Abstract
The ability to mature oocytes in vitro provides a tool for creating embryos by parthenogenesis, fertilization, and cloning. Unfortunately the quality of oocytes matured in vitro falls behind that of in vivo matured oocytes. To address this difference, transcriptional profiling by deep sequencing was conducted on pig oocytes that were either matured in vitro or in vivo. Alignment of over 18 million reads identified 1,316 transcripts that were differentially represented. One pathway that was overrepresented in the oocytes matured in vitro was for Wingless-type MMTV integration site (WNT) signaling. In an attempt to inhibit the WNT pathway, Dickkopf-related protein 1 was added to the in vitro maturation medium. Addition of Dickkopf-related protein 1 improved the percentage of oocytes that matured to the metaphase II stage, increased the number of nuclei in the resulting blastocyst stage embryos, and reduced the amount of disheveled segment polarity protein 1 protein in oocytes. It is concluded that transcriptional profiling is a powerful method for detecting differences between in vitro and in vivo matured oocytes, and that the WNT signaling pathway is important for proper oocyte maturation.
Introduction
In vitro maturation (IVM) of mammalian oocytes has long been a problematic step for the in vitro production of embryos. While there have been major advances in improving both oocyte maturation and embryo culture over the past decades, and oocytes can successfully be matured in vitro, the rate of maturation and extent of maturation is lower than in vivo matured oocytes. Maturation of pig oocytes in vitro is even lower as compared to other livestock species [1], [2]. While IVM pig oocytes are capable of being fertilized, developing to the blastocyst stage and producing offspring after transfer to a surrogate, the developmental competence of these oocytes is lower than in vivo produced oocytes; partially due to a lower number of nuclei in the resulting blastocyst [3], implying that the quality of the blastocyst is lower. Incomplete oocyte maturation can cause abnormal chromosome segregation and reduce the developmental capacity of the embryos [4], [5]. While much of the focus trying to manipulate and understand culture conditions has been post-fertilization, the area for the greatest gain might be during IVM. By improving the oocyte's overall health in vitro, the stage could be set for improved development by providing the oocytes most of what they need to be developmentally competent.
To address the discrepancy of the quality of in vitro versus in vivo matured oocytes, we transcriptionally profiled IVM and in vivo matured oocytes by using next generation Illumina sequencing. One of the pathways that was overrepresented in the IVM oocytes was the Wingless-type MMTV integration site (WNT) pathway (see data below).
The WNT pathway and WNT related proteins are associated with embryogenesis, cancer, embryonic stem cells, and signal transduction. There are many members of the WNT signaling pathway that are expressed in ovaries, oocytes and early embryos of mice [6]. Indeed, WNT related proteins Frizzled Family Receptors (FZD1, FZD4, and FZD5) are present at the oocyte to embryo transition [6] and WNT ligands (WNT1, 3, 5A and 7A/B) are present in the cytoplasm of early mouse embryos. In addition, the WNT pathway plays a role in the fate of stem cells [7] as stimulation of this pathway acts as stem cell growth factors [8]. Furthermore, WNT pathway components are responsible for meiotic spindle orientation in Caenorhabditis elegans [9]. Inhibition of the WNT signaling pathway during early embryo development is deleterious in the cow [10], but not the mouse [11]. Interestingly, Dickkopf-related protein 3 (DKK3: thought to be a negative regulator of the WNT signaling pathway) is present in the oocyte and early mouse embryo [6], but it is controversial as to whether or not it is directly involved in the WNT pathway [12], [13]. Since the FZD1 receptor is located on the mouse oocyte as it matures, WNT ligands may control gene expression through a WNT-initiated signaling cascade during oocyte maturation [6]. Harwood et al [6] maintain that all the molecules needed for activation of alternative WNT pathways exist in the growing oocyte; therefore, repression of WNT and regulation of WNT related gene expression can be controlled by cross-talk between the oocyte and granulosa cells to aid in the maturation of the oocyte. Little more is known about WNT pathways during oocyte growth and maturation. We propose that WNT ligands might play a role in regulating oocyte maturation in the pig. Here we characterize the response to inhibiting the WNT pathway by adding Dickkopf-related protein 1 (DKK1) to the maturation system.
Results
Illumina sequencing of MII oocytes produced through IVM or in vivo
In vitro and in vivo produced metaphase II oocytes were analyzed by Illumina deep sequencing that generated 100 bp reads. The reads were aligned to our porcine custom database [14], yielding 18,141,082 reads aligning to the genome. Construction of a 95% confidence interval determined that a mean number of 7 reads or more were needed to be statistically greater than that of a mean of 0. A search for aromatase (CYP19A1), a transcript that is highly abundant in cumulus cells, was made to validate that there was no cumulus cell contamination. The mean number of normalized reads was 0.53 for the IVV oocytes and 0.08 for the IVM oocytes. The inability to detect more than 7 reads implies that there was little, if any, contamination by the cumulus cells. Those alignments with at least 7 reads, a p value of 0.1 (Students t-test) or lower, and at least 2 fold difference in abundance resulted in 1,316 transcripts that were more or less abundant in the two groups of oocytes. DAVID [15], [16] analysis showed a number of pathways that were over- or under-represented in the two groups (Table 1). After mining through this data we noted that there were several genes related to the WNT pathway that were over represented in IVM oocytes; specifically, WNT7A, FZD4, and DVL1. These are three of the early gene members in the WNT signaling pathway. We then decided to perturb this pathway during in vitro maturation and possibly effect oocyte maturation, subsequent embryo development, and embryo quality.
Table 1. David gene pathways that were mis-represented in the in vitro matured oocytes.
| Gene Pathways That Were Less Represented in the In Vitro Matured Oocytes | |
| Term | P-Value |
| T cell receptor signaling pathway | 0.002 |
| Regulation of actin cytoskeleton | 0.026 |
| Progesterone-mediated oocyte maturation | 0.029 |
| Ubiquitin mediated proteolysis | 0.037 |
Real time validation for WNT7A, FZD4, and DVL1
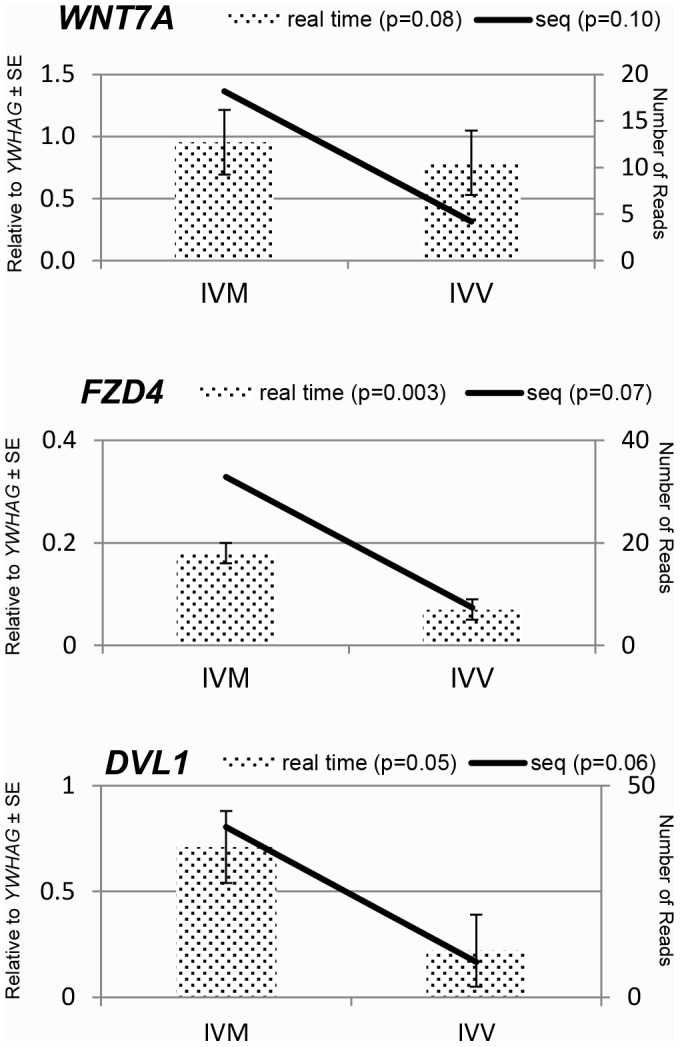
Real time PCR did validate the Illumina sequencing data with all three transcripts showing a higher abundance in the IVM oocytes (Fig. 1). The abundance of message for eight additional genes was determined. These genes were selected based on their similar or differential representation in the Illumina sequencing. Of the other three transcripts that appeared more abundant based on the Illumina sequencing, two (solute carrier family 7 (cationic amino acid transporter, y+ system), member 3 (SLC7A3) & zona pellucida protein 2 (ZP2)) were validated by real time PCR and the third (SOX4), while not different, showed the same directionality. Of the other five that were not significant by Illumina sequencing, four (heat shock protein 90 (HSP90AA1), mitochondrial ribosomal protein S36 (MRPS36), pyruvate dehydrogenase complex component X (PDHX), ubiquitin associated protein 2 (UBAP2)) were determined to be significant by real time PCR.
Figure 1. Real time PCR validation of Illumina sequencing data for WNT7A, FZD4 and DVL1.
Maturation of oocytes with DKK1
After maturing the oocytes with DKK1 there was a higher percentage of oocytes that developed to the metaphase II stage (Table 2). While there was no difference between the percentage cleavage (38.2% vs 35.0%) or the percentage of blastocysts (31.8% vs 27.8%), there was a significant difference in that DKK1 treated oocytes had more nuclei when they developed to the blastocyst stage compared to the controls (Table 2).
Table 2. Maturation of oocytes in the presence or absence of DKK1 (data represents four replications).
| Treatment | Total | LS Means Percent Metaphase II ±SE, n | LS Means Percent Cleavage ±SE, n | LS Mean Percent Blastocysts ±SE, n | LS Mean Number of Nuclei in Blastocysts ± SE, n |
| CONTROL | 661 | 73.1a±0.05, 483 | 35.0±2.1, 173 | 27.8±1.6, 145 | 28.6a±1.0, 67 |
| DKK1 | 543 | 82.1b±0.06, 446 | 38.2±2.2, 166 | 31.8±1.6, 142 | 32.7b±0.9, 83 |
Within a column P<0.05.
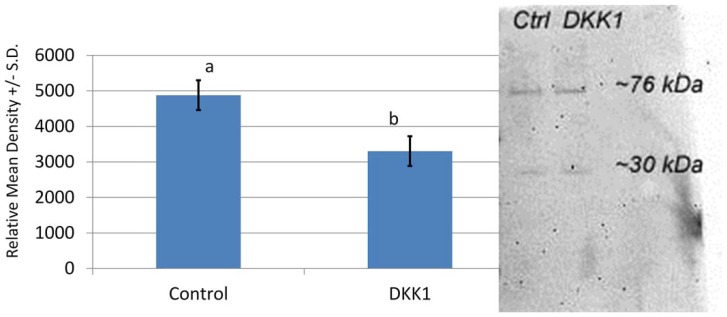
We were unable to identify an antibody that recognized pig WNT7A or FZD4, so anti-DVL1 was custom made based on the pig sequence. Western blot analysis showed a decrease in DVL1 in response to culture with DKK1 (Fig. 2).
Figure 2. Western blot of DVL1 in oocytes matured in the presence or absence of DKK1.
The mean densitometry of the two bands (30 and 76 kDa) was different between the two treatments a,bp = 0.01.
Discussion
In a previous study evaluating transcript abundance in oocytes from cyclic versus prepubertal females, it was shown that development was lower from the prepubertal derived oocytes and about 10% of the transcriptome was different between the two groups of oocytes [17]. Message associated with metabolism and biological processes were higher in oocytes from cyclic females, while oocytes from prepubertal females had an increase in the abundance of message for translation [17]. A more targeted approach identified a number of transcripts related to apoptosis, fatty acid oxidation, and glycolysis that were higher in oocytes from prepubertal animals as compared to adult animals [18]. In contrast, many transcripts associated with redox homeostasis were lower in the prepubertal oocytes compared to the cyclic oocytes [19].
While the list of pathways that were identified as different between the in vitro and in vivo matured oocytes somewhat overlaps with the adult versus prepubertal comparison above, the list provides a number of unique directions to pursue; here we focused on the WNT pathway.
After a misrepresentation of the three WNT pathway related transcripts was discovered in our Illumina sequencing data, and subsequently validated via real-time PCR, we wanted to find a way to manipulate or correct the over representation. One such idea was to add DKK1 to the maturation medium. DKK1 is a member of the DKK family that competitively binds to LRP. When bound to LRP it prevents LRP from interacting with FZD4 which is needed along with Frizzled to bind to WNT; thus inhibiting the WNT pathway. When added to the maturation medium the abundance of DVL1 decreased. By manipulating WNT signaling pathway related transcripts by competitive inhibition it appears as though IVM oocytes are more like their in vivo produced counterparts. In addition, there was a clear increase in maturation percentage of oocytes that are matured in the presence of 200 mM DKK1 compared to control oocytes. Subsequent development of the DKK1 treated embryos to the blastocyst stage was not significant; however, the increase in total cell number lends to the belief that adding DKK1 may be encouraging the production of higher quality oocytes. While the increase in the number of nuclei represents an advancement, the number of nuclei in blastocysts produced in vivo is still far greater than the 30 or so nuclei reported here. Clearly, more work needs to be done to improve the quality of the oocyte maturation, fertilization, and embryo culture systems. The importance of the WNT signaling pathway during oocyte maturation appears to be similar in species ranging from zebrafish [20] to rhesus monkeys [21].
While our direction of inquiry was toward the canonical signaling pathway because of the change in abundance of transcripts, it is possible that DKK1 may activate a non-canonical pathway as well as the active planar cell polarity pathway; generally the result of activation of this pathway is a calcium release that could act in the oocyte. It is possible that a calcium signal could alter message or protein abundance during maturation. Since calcium signaling is integral to sperm signaling at fertilization, it is not clear if there would be an advantage to calcium release in the oocyte prior to fertilization.
It is of interest to note that the Illumina sequencing and the real time PCR apparently did not have the same sensitivity. For example, Illumina sequencing did not detect a difference between maturation conditions for HSP90AA1, MRPS36, PHDX or UBAP2, while real time PCR detected a difference (P<0.09) for each message. While this observation may appear incongruous, a more serious concern would be raised if both methods detected a difference and those differences were in a different direction, e.g. the abundance was higher via real time PCR and lower in the Illumina sequencing dataset.
Another pathway that was over-represented in both the in vitro and in vivo matured oocytes was for ubiquitin mediated proteolysis. At first glance this observation appears incongruent. However closer examination shows that different transcripts are represented in each pathway. For example, both protein inhibitor of activated STAT3 (PIAS3) and PIAS4 are represented as part of the ubiquitin mediated proteolysis and both are higher in IVM oocytes, while PIAS2 is higher in the in vivo matured oocytes. Since PIAS4 is also a repressor of WNT [22] it might imply that the IVM oocyte is attempting to compensate for inadequate culture conditions by up-regulating or stabilizing message for PIAS4. Similarly, other ubiquitin mediated proteolysis pathway members were differentially represented in the two groups of oocytes. These include ubiquitin-conjugating enzymes (UBE) UBE2J2, UBE2C, UBE2K, and UBE2O which were higher in the IVM oocytes, while UBE2D3, UBE2N, and UBE2W were higher in the in vivo matured oocytes.
These data also show that global transcriptional profiling using Illumina deep sequencing data can be a useful tool to improve in vitro maturation. This was just one altered pathway from the sequence data set and just a few misrepresented transcripts. Thus, we believe this is only a start of what could be done using the sequence data to improve porcine oocyte maturation and in vitro development. With the DKK1 experiments we showed that perturbing the WNT pathway does make IVM oocytes behave more like in vivo matured oocytes. Although, another direction we propose to pursue would be trying to increase WNT representation during maturation and determine if that further harms oocyte and embryo development.
Materials and Methods
Animal Research
This study was carried out under the approval of the University of Missouri Animal Care and Use Committee (approval #6523). Euthanasia was carried out via injection of pentobarbital sodium and phenytoin sodium as approved by the American Veterinary Medical Association Panel on Euthanasia. The animals were treated humanely.
Oocyte collection for Illumina sequencing of in vitro and in vivo matured oocytes
Oocytes were matured in vitro or in vivo. Three gilts were monitored twice daily for standing estrus. Two days after first detection of estrus, in vivo matured oocytes were collected immediately after euthanizing the gilts by flushing the oviducts with TL-HEPES supplemented with 0.1% polyvinyl alcohol [23]. The metaphase II oocytes were quickly collected into pools of 10 and snap frozen in liquid nitrogen for RNA isolation. The IVM oocytes were obtained by euthanizing three prepubertal gilts, from a similar genetic background, and recovering the ovaries. Follicles 2–8 mm in diameter were aspirated by using an 18 gauge needle and syringe. Oocytes with multiple layers of cumulus cells and uniform cytoplasm were placed in M199 supplemented with LH, FSH, and EGF for 42 hours [24]. Upon maturation, cumulus cells were stripped and healthy metaphase II oocytes were collected in pools of 10 from each female and snap frozen. Total RNA was extracted from three pools of 10 oocytes for both treatments with an AllPrep DNA/RNA micro isolation kit (Qiagen). cDNA was synthesized using oligo (dT′) primed reverse transcriptase with Superscript III (Invitrogen, Grand Island, NY). Second strand cDNA was synthesized using DNA polymerase I and each sample was sequenced twice using an Illumina Genome Analyzer II. Data were deposited at NCBI (RNA-seq data for IVV (n = 3) and IVM (n = 3) samples were submitted to the NCBI Sequence Read Archive (SRA) and are available under accessions SAMN02640035, SAMN02640036, SAMN02640037, SAMN02640038, SAMN02640039, and SAMN02640040, respectively). All reads were aligned to a 34,433 member custom built porcine transcriptome [14]. If any reads aligned with more than one transcript, they were discarded from the analysis due to ambiguity. A normalization factor for each sample was computed to correct for variations between samples by taking the sample with the smallest number of alignable reads and then deriving a ratio using the total number of reads for each of the remaining samples. After normalization, the two runs for each sample were summed to get a representative number of aligned reads for each biological replicate. A mean for each transcript was calculated for the three biological replicates for each treatment group. A 95%-confidence interval was constructed around the mean number of reads based on the calculated pooled variance of the two sequencing runs to identify the average number of reads for each transcript that was statistically greater than zero. A Student's t-test and an added stringency of at least a 2-fold change between the means for each transcript was then used to determine if the means of the two groups for each transcript were statistically different. We then used DAVID (http://david.abcc.ncifcrf.gov/) to find pathways that were mis-represented in the IVM oocytes.
Real time validation
Real-time PCR was conducted for 11 candidate genes and a housekeeping gene (Table 3), using IQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA) and the amplified cDNA from each biological replicate (diluted to 5 ng/µL) as template. Primers (Table 4) were designed by using Integrated DNA Technology (Coralville, Iowa) software, and real time PCR was completed in triplicate for every biological replicate on the MyiQTM Single-Color Real-Time PCR Detection System (Bio-Rad Laboratories) to verify the differential expression of the chosen transcripts between IVM oocytes and in vivo derived oocytes. Primer efficiency tests were completed for each primer set by generating a standard curve using 10 ng dilutions of our 5 ng/µL reference cDNA pool. Real-time PCR was run at each concentration (5 ng/µL, 0.5 ng/µL, 0.05 ng/µL) to validate each primer set. Expression levels for each mRNA transcript were calculated in the same manner as we reported previously [14]. The abundance of each transcript was calculated relative to the reference sample and the housekeeping gene, tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide (YWHAG) [14], [25]. The reference sample used for DVL1 contained four biological replicates of in vivo fertilized/in vivo cultured blastocysts and in vivo fertilized/in vitro cultured blastocysts that were pooled together [14]. The reference sample used for FZD4 and WNT7A contained pools of reproductive and non-reproductive tissues across different stages of development [25]. Expression levels between treatments were determined using the comparative threshold cycle (CT) method for each gene. The 2-ΔΔCT values were first analyzed for normality and skewness. The resulting values were then analyzed using the general linear model (PROC GLM) in the Statistical Analysis System (SAS; SAS Institute, Cary, NC). Differences in expression were found by using the Least Squares Means (LSMeans) generated by PROC GLM (P<0.05) [26].
Table 3. Transcript abundance of selected genes as determined by Illumina sequencing and real time PCR.
| Illumina Sequencing | Real Time PCR | |||||
| Gene | In Vivo Matured # Reads ± SE | In Vitro Matured # Reads ± SE | P-Value | In Vivo Matured Relative Amount ± SE | In Vitro Matured Relative Amount ± SE | P-Value |
| ATG4 | 7.8 | 17.0 | NS | 3.2±0.5 | 3.3±0.5 | NS |
| HSP90AA1 | 4.5 | 13.9 | NS | 7.7±0.6 | 5.8±0.6 | <0.06 |
| MRPS36 | 43.7 | 23.8 | NS | 3.85±0.4 | 0.7±0.4 | <0.001 |
| PDHX | 99.8 | 69.1 | NS | 7.9±1.1 | 4.2±1.1 | <0.03 |
| SLC7A3 | 15.7 | 43.6 | <0.02 | 1.8±0.4 | 3.6±0.5 | <0.02 |
| SOX4 | 62.2 | 21.6 | <0.06 | 4.7±0.2 | 4.2±0.2 | NS |
| UBAP2 | 26.5 | 86.0 | NS | 5.3±0.5 | 3.8±0.6 | <0.09 |
| ZP2 | 141.2 | 1,134.2 | <0.06 | 84.1±16.8 | 272.0±16.8 | <0.001 |
Table 4. Accession number and primer sequences used for transcript amplification.
| Transcript Annotation | Accession # | Sense Primer | Anti-Sense Primer |
| DVL1 | XM_003127500 | TGGCTCAAGATCACCATCGCCAAT | AAGACGTAGTAGCACTGCTCGGAGA |
| FZD4 | NM_001243330 | CAGCACTCAGGCCAACGCCAT | AGAGCAGATCCGGGCTGCAGT |
| WNT7A | XM_003132396 | AGCGATGCTGAGCTTGTCTTGTCT | TATTGCTATGTTGAGGCCCAGGGT |
| YWHAG | NM_012479 | TCCATCACTGAGGAAAACTGCTAA | TTTTTCCAACTCCGTGTTTCTCTA |
| ZP2 | NM_003460.1 | TCCTGTGACCTGTAATGCCACACA | AACCTCAGGCCGTTTGTTGTTTCC |
| HSP90AA1 | NM_005348.3 | ACTCCGGGAAAGAGCTGCACATTA | AGACTTGGCGATCGTACCAAGGTT |
| UBAP2 | NM_018449.2 | GGCCAAGCCCTTGTCTTCACAAAT | AAATCCTGAGCCAAGGACGGATGA |
| SLC7A3 | NM_032803.5 | GCAGCGTTCATGGCATTCCTCTTT | ACACAAACAGCCACCAGGGAGTAA |
| MRPS36 | NM_033281.5 | AGGGTCGTTCAGGTAGTCAAACCA | TTGTTGACCCTGATACATCAGCAA |
| PDHX | NM_001135024.1 | TGGTATCACAGCACGCAGTCTCTT | AGCACTCACAGCTTCACCTTCCTT |
| SOX4 | NM_003107.2 | AGGGCGGCTGGTTAATATCTCACA | AAGGATGGATACTGGTGGCAGGTT |
| ATG4A | XR_113562.1 | CTTCTCCTACACCCATTTGTGCCA | TGACAGCTGGAGAATGGAGTCAGT |
In Vitro Maturation and DKK1 treatment
Ovaries were obtained from a Farmland, Incorporated slaughterhouse in Milan, MO and transported to the laboratory at ambient temperature. Ovaries were washed with physiological saline with 10 µg/mL gentamicin at 37°C. Follicles between 2–8 mm were aspirated using a 10 mL syringe and an 18 gauge needle. The aspirate then was washed in TL- HEPES [23] three times and oocytes with multiple layers of cumulus cells and homogeneous cytoplasm were then placed into maturation media either with or without 200 ng/mL DKK1 as recommended by the manufacturer (PeproTech 120–30). Cumulus cells were stripped after 42 hours by vortexing in 1 mg/mL hyaluronidase for 4 min. The cumulus cell-free oocytes were selected for the presence of an extruded polar body, thus considered to be at metaphase II, and data was collected for both treatments and recorded.
In Vitro Fertilization and Embryo Culture
Metaphase II oocytes were washed in Modified Tris Buffered Media (MTBM) [24] and co-incubated in 100 µL of MTBM with 0.25×106 frozen-thawed sperm for 5 hours [24]; this was considered Day 0. Five hours after fertilization the presumptive zygotes were washed in Porcine Zygote Media 3 (PZM3: [27]), transferred to 4 well Nunc plates and then cultured in 5% CO2 at 38.5°C for 28–30 hours, at which point cleavage data was obtained. The embryos were cultured in 5% O2, 5% CO2, 90% N2 at 38.5°C until day 6 at a density of 35–43 per 500 µL under an oil overlay. Development to blastocyst was recorded and total cell number was determined after staining with Hoechst 33342 (0.6 µg/mL) and visualized under UV light.
Western blots of oocytes for DVL1
Fifty MII oocytes from either control or DKK1 treatments were collected washed in diethyl pyrocarbonate treated PBS with polyvinyl alcohol, snap frozen and stored at −80°C until ready for use in a western blot. The samples were thawed on ice, at which time 10 µL of 6x Laemmli's loading buffer (Boston Bioproducts, Ashland, MA. BP-110R) was added to each sample. The samples were then heated to 96°C for 5 minutes, centrifuged and run on a mini protein gel 4–20% (BioRad 4561093s). Proteins were then transferred to polyvinylidene fluoride membrane via an iBlot transfer apparatus (Invitrogen) and iBlot gel transfer stacks (Invitrogen). The membrane was blocked for 4 hours in tris buffered saline StarlingBlock blocking buffer (Thermo 37542) with 0.1% tween on a rocker. The membrane was then washed one time in tris buffered saline (Fisher BP2471-1) with 0.1% tween. Goat polyclonal primary antibody to DVL1 (custom made by Everest Biotech EB11466 by using peptide KLPVAPERVTLAD) was added in 1∶500 dilution and incubated overnight at 4°C. It was washed 5 times then placed in 1∶20,000 dilution of horseradish peroxidase conjugated rabbit anti goat secondary antibody (Pierce Scientific 31402) for 1.5 hours, washed again 5 times and then a 1∶1 mix of supersignal peroxide solution and Luminol/enhancer solution was added to cover the membrane for 3 minutes. The excess was drained off and the membrane was visualized on a Fotodyne imagining system (Highland, WI, USA) with a 5 min exposure. Later, the densitometry of the blots was analyzed using Foto/Analyst and total lab quant ID western blot analysis. The densitometry readings of both bands were summed together for the calculations. Three replications were completed and a paired one-tailed Student's t-test was used to detect differences.
Funding Statement
Funding from Food for the 21st Century at the University of Missouri, and by the University of Missouri grant 13321 from the Missouri Life Science Research Board via Dr. Jerry Taylor. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Hirao Y (2011) Conditions affecting growth and developmental competence of mammalian oocytes in vitro. Anim Sci J 82: 187–197. [DOI] [PubMed] [Google Scholar]
- 2. Yoshioka K (2011) Development and application of a chemically defined medium for the in vitro production of porcine embryos. J Reprod Dev 57: 9–16. [DOI] [PubMed] [Google Scholar]
- 3. Alvarez GM, Dalvit GC, Achi MV, Miguez MS, Cetica PD (2009) Immature oocyte quality and maturational competence of porcine cumulus-oocyte complexes subpopulations. Biocell 33: 167–177. [PubMed] [Google Scholar]
- 4. Jones KT, Lane SI (2012) Chromosomal, metabolic, environmental, and hormonal origins of aneuploidy in mammalian oocytes. Exp Cell Res 318: 1394–1399. [DOI] [PubMed] [Google Scholar]
- 5. Fragouli E, Wells D, Delhanty JD (2011) Chromosome abnormalities in the human oocyte. Cytogenet Genome Res 133: 107–118. [DOI] [PubMed] [Google Scholar]
- 6. Harwood BN, Cross SK, Radford EE, Haac BE, De Vries WN (2008) Members of the WNT signaling pathways are widely expressed in mouse ovaries, oocytes, and cleavage stage embryos. Dev Dyn 237: 1099–1111. [DOI] [PubMed] [Google Scholar]
- 7. ten Berge D, Kurek D, Blauwkamp T, Koole W, Maas A, et al. (2011) Embryonic stem cells require Wnt proteins to prevent differentiation to epiblast stem cells. Nat Cell Biol 13: 1070–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brown NL, Knott L, Halligan E, Yarram SJ, Mansell JP, et al. (2003) Microarray analysis of murine palatogenesis: temporal expression of genes during normal palate development. Dev Growth Differ 45: 153–165. [DOI] [PubMed] [Google Scholar]
- 9. Schlesinger A, Shelton CA, Maloof JN, Meneghini M, Bowerman B (1999) Wnt pathway components orient a mitotic spindle in the early Caenorhabditis elegans embryo without requiring gene transcription in the responding cell. Genes Dev 13: 2028–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Denicol AC, Dobbs KB, McLean KM, Carambula SF, Loureiro B, et al. (2013) Canonical WNT signaling regulates development of bovine embryos to the blastocyst stage. Sci Rep 3: 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xie H, Tranguch S, Jia X, Zhang H, Das SK, et al. (2008) Inactivation of nuclear Wnt-beta-catenin signaling limits blastocyst competency for implantation. Development 135: 717–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guder C, Pinho S, Nacak TG, Schmidt HA, Hobmayer B, et al. (2006) An ancient Wnt-Dickkopf antagonism in Hydra. Development 133: 901–911. [DOI] [PubMed] [Google Scholar]
- 13. Niehrs C (2006) Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene 25: 7469–7481. [DOI] [PubMed] [Google Scholar]
- 14. Bauer BK, Isom SC, Spate LD, Whitworth KM, Spollen WG, et al. (2010) Transcriptional profiling by deep sequencing identifies differences in mRNA transcript abundance in in vivo-derived versus in vitro-cultured porcine blastocyst stage embryos. Biol Reprod 83: 791–798. [DOI] [PubMed] [Google Scholar]
- 15. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 16. Huang da W, Sherman BT, Lempicki RA (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paczkowski M, Yuan Y, Fleming-Waddell J, Bidwell CA, Spurlock D, et al. (2011) Alterations in the transcriptome of porcine oocytes derived from prepubertal and cyclic females is associated with developmental potential. J Anim Sci 89: 3561–3571. [DOI] [PubMed] [Google Scholar]
- 18. Yuan Y, Ida JM, Paczkowski M, Krisher RL (2011) Identification of developmental competence-related genes in mature porcine oocytes. Mol Reprod Dev 78: 565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan Y, Wheeler MB, Krisher RL (2012) Disrupted redox homeostasis and aberrant redox gene expression in porcine oocytes contribute to decreased developmental competence. Biol Reprod 87: 78. [DOI] [PubMed] [Google Scholar]
- 20. Abrams EW, Mullins MC (2009) Early zebrafish development: it's in the maternal genes. Curr Opin Genet Dev 19: 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng P, Vassena R, Latham K (2006) Expression and downregulation of WNT signaling pathway genes in rhesus monkey oocytes and embryos. Mol Reprod Dev 73: 667–677. [DOI] [PubMed] [Google Scholar]
- 22. Sachdev S, Bruhn L, Sieber H, Pichler A, Melchior F, et al. (2001) PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev 15: 3088–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hagen DR, Prather RS, Sims MM, First NL (1991) Development of one-cell porcine embryos to the blastocyst stage in simple media. J Anim Sci 69: 1147–1150. [DOI] [PubMed] [Google Scholar]
- 24. Abeydeera LR, Day BN (1997) In vitro penetration of pig oocytes in a modified Tris-buffered medium: effect of BSA, caffeine and calcium. Theriogenology 48: 537–544. [DOI] [PubMed] [Google Scholar]
- 25. Whitworth KM, Agca C, Kim JG, Patel RV, Springer GK, et al. (2005) Transcriptional profiling of pig embryogenesis by using a 15-K member unigene set specific for pig reproductive tissues and embryos. Biol Reprod 72: 1437–1451. [DOI] [PubMed] [Google Scholar]
- 26. Redel BK, Brown AN, Spate LD, Whitworth KM, Green JA, et al. (2012) Glycolysis in preimplantation development is partially controlled by the Warburg Effect. Mol Reprod Dev 79: 262–271. [DOI] [PubMed] [Google Scholar]
- 27. Im GS, Lai L, Liu Z, Hao Y, Wax D, et al. (2004) In vitro development of preimplantation porcine nuclear transfer embryos cultured in different media and gas atmospheres. Theriogenology 61: 1125–1135. [DOI] [PubMed] [Google Scholar]