our study reports, for the first time, a crucial role for the Sonic Hedgehog pathway in diosgenin-induced megakaryocytic differentiation in HEL cells.
Introduction
Blockage of differentiation or maturation arrest generates myeloid leukemia with genetic lesions in cells. In leukemic cells, this blockage results continuous proliferation, prevention of terminal differentiation and protection from cell death observed in normal blood cells [1]. Differentiation therapy is a potentially less toxic cancer therapy that involves the use of agents, alone or in combination, that modify differentiation and growth of cancer cells [2].
Diosgenin, a plant steroid, has various actions including anti-inflammatory and anti-thrombotic activities as well as anticancer properties [3]–[5]. Previously, Beneytout et al. demonstrated that 10µM diosgenin induced megakaryocytic differentiation of HEL cells with increased cell size, nuclear complexity and glycoprotein Ib (GpIb) expression [6]. Recently, we demonstrated that diosgenin-differentiated cells showed nuclear polyploidization and increased expression of the platelet marker CD41 associated with diminution of the erythroid marker glycophorin A (GpA) [7], [8]. Diosgenin induced megakaryocytic differentiation of HEL cells through combined ERK activation and inhibition of the p38 MAPK pathways [7], [9]. Inhibition of ERK activation by a MEK inhibitor abrogated diosgenin-induced differentiation [7].
The Hedgehog (Hh) family of secreted intercellular signaling proteins is essential for the development of many tissues during embryogenesis and is also involved in homeostasis of adult tissues, including skin, gut, bone, and thymus [10]–[14].
In vertebrates, there are three Hh-ligands, Sonic (SHh), Indian (IHh) and Desert Hh (DHh) all closely related to Hh in Drosophila, that regulate a well-defined molecular-genetic signal transduction pathway [15], [16]. The Hh proteins share a common signaling pathway which is initiated when the Hh ligand binds to its cell-surface receptor PATCHED (PTC) which abrogates suppression of the 7-transmembrane-helix protein Smoothened (SMO), the key player for signal transduction of the Hh pathway [17]. SMO activation leads to production of activating forms of the glioma-associated oncoproteins 1–3 (GLI1-3), the Hh transcription factors. Studies on the role of the Hh signaling pathway in hematopoiesis have led to conflicting results. In zebrafish, mutants of the Hh pathway have defects in hematopoietic stem cell (HSC) formation and definitive hematopoiesis [18]. It has also been reported that Hh is implicated in lymphocytic lineage commitment. In fact when PTC1 is suppressed, a defect in the population of the common lymphoid progenitors (CLP) is observed [19].
Given the importance of Hh signaling in tumor biology, a range of drugs has recently emerged to block this signaling pathway. Cyclopamine and jervine, two steroidal alkaloids, extracted from Veratrum californicum have a potent inhibitory activity against SMO [20], [21]. Other synthetic compounds have been developed against GLI transcription factors such as GANT-61 which acts downstream of SMO [22]. In a previous study, we showed that cyclopamine, but not jervine, inhibited cell proliferation and induced apoptosis in human erythroleukelia cell lines [23]. Clinical trials investigating the use of Hh inhibitors in patients have recently been initiated for different cancers such as prostate cancer, BCR-ABL–positive acute myeloid leukemia, pancreatic and ovarian cancer [24]–[27]. Because drug resistance appears after long-term Hh inhibition, combining Hh inhibitors with ionizing radiation, chemotherapy or other molecular targeted agents could represent an alternative therapeutic strategy. In fact, different studies suggested that vismodegib (selective hedgehog pathway inhibitor) combined with anticancer drugs increased their respective activities [24], [25].
In this study, we studied the role of SHh during diosgenin-induced megakaryocytic differentiation in the human erythroleukemia cell line HEL. Diosgenin activated SHh production leading to SMO expression and GLI activation. Inhibition of the SHh pathway confirmed that this pathway is involved during diosgenin-induced megakaryocytic differentiation.
Materials and Methods
Materials
RPMI 1640 medium, fetal calf serum (FCS) and penicillin/streptomycin were supplied by Gibco BRL (Cergy Pontoise, France). Diosgenin ((25R)-5-spirosten-3β-ol), siSMO and primers for RT-PCR were purchased from Sigma Aldrich (Saint Quentin Fallavier, France). GANT-61 was provided by Calbiochem (Fontenay-sous-bois, France).
Phospho-ERK1/ERK2 DuoSet IC ELISA was purchased from R&D Systems (Lille, France). Sonic Hedgehog Human ELISA Kit and SMO antibody were supplied from Abcam (Cambridge, MA). GAPDH antibody was purchased from Santa Cruz Biotechnology (Tebu-Bio, Le Perray en Yvelines, France).
Cell lines, culture and treatment
The HEL cell line was kindly provided by Professor J.P. Cartron (INSERM U76, Paris, France) and came from ATCC-LGC Standards (Molsheim, France). The TF1a cell line was also purchased from ATCC-LGC Standards. Cells were seeded at 105 cells/ml in tissue culture flasks, grown in RPMI-1640 medium supplemented with 10% FCS, 1% sodium pyruvate, 1% HEPES, 100 U/ml penicillin and 100 µg/ml streptomycin. Cultures were maintained in a humidified 5% CO2 atmosphere with at 37 °C. Cells grown for 24 h in culture medium prior to exposure or not to 10 µM diosgenin. For pretreatments 5 µM GANT-61 were added in culture medium for 48 h before subsequent drug treatment. The same amount of vehicle (<0.1% DMSO or ethanol) was added to control cells. Cell viability was determined by trypan blue dye exclusion.
RNA extraction and semi-quantitative RT-PCR analysis
Total RNA was extracted by RNeasy Mini Kit (Qiagen, Courtaboeuf, France) from treated and control cells. 2 µg of total RNA were transcribed into cDNA using the Omniscript RT kit (Qiagen), and 2 µl of the reverse-transcribed cDNA were used for PCR using the HotStarTaq DNA polymerase mix kit (Qiagen) with 20 pmol of human sense and antisense primers (Table 1).
Table 1. Characteristics of RT-PCR primers used for SHh signaling studies.
| Target gene | Primer sequence | Size of PCR product | Temperature of hybridation |
| Human 18S | Forward: GCTGGAATTACCGCGGCTGCT Reverse: CGGCTACCACATCCAAGGAAGG | 186 | 58°C |
| Human SHh | Forward: CAGTGGACATCACCACGTCT Reverse: CCGAGTTCTCTGCTTTCACC | 138 | 60°C |
| Human SMO | Forward: CCCATCCCTGACTGTGAGAT Reverse: TTTGGCTCATCGTCACTCTG | 176 | 64°C |
| Human GLI1 | Forward: ACAGCCAGTGTCCTCGACTT Reverse: ATAGGGGCCTGACTGGAGAT | 197 | 60°C |
| Human GLI2 | Forward: GCGTGTTTACCCAATCCTGT Reverse: GATGCTCCCTCAGAGTCCTG | 265 | 60°C |
The number of amplification cycles was selected for each gene according to qPCR results, after the Ct value and before plateau levels e.g. during the exponential phase of amplification.
PCR resulting fragments were visualized by electrophoresis on a 1% agarose gel containing ethidium bromide.
PCR Microarray
Total RNA was extracted by RNeasy Mini Kit (Qiagen, Courtaboeuf, France) from treated and control cells. 1µg of total RNA was transcribed into cDNA using RT2 First Strand Kit (QIAGEN) and used for quantitative-PCR according to the RT2 profiler PCR array « human hedgehog signaling pathway » (QIAGEN). Relative levels of mRNA gene expression were calculated using the 2−ΔΔCt method [28].
Sonic Hedgehog immunoassay
Sonic Hedgehog production was assessed in cell culture supernatants using the “Sonic Hedgehog Human ELISA kit” (Abcam) according to the manufacturer's instructions.
Protein expression
After treatment, cells were washed and lysed in RIPA lysis buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1% deoxycholate, 1% NP-40, 0.1% SDS, 20 µg/ml aprotinin) containing protease inhibitors (Complete Mini, Roche Diagnostics, Meylan, France). Briefly, as previously described [29], proteins (20–50 µg) were separated by electrophoresis on SDS-polyacrylamide gels, transferred to PVDF membranes (Amersham Pharmacia Biotech, Saclay, France) and probed with respective human antibodies against SMO (Abcam, Cambridge, MA), and GAPDH (Santa Cruz Biotechnology, Tebu-Bio, Le Perray en Yvelines, France). After incubation with secondary antibodies (Dako France S.A.S., Trappes, France), blots were developed using the ECL Plus Western Blotting Detection System (Amersham Pharmacia Biotech) and visualized with the G: BOX system (Syngene, Ozyme, Saint-Quentin en Yvelines, France). Membranes were then reblotted with anti-GAPDH used as a loading control.
Subcellular protein fractionation
After treatment, HEL and TF1a cells were incubated alone or with diosgenin, or GANT-61. Cytosolic and nuclear fractions were obtained using the Subcellular Protein Fractionation Kit according to the manufacturer's protocol (Thermo Fischer Scientific, Rockford, IL, USA) as previously described [30].
Electromobility shift assay (EMSA)
EMSA experiments were performed using the DIG Gel Shift Kit (Roche Diagnostics). Briefly, nuclear extracts were prepared from cells treated or not with 10µM diosgenin or 5 µM GANT-61. GLI binding reactions were carried out with 5 µg nuclear proteins incubated with digoxigenin (DIG) labeled GLI probe according to the manufacturer's protocol. GLI consensus probe sequence was: forward: 5′-CTCCCGAAGACCACCCACAATGAT-3′, and reverse: 5′-ATCATTGTGGGTGGTCTTCGGGAG-3′. The samples were loaded on a 5% native polyacrylamide gel in Tris-Borate-EDTA buffer. After transfer to nylon membranes and incubation with anti-DIG antibody conjugated with alkaline phosphatase, gel mobility shift was visualized by incubation with CSPD chemiluminescence reagent and detected by the G: BOX system. Quantification of each band was performed by densitometry analysis software with respect to band intensity and band area. Results were expressed relative to controls in arbitrary units.
Evaluation of nuclear ploidy
For DNA content analysis, after treatment, cells were fixed and permeabilized in 70% ethanol in phosphate-buffered saline (PBS) at −20 °C overnight, washed in PBS, treated with RNase (40 U/μl, Boehringer Mannheim, Meylan, France) for 1 h at room temperature and stained with propidium iodide (PI) (50 µg/ml). Flow cytometry analyses (FC) were performed as previously described [31].
SMO-specific short-interfering RNA (siRNA) transfection
SMO-specific siRNA (siSMO) were obtained from Sigma Aldrich (Saint-Quentin Fallavier, France) and transfected by electroporation using the AMAXA Nucleofactor system (Lonza, Basel, Switzerland) at 100 nM for 72 h as recommended. SMO silencing was confirmed by western blot.
Statistical analysis
Data are expressed as the arithmetic means ± standard error of mean (SEM) of separate experiments. The statistical significance of results obtained from in vitro studies was evaluated by the two tailed unpaired Student's t-test, with P<0.05 being considered as significant.
Results
Diosgenin modulates Hedgehog gene expression during megakaryocytic differentiation in HEL cells
First, we analyzed gene expression of major actors of the Hh pathway using the RT2 profiler PCR array « human hedgehog signaling pathway » (QIAGEN) (Fig. 1). We observed that transcription factors GLI1 and GLI3 were upregulated at 24 and 48 h in diosgenin-treated cells. However, GLI1 upregulation was more pronounced than GLI3 at 48 h, while GLI2 upregulation was not statistically significant. We also found that Hh ligands, DHh, IHh and SHh, were overexpressed at 24 and 48 h. Of note, SHh expression was higher compared to DHh and IHh after 48 h treatment. Receptor genes, including SMO and PTC, were overexpressed except for PTCD3 at both times. As for the negative regulator of the Hh pathway, SUFU, we observed strong upregulation after 48 h treatment.
Figure 1. Effect of diosgenin on human Hedgehog signaling pathway gene expression after 24 and 48 h treatment.
Cells were treated or not (control) with 10 µM diosgenin for 24 and 48 h then total RNA was extracted. 1µg of total RNA were transcribed into cDNA and used for quantitative-PCR according to the RT2 profiler PCR array « human hedgehog signaling pathway ». Relative levels of mRNA gene expression were calculated using the 2−ΔΔCt method versus untreated cells. Each value represents the mean ± SEM of three separate experiments.
Taken together, these results suggest that during diosgenin-induced megakaryocytic differentiation in HEL cells, there was an overall upregulation of gene expression of the main actors in the Hh pathway, especially GLI1, SMO, SHh and SUFU.
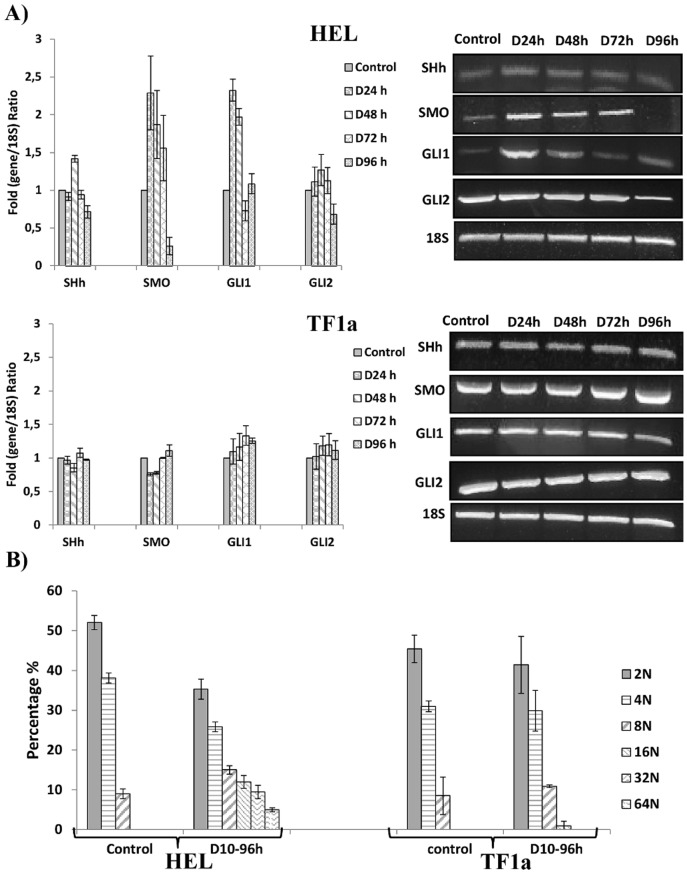
In order to confirm results from the PCR array and to determine whether the SHh pathway was involved throughout diosgenin-induced megakaryocytic differentiation in HEL cells, we analyzed gene expression of SHh, SMO, GLI1 and GLI2 in HEL and in TF1a cell lines after 24 h to 96 h diosgenin treatment (Fig. 2A) by semi-quantitative RT-PCR.
Figure 2. Effect of diosgenin on SHh, SMO and GLI1 gene expression during megakaryocytic differentiation in HEL and TF1a cell lines.
(A) Cells were treated or not with 10 µM diosgenin for 24, 48, 72 and 96 h then SHh, SMO and GLI1 genes expressions were evaluated. Total RNA was extracted and 2µg of total RNA were transcribed into cDNA and used for PCR. PCR resulting fragments were visualized by electrophoresis on a 1% agarose gel containing ethidium bromide. Quantification of SHh, SMO and GLI1 transcripts were normalized to 18S as an internal control. The agarose gels shown are representative of six separate experiments. (B) Cells were treated or not with 10µM diosgenin for 96 h and megakaryocytic differentiation was evaluated by analyzing nuclear ploidy. Cells were fixed and permeabilized in 70% ethanol in PBS at −20 °C overnight, washed in PBS, treated with RNase and stained with PI. Then, flow cytometric analyses (FC) were performed to analyze DNA content.
In diosgenin-treated HEL cells, we showed that SHh was overexpressed at 48 h. For other treatment times we did not detect significant changes in gene expression. As for SMO, we found that it was overexpressed from 24 h to 72 h and repressed after 96 h treatment. Furthermore, GLI1 was overexpressed from 24 to 48 h treatment. Of note no significant changes in GLI2 expression were observed.
On the other hand, in diosgnenin-treated TF1a cells, we did not observe any significant changes in SHh, SMO or GLI1-2 gene expression.
Next, we analyzed megakaryocytic differentiation of HEL and TF1a cells after diosgenin treatment by studying nuclear ploidy. Mekagaryocytic differentiation is characterized by progressive polyploidization that ranges from 8N to 128N in normal megakaryocytes. As shown in Fig. 2B, diosgenin increased nuclear ploidy up to 64N in HEL cells after 96 h. In contrast, diosgenin did not induce increased nuclear ploidy in TF1a cells.
Our data document that only HEL cells were able to differentiate after diosgenin treatment with activation of the SHh pathway. In TF1a cells, diosgenin did not induce mergakaryocytic differentiation or SHh pathway activation. We speculated then, that the SHh pathway was involved in diosgenin-induced megakaryocytic differentiation.
Diosgenin induces SHh-N production, SMO overexpression and GLI activation
SHh protein is synthesized as a precursor of about 45 kDa, followed by autoproteolytic cleavage which generates an amino-terminal peptide that is linked to cholesterol at its carboxyl terminus. All signaling activities are mediated by this N-terminal peptide, but the C-terminal region is necessary to catalyse the autoproteolytic cleavage event [32]. We checked the effect of diosgenin on SHh-N production using the ‘Sonic Hedgehog Human ELISA Kit’. As shown in Fig. 3A, 10 µM diosgenin induced strong and sustained SHh-N production and secretion, starting at 12 h. This result suggests that the SHh pathway is activated during diosgenin-induced megakaryocytic differentiation in HEL cells.
Figure 3. Effect of diosgenin on SHh-N production, SMO expression and GLI1 activation in HEL cells.
Cells were treated with 10 µM diosgenin for 12, 24, 48, 72 and 96 h. (A) SHh-N production was evaluated in cell culture supernatanst using the “Sonic Hedgehog Human ELISA Kit”. Each value represents the mean ± SD of three separate experiments, * P< 0.05 diosgenin vs. control. (B) SMO expression was evaluated by Western blot analysis after diosgenin treatment. GAPDH was used as a loading control. (C) GLI1 activation was evaluated by electromobility shift assay using the DIG Gel Shift Kit. The blots shown are representative of five separate experiments.
Because secreted SHh-N can have autocrine or paracrine activity in physiological conditions we studied the activation of the SHh signaling pathway at different stages in diosgenin-treated cells. It is well known that in the presence of Hh ligands, the inhibitory activity of PTC on the positive transmembrane effector SMO is lost and ultimately results in modulation of the activity of the three GLI zinc finger transcription factors at target promoters [33]. SMO overexpression was confirmed at the protein level by western blotting as shown in Fig. 3B. This overexpression lasted from 12 h to 72 h after diosgenin treatment. Next, we examined the effect of diosgenin on GLI1 nuclear activation by gel shift (Fig. 3C). Our results demonstrated that 10 µM diosgenin increased GLI transcriptional activity starting at 12 h as shown by increased DNA binding to a consensus probe. This activation was prolonged until 48 h and then decreased at the end of HEL differentiation.
SHh pathway is implicated in megakaryocytic differentiation induced by diosgenin
As we showed that the SHh pathway was activated by diosgenin, we wanted to determine whether diosgenin-induced megakaryocytic differentiation of HEL cells was SHh-dependent. We used SMO siRNA or 5 µM GANT-61 during 48 h as a pretreatment. GANT-61 is a small molecule that specifically inhibits GLI [34]. Inhibition of SMO expression after silencing with siRNA or GANT-61 pretreatment was confirmed by western blotting (Fig. 4A) and inhibition of GLI1 activation after GANT-61 pretreatment was assessed by gel shift (Fig. 4B). After confirmation of SMO silencing and GLI inhibition we showed that, in these conditions, diosgenin failed to increase nuclear ploidy to more than 8N, and that cell ploidy remained principally 2N and 4N whereas diosgenin alone increased nuclear ploidy up to 64N (Fig.4C). As polyploidization is a hallmark of megakaryocytic differentiation, this illustrated that sustained SMO and GLI activation was involved in diosgenin-induced megakaryocytic differentiation.
Figure 4. Effects of SMO and GLI1 inhibition on diosgenin-induced megakaryocytic differentiation in HEL cells.
(A) Cells were transfected with siSMO or pretreated with 5µM GANT-61 for 48 h (G5) then treated with 10 µM diosgenin for 12, 24, 48, 72 and 96 h.SMO expression was evaluated by Western blot. GAPDH was used as a loading control. (B) Cells were pretreated with 5µM GANT-61 (G5) then treated with 10 µM diosgenin for 12, 24, 48, 72 and 96 h. GLI1 activation was evaluated by electromobility shift assay using the DIG Gel Shift Kit. The blots shown are representative of five separate experiments. (C) Cells were transfected with siSMO or pretreated with 5µM GANT-61 (G5) then treated with 10 µM diosgenin for 96 h. Megakaryocytic differentiation was assessed by analyzing nuclear ploidy. (D) Cells were transfected with siSMO or pretreated with 5µM GANT-61 (G5) then treated with 10 µM diosgenin for 5, 20 min, 1, 3, 6, 12 and 24 h. ERK1/2 phosphorylation was quantified using DuoSet IC assay kit. Each value represents the mean ± SEM of three separate experiments, * P< 0.05 siSMO+diosgenin or GANT-61+ diosgenin vs. diosgenin, # P< 0.05 diosgenin vs. control.
SMO silencing or GANT-61 pretreatment inhibit ERK1/2 activation
A recent study provided evidence for GLI3 and GLI1 as novel substrates of MAP-kinases (MAPKs) including the MEK substrate ERK2 [35]. On the other hand, it has been shown that Hh-binding to PTC1 stimulates ERK1/2 activation [36] Furthermore, it is also known that ERK1/2 activation is essential in normal mekacaryocytopoeisis and during its induction by diosgenin on HEL cells [7]. As shown in Fig. 4D, SMO silencing or GANT-61 pretreatment inhibited ERK1/2 activation induced by diosgenin. This result indicated that in our model SHh pathway activation acted upstream upstream to ERK.
Discussion
Differentiation therapy has emerged as a powerful method to target specific hematologic malignancies. One of the best examples is the use of retinoic acid which was one of the first substances used [37]. The mode of action of differentiating agents, whether chemical or natural, is generally to enable or activate signal transduction pathways normally activated by the binding of hematopoietic factors and promote transcription of genes regulating hematopoiesis [38]. Diosgenin is a natural product able to induce megakaryocytic differentiation in the erythroleukemia cell line HEL [4]–[6], but its mode of action is still unclear.
In the present report, we focused our interest on Hh pathways. These signaling pathways represent strong candidate targets to eradicate leukemia, as they convergently regulate hematopoietic stem cell proliferation and have been identified as putative modulators of leukemia stem cell behavior [39], [40]. Of these pathways, SHh signaling is gaining considerable attention as a therapeutic target for myeloid malignancies [40], [41], despite the fact that its role in normal and malignant human hematopoiesis remains poorly defined and controversial [42]–[44].
In our study, we showed that diosgenin induced SHh-N production in HEL cells which activated the SHh pathway. Indeed, this was confirmed by SMO overxpression and GLI activation. It is well known that in the presence of Hh ligands, the transmembrane receptor PTC releases its inhibition of another transmembrane protein SMO, allowing SMO to assume an active conformation. In this activated state, SMO transduces signals that cause the nuclear translocation of the GLI family of transcription factors, with ultimate influences on cell cycling [45].
The SHh protein undergoes an autocatalytic processing reaction that involves internal cleavage, the amino-terminal product of this cleavage receives a covalent cholesterol adduct and becomes active in signaling [46]. The diosgenin structure is related to that of cholesterol. It can be suggested that diosgenin could mimic the effect of physiological cholesterol and bind to SHh-N which in turn would active signal transduction. However, to date no direct binding of diosgenin to SHh-N has been demonstrated, further studies should address this point.
Megakaryocytopoiesis is a highly regulated phenomenon that involves a wide spectrum of cytokines and growth factors in physiological conditions. The role of Hh proteins in the regulation of hematopoiesis has proven controversial, with different experimental models supporting opposing interpretations. A role for IHh in erythropoiesis has been proposed based on ex vivo assays: the addition of recombinant IHh to visceral endoderm-depleted epiblasts reinstated vasculogenesis and primitive erythropoiesis, suggesting that IHh may be responsible for haematopoietic stem cell activation and differentiation during primitive haematopoiesis [47]. In adults, SHh has already been implicated in diverse types of differentiation such as osteogenesis [48]–[50].
There is a little evidence to demonstrate the importance of the SHh pathway in differentiation therapy for the treatment of hematological neoplasms. Here we characterized the blockage of megacaryocyctic differentiation induced by diosgenin in HEL cells after blocking the SHh pathway by siSMO. An antagonist of GLI (GANT-61) was also used to further examine SHh implication in diosgenin-induced megakaryocytic differentiation. Inhibition of two different targets of the SHh pathway blocked megakaryocytic differentiation induced by diosgenin in HEL cells.
In different studies, treatment with cyclopamine, a specific SMO inhibitor, induces monocytic differentiation [51] or eosinophilic differentiation [52] of HL-60 cells, but inhibition of the SHh pathway by cyclopamine was also implicated in blockage of erythroid differentiation [53].
As previously described, diosgenin induced megakaryocytic differentiation of the HEL cell line through sustained ERK activation and inhibition of p38 MAPK pathways [7]. In this study, we showed that SHh inhibition blocked diosgenin induced megakaryocytic differentiation and down-regulated ERK1/2 phosphorylation. It is well known that SHh activation leads to MAPK MEK-1/ERK pathway activation [54]. In addition, MEK-1 activation was shown to synergize with the canonical Hh pathway resulting in significant enhancement of GLI-dependent transcriptional activation [54].
Here, SHh pathway inhibition appears to affect megakaryocytic differentiation induced by diosgenin in HEL cells. Further investigations should determine the clinical applications of modulating the SHh pathway in treatment of hematological malignancies.
Conclusion
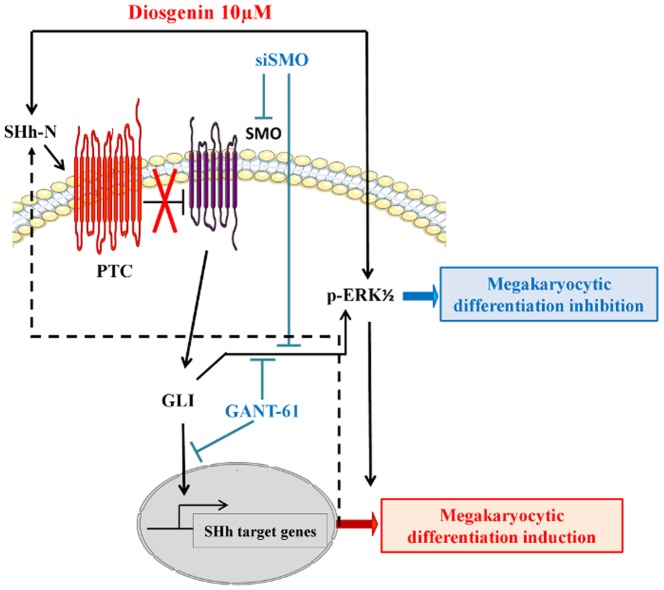
Our data document, for the first time, a crucial role for the Sonic Hedgehog pathway in diosgenin-induced megakaryocytic differentiation in HEL cells. Inhibiting key actors of this pathway, SMO and GLI, blocked diosgenin-induced megakaryocytic differentiation (Fig. 5). Furthermore, SHh pathway inhibition led to ERK1/2 inhibition (Fig. 5) whose activation was absolutely required to induce megakaryocytic differentiation as previously described [9].
Figure 5. Megakaryocytic differentiation mechanisms in diosgenin-treated cells in relation to the SHh pathway.
Diosgenin induced megakaryocytic differentiation in HEL cells. The SHh pathway is involved in diosgenin-induced megakaryocytic differentiation since its inhibition blocks this phenomenon. SMO inhibition by total silencing using siRNA or GLI1 inhibition using GANT-61, a specific inhibitor, blocked megakaryocytic differentiation induced by diosgenin. In addition, SHh pathway inhibition leads to ERK1/2 inhibition whose activation is absolutely required to induce megakaryocytic differentiation.
Acknowledgments
Authors are grateful to Pr J.P. Cartron (INSERM U76, Paris, France) for providing the HEL cell line.
Funding Statement
This research was supported by grants from the French Ministry of Education and Research and from the Conseil Régional du Limousin. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sell S (2005) Leukemia: stem cells, maturation arrest, and differentiation therapy. Stem Cell Rev 1: 197–205. [DOI] [PubMed] [Google Scholar]
- 2. Maeda S, Sachs L (1978) Control of normal differentiation of myeloid leukemic cells. XIII. Inducibility for some stages of differentiation by dimethylsulfoxide and its disassociation from inducibility by MGI. J Cell Physiol 94: 181–185. [DOI] [PubMed] [Google Scholar]
- 3. Gao M, Chen L, Yu H, Sun Q, Kou J, et al. (2012) Diosgenin down-regulates NF-kappaB p65/p50 and p38MAPK pathways and attenuates acute lung injury induced by lipopolysaccharide in mice. Int Immunopharmacol 15: 240–245. [DOI] [PubMed] [Google Scholar]
- 4. Das S, Dey KK, Dey G, Pal I, Majumder A, et al. (2012) Antineoplastic and apoptotic potential of traditional medicines thymoquinone and diosgenin in squamous cell carcinoma. PLoS One 7: e46641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen PS, Shih YW, Huang HC, Cheng HW (2011) Diosgenin, a steroidal saponin, inhibits migration and invasion of human prostate cancer PC-3 cells by reducing matrix metalloproteinases expression. PLoS One 6: e20164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beneytout JL, Nappez C, Leboutet MJ, Malinvaud G (1995) A plant steroid, diosgenin, a new megakaryocytic differentiation inducer of HEL cells. Biochem Biophys Res Commun 207: 398–404. [DOI] [PubMed] [Google Scholar]
- 7. Leger DY, Liagre B, Beneytout JL (2006) Role of MAPKs and NF-kappaB in diosgenin-induced megakaryocytic differentiation and subsequent apoptosis in HEL cells. Int J Oncol 28: 201–207. [PubMed] [Google Scholar]
- 8. Leger DY, Battu S, Liagre B, Beneytout JL, Cardot PJ (2006) Megakaryocyte cell sorting from diosgenin-differentiated human erythroleukemia cells by sedimentation field-flow fractionation. Anal Biochem 355: 19–28. [DOI] [PubMed] [Google Scholar]
- 9. Cailleteau C, Micallef L, Lepage C, Cardot PJ, Beneytout JL, et al. (2010) Investigating the relationship between cell cycle stage and diosgenin-induced megakaryocytic differentiation of HEL cells using sedimentation field-flow fractionation. Anal Bioanal Chem 398: 1273–1283. [DOI] [PubMed] [Google Scholar]
- 10. Crompton T, Outram SV, Hager-Theodorides AL (2007) Sonic hedgehog signalling in T-cell development and activation. Nat Rev Immunol 7: 726–735. [DOI] [PubMed] [Google Scholar]
- 11. Outram SV, Hager-Theodorides AL, Shah DK, Rowbotham NJ, Drakopoulou E, et al. (2009) Indian hedgehog (Ihh) both promotes and restricts thymocyte differentiation. Blood 113: 2217–2228. [DOI] [PubMed] [Google Scholar]
- 12. Zacharias WJ, Madison BB, Kretovich KE, Walton KD, Richards N, et al. (2011) Hedgehog signaling controls homeostasis of adult intestinal smooth muscle. Dev Biol 355: 152–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gritli-Linde A, Hallberg K, Harfe BD, Reyahi A, Kannius-Janson M, et al. (2007) Abnormal hair development and apparent follicular transformation to mammary gland in the absence of hedgehog signaling. Dev Cell 12: 99–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hwang S, Thangapandian S, Lee KW (2013) Molecular dynamics simulations of sonic hedgehog-receptor and inhibitor complexes and their applications for potential anticancer agent discovery. PLoS One 8: e68271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Echelard Y, Epstein DJ, St-Jacques B, Shen L, Mohler J, et al. (1993) Sonic hedgehog, a member of a family of putative signaling molecules, is implicated in the regulation of CNS polarity. Cell 75: 1417–1430. [DOI] [PubMed] [Google Scholar]
- 16. Krauss S, Concordet JP, Ingham PW (1993) A functionally conserved homolog of the Drosophila segment polarity gene hh is expressed in tissues with polarizing activity in zebrafish embryos. Cell 75: 1431–1444. [DOI] [PubMed] [Google Scholar]
- 17. Ingham PW, McMahon AP (2001) Hedgehog signaling in animal development: paradigms and principles. Genes Dev 15: 3059–3087. [DOI] [PubMed] [Google Scholar]
- 18. Gering M, Patient R (2005) Hedgehog signaling is required for adult blood stem cell formation in zebrafish embryos. Dev Cell 8: 389–400. [DOI] [PubMed] [Google Scholar]
- 19. Uhmann A, Dittmann K, Nitzki F, Dressel R, Koleva M, et al. (2007) The Hedgehog receptor Patched controls lymphoid lineage commitment. Blood 110: 1814–1823. [DOI] [PubMed] [Google Scholar]
- 20. Bryden MM, Evans HE, Keeler RF (1971) Cyclopia in sheep caused by plant teratogens. J Anat 110: 507. [PubMed] [Google Scholar]
- 21. Chen JK, Taipale J, Cooper MK, Beachy PA (2002) Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev 16: 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lauth M, Bergstrom A, Shimokawa T, Toftgard R (2007) Inhibition of GLI-mediated transcription and tumor cell growth by small-molecule antagonists. Proc Natl Acad Sci U S A 104: 8455–8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghezali L, Leger DY, Limami Y, Cook-Moreau J, Beneytout JL, et al. (2013) Cyclopamine and jervine induce COX-2 overexpression in human erythroleukemia cells but only cyclopamine has a pro-apoptotic effect. Exp Cell Res 319: 1043–1053. [DOI] [PubMed] [Google Scholar]
- 24. Gonnissen A, Isebaert S, Haustermans K (2013) Hedgehog signaling in prostate cancer and its therapeutic implication. Int J Mol Sci 14: 13979–14007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katagiri S, Tauchi T, Okabe S, Minami Y, Kimura S, et al. (2013) Combination of ponatinib with Hedgehog antagonist vismodegib for therapy-resistant BCR-ABL1-positive leukemia. Clin Cancer Res 19: 1422–1432. [DOI] [PubMed] [Google Scholar]
- 26. Rubin LL, de Sauvage FJ (2006) Targeting the Hedgehog pathway in cancer. Nat Rev Drug Discov 5: 1026–1033. [DOI] [PubMed] [Google Scholar]
- 27. Lin TL, Wang QH, Brown P, Peacock C, Merchant AA, et al. (2010) Self-renewal of acute lymphocytic leukemia cells is limited by the Hedgehog pathway inhibitors cyclopamine and IPI-926. PLoS One 5: e15262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29. Pinon A, Limami Y, Micallef L, Cook-Moreau J, Liagre B, et al. (2011) A novel form of melanoma apoptosis resistance: melanogenesis up-regulation in apoptotic B16-F0 cells delays ursolic acid-triggered cell death. Exp Cell Res 317: 1669–1676. [DOI] [PubMed] [Google Scholar]
- 30. Corbiere C, Liagre B, Terro F, Beneytout JL (2004) Induction of antiproliferative effect by diosgenin through activation of p53, release of apoptosis-inducing factor (AIF) and modulation of caspase-3 activity in different human cancer cells. Cell Res 14: 188–196. [DOI] [PubMed] [Google Scholar]
- 31. Corbiere C, Liagre B, Bianchi A, Bordji K, Dauca M, et al. (2003) Different contribution of apoptosis to the antiproliferative effects of diosgenin and other plant steroids, hecogenin and tigogenin, on human 1547 osteosarcoma cells. Int J Oncol 22: 899–905. [PubMed] [Google Scholar]
- 32. Perler FB (1998) Protein splicing of inteins and hedgehog autoproteolysis: structure, function, and evolution. Cell 92: 1–4. [DOI] [PubMed] [Google Scholar]
- 33. Taipale J, Beachy PA (2001) The Hedgehog and Wnt signalling pathways in cancer. Nature 411: 349–354. [DOI] [PubMed] [Google Scholar]
- 34. Stanton BZ, Peng LF (2010) Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol Biosyst 6: 44–54. [DOI] [PubMed] [Google Scholar]
- 35. Whisenant TC, Ho DT, Benz RW, Rogers JS, Kaake RM, et al. (2010) Computational prediction and experimental verification of new MAP kinase docking sites and substrates including Gli transcription factors. PLoS Comput Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chang H, Li Q, Moraes RC, Lewis MT, Hamel PA (2010) Activation of Erk by sonic hedgehog independent of canonical hedgehog signalling. Int J Biochem Cell Biol 42: 1462–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Leszczyniecka M, Roberts T, Dent P, Grant S, Fisher PB (2001) Differentiation therapy of human cancer: basic science and clinical applications. Pharmacol Ther 90: 105–156. [DOI] [PubMed] [Google Scholar]
- 38. Tsiftsoglou AS, Pappas IS, Vizirianakis IS (2003) Mechanisms involved in the induced differentiation of leukemia cells. Pharmacol Ther 100: 257–290. [DOI] [PubMed] [Google Scholar]
- 39. Sengupta A, Banerjee D, Chandra S, Banerji SK, Ghosh R, et al. (2007) Deregulation and cross talk among Sonic hedgehog, Wnt, Hox and Notch signaling in chronic myeloid leukemia progression. Leukemia 21: 949–955. [DOI] [PubMed] [Google Scholar]
- 40. Lin TL, Matsui W (2012) Hedgehog pathway as a drug target: Smoothened inhibitors in development. Onco Targets Ther 5: 47–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Irvine DA, Copland M (2012) Targeting hedgehog in hematologic malignancy. Blood 119: 2196–2204. [DOI] [PubMed] [Google Scholar]
- 42. Gao J, Graves S, Koch U, Liu S, Jankovic V, et al. (2009) Hedgehog signaling is dispensable for adult hematopoietic stem cell function. Cell Stem Cell 4: 548–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hofmann I, Stover EH, Cullen DE, Mao J, Morgan KJ, et al. (2009) Hedgehog signaling is dispensable for adult murine hematopoietic stem cell function and hematopoiesis. Cell Stem Cell 4: 559–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mar BG, Amakye D, Aifantis I, Buonamici S (2011) The controversial role of the Hedgehog pathway in normal and malignant hematopoiesis. Leukemia 25: 1665–1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hooper JE, Scott MP (2005) Communicating with Hedgehogs. Nat Rev Mol Cell Biol 6: 306–317. [DOI] [PubMed] [Google Scholar]
- 46. Mann RK, Beachy PA (2004) Novel lipid modifications of secreted protein signals. Annu Rev Biochem 73: 891–923. [DOI] [PubMed] [Google Scholar]
- 47. Dyer MA, Farrington SM, Mohn D, Munday JR, Baron MH (2001) Indian hedgehog activates hematopoiesis and vasculogenesis and can respecify prospective neurectodermal cell fate in the mouse embryo. Development 128: 1717–1730. [DOI] [PubMed] [Google Scholar]
- 48. Wu X, Walker J, Zhang J, Ding S, Schultz PG (2004) Purmorphamine induces osteogenesis by activation of the hedgehog signaling pathway. Chem Biol 11: 1229–1238. [DOI] [PubMed] [Google Scholar]
- 49. Beloti MM, Bellesini LS, Rosa AL (2005) Purmorphamine enhances osteogenic activity of human osteoblasts derived from bone marrow mesenchymal cells. Cell Biol Int 29: 537–541. [DOI] [PubMed] [Google Scholar]
- 50. Cai JQ, Huang YZ, Chen XH, Xie HL, Zhu HM, et al. (2012) Sonic hedgehog enhances the proliferation and osteogenic differentiation of bone marrow-derived mesenchymal stem cells. Cell Biol Int 36: 349–355. [DOI] [PubMed] [Google Scholar]
- 51. Bai LY, Weng JR, Lo WJ, Yeh SP, Wu CY, et al. (2012) Inhibition of hedgehog signaling induces monocytic differentiation of HL-60 cells. Leuk Lymphoma 53: 1196–1202. [DOI] [PubMed] [Google Scholar]
- 52. Takahashi T, Kawakami K, Mishima S, Akimoto M, Takenaga K, et al. (2011) Cyclopamine induces eosinophilic differentiation and upregulates CD44 expression in myeloid leukemia cells. Leuk Res 35: 638–645. [DOI] [PubMed] [Google Scholar]
- 53. Detmer K, Walker AN, Jenkins TM, Steele TA, Dannawi H (2000) Erythroid differentiation in vitro is blocked by cyclopamine, an inhibitor of hedgehog signaling. Blood Cells Mol Dis 26: 360–372. [DOI] [PubMed] [Google Scholar]
- 54. Riobo NA, Saucy B, Dilizio C, Manning DR (2006) Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci U S A 103: 12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]