Abstract
Leucine-rich repeat kinase 2 (LRRK2) is enriched in the striatal projection neurons (SPNs). Here we show that LRRK2 negatively regulates protein kinase A (PKA) activity in the SPNs during synaptogenesis and in response to dopamine receptor Drd1 activation. LRRK2 interacted with PKA regulatory subunit IIβ (PKARIIβ). A lack of LRRK2 promoted the synaptic translocation of PKA and increased PKA-mediated phosphorylation of actin-disassembling enzyme cofilin and glutamate receptor GluR1, resulting in abnormal synaptogenesis and transmission in the developing SPNs. Furthermore, PKA-dependent phosphorylation of GluR1 was also aberrantly enhanced in the striatum of young and aged LRRK2-null mice after treatment with a Drd1 agonist. Notably, a Parkinson’s disease-related LRRK2 R1441C missense mutation that impaired the interaction of LRRK2 with PKARIIβ also induced excessive PKA activity in the SPNs. Our findings reveal a new regulatory role of LRRK2 in PKA signaling, and provide a new pathogenic mechanism of SPN dysfunction in Parkinson’s disease.
INTRODUCTION
The impairment of midbrain nigrostriatal dopaminergic innervation at the striatum and the resulting imbalance of striatal output via the direct and indirect pathways underlie the neurological basis of Parkinson’s disease-related movement disorders1. Parkinson’s disease also causes substantial alterations of the dendritic spine morphology and function of SPNs2,3. The striatum consists of more than 90% of SPNs that are located in two complementary compartments named striosomes and matrix4. Notably, LRRK2, one of the most common genetic factors associated with both familial and sporadic Parkinson’s disease5, is particularly enriched in the striosomal SPNs in contrast to a low expression in the nigrostratal dopaminergic neurons6. This distinct expression pattern suggests that LRRK2 may play an important role in the SPNs in response to dopamine stimulation. However, little is known about the pathophysiological functions of LRRK2 in the SPNs under either the normal physiological or disease conditions.
The onset of LRRK2 expression is also coincided with the formation of dendritic spines during neuron development7. LRRK2 is involved in actin dynamics that regulates the extension of dendrites and axons in neuron morphogenesis8. Dendritic spine formation critical for the connectivity and plasticity in the brain is also determined by the actin cytoskeleton9. Spine morphogenesis involves the transition from the initial long, thin, and highly flexible filopodia to the more stable spines that are considered mature when showing characteristic bulbous enlargements (spine head) and a distinct neck10. Cofilin, a modulator of actin filament turnover, critically regulates the dynamics of spine formation11. Cofilin activity are inhibited by LIM kinase-mediated phosphorylation and stimulated by Slingshot-induced de-phosphorylation on a highly conserved Serine at residue three (S3) 12. However, alternative regulatory mechanisms have been described, including a PKA-dependent pathway13.
PKA-mediated signaling pathways are critical for neuron development and function14. PKA holoenzyme contains two catalytic subunits and two regulatory subunits. Cyclic AMP (cAMP) activates the kinase activity of PKA through disrupting the association between the catalytic and regulatory subunits15. The mammalian PKA family can be subdivided into types I and II based on their regulatory subunits16. The PKARIIβ regulatory subunit is particularly abundant in the SPNs17,18. Dopamine can activate PKA signaling in the SPNs via binding with Drd1-like dopamine receptors19. The specificity and efficacy of PKA signaling in neurons can also be regulated by A kinase anchoring proteins (AKAPs), which determines the subcellular distribution of PKA holoenzyme by interacting with the regulatory subunits20.
In this study we demonstrate that LRRK2 is a new AKAP-like regulator of the subcellular distribution of PKARIIβ in neurons, and assign a new physiological role for LRRK2 as a negative modulator of PKA activity in the SPNs during synaptogenesis and in response to dopamine receptor activation. We further investigated the impact of Parkinson’s disease-related LRRK2 missense mutations on the PKA activity and reveal a new pathogenic mechanism of LRRK2 that alters the response of SPNs to dopamine stimulation in the Parkinson’s disease-related LRRK2 R1441C knock-in mice.
RESULTS
LRRK2 regulates the SPN synaptogenesis and transmission
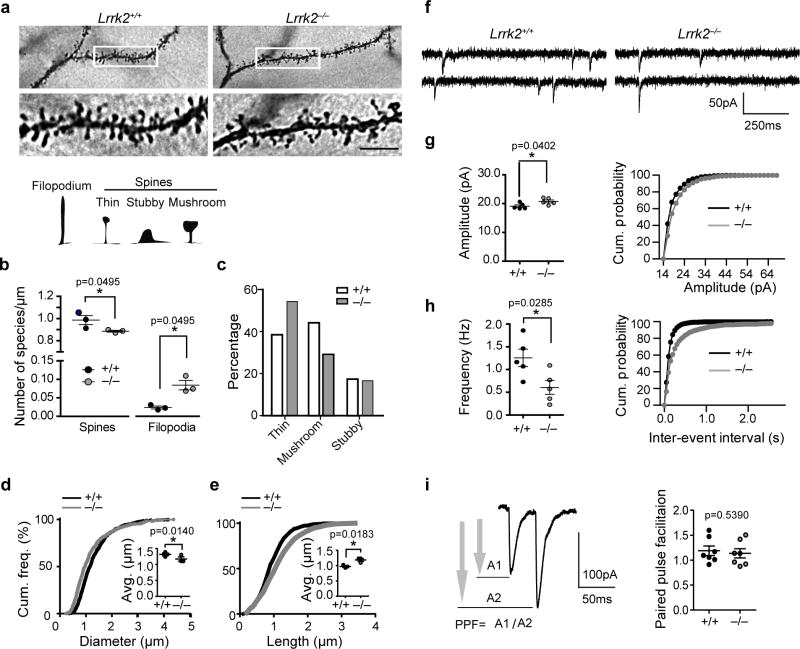
To extend our previous study on the role of LRRK2 in neuron development8, we investigated whether LRRK2 affects the dendritic spine formation in the SPNs. Notably, Golgi-COX staining revealed a significant decrease of dendritic spines and increase of dendritic filopodia in the dorsal SPNs of postnatal day 15 (P15) LRRK2 knockout (LRRK2−/−) pups compared to the age-matched wild-type (LRRK2+/+) littermate controls (Fig. 1a,b). When the dendritic spines were further categorized as thin, mushroom or stubby based on their maturation stages, a higher proportion of thin and less mushroom spines were found in the LRKK2−/− neurons (Fig. 1c). The cumulative frequency distribution analysis showed LRRK2−/− SPNs having significantly longer dendritic spines while the spine heads were smaller (Fig. 1d,e). The critical involvement of LRRK2 in the maturation of dendritic spines is also supported biochemically by the reduction of postsynaptic density (PSD) protein PSD95 expression in striatal extracts of P15 and P21 LRRK2−/− mice (Supplementary Fig. 1a,b). We also measured the morphological changes of SPN dendritic spines in 1-month-old LRRK2−/− mice. The density and length of the dendritic spines were comparable between LRRK2+/+ and LRRK2−/− neurons, but the diameter of spine heads remains significant smaller in the LRRK2−/− neurons (Supplementary Fig. 1c–e). Overall, the loss of LRRK2 significantly alters the dendritic spine morphology in the SPNs, especially during the developmental stages.
Figure 1. Loss of LRRK2 leads to decreased number of mature spines and altered synaptic transmission.
(a) Golgi-Cox staining of SPNs from P15 littermate LRRK2+/+ and LRRK2−/− pups. The bottom panels are enlargement of boxed areas of the upper panels. Scale bar: 5μm. (b) The density of dendritic spines and filopodia (n=3 three pups per genotype). Data represent mean ± SEM. Mann-Whitney Test (spine: z=1.964, filopodia: z=−1.964). *P<0.05. The inset shows schematic representation of filopodium and different types of dendritic spines. (c) The distribution of different types of dendritic spines in LRRK2+/+ and LRRK2−/− SPNs (nLRRK2+/+=839 spines, nLRRK2−/− =835 spines from six neurons of three pups per genotype, Chi-Squared test (χ2(2)=50.13), p<0.0001. (d,e) Cumulative frequencies of spine head diameter (d, KS-test, p<0.001) and spine length (e, KS-test, p<0.001) in the spines of SPNs described above. Scatter blots show average head diameter and length. Data represent mean ± SEM. Unpaired t-test, t(4)=1.839 and 3.853, respectively. *P<0.05. (f) Traces of mEPSCs recorded from P15 LRRK2+/+ and LRRK2−/− SPNs. (g, h) Mean values and cumulative distribution for amplitude (g, Unpaired t-test, t(8)=2.446; KS-test, *p<0.01) and frequency (h, Unpaired t-test, t(8)=2.667; KS-test, p<0.001) of mEPSCs from P15 pups (nLRRK2+/+=11 neurons, nLRRK2−/− =13 neurons of five pups per genotype). Data represent mean ± SEM. *P<0.05. (i) Paired-pulse facilitation measured at a 50ms inter-pulse interval and summary data for nLRRK2+/+=5 neurons, nLRRK2−/−=7 neurons (t(12)=0.6323).
To further examine whether the alteration of spine number and morphology in the LRRK2−/− SPNs affects synaptic transmission, we measured glutamatergic miniature excitatory postsynaptic currents (mEPSCs) in striatal slices from P15 LRRK2−/− and littermate control LRRK2+/+ mice by whole-cell voltage clamp recordings (Fig. 1f). While the amplitude of mEPSCs was moderately but significantly increased in LRRK2−/− neurons (Fig. 1g), the frequency of mEPSCs was significantly reduced (Fig. 1h). To probe the pre- versus postsynaptic nature of these changes, we recorded the responses of SPNs to paired stimuli, and found that the paired-pulse facilitation (PPF) showed no difference between the genotypes (Fig. 1i). These findings suggest that LRRK2 mainly regulates the postsynaptic response of SPNs.
LRRK2−/− neurons show aberrant phosphorylation of cofilin
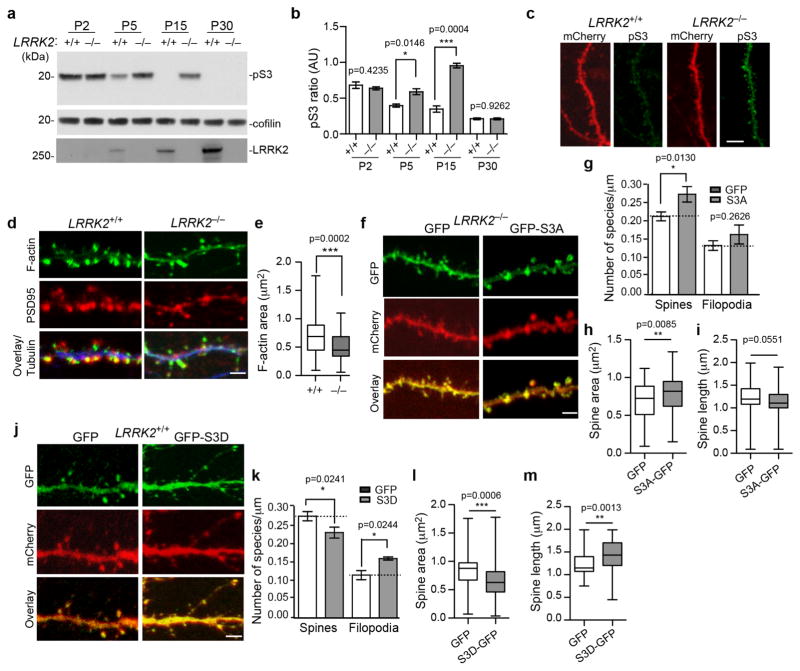
We examined the expression level and activity of actin binding proteins in the striatal homogenate of LRRK2−/− and age-matched littermate control mice (Supplementary Fig. 2a). We found that while the expression level of total cofilin was comparable, the phosphorylation of cofilin at the conserved residue Serine three (S3) was significantly increased in the P5 to P21 LRRK2−/− mouse brains compared to the controls., (Fig. 2a,b; Supplementary Fig. 2a).
Figure 2. Abnormal phosphorylation of cofilin in LRRK2−/− neurons.
(a) Western blot analysis of forebrain homogenates from LRRK2+/+ and LRRK2−/− mice. The full-length images are in shown in Supplementary Fig. 2e. (b) The pS3 cofilin levels normalized against total cofilin levels (n=3 per genotype). Data represent mean±SEM, Unpaired t-test, t(4)=0.8906, 0.1047, 11.11, and 4.117, respectively. *p<0.05, ***p<0.001. (c) Representative images of spines and their parental dendrites of cultured LRRK2+/+ and LRRK2−/− hippocampal neurons transfected with mCherry (red) and stained with pS3 cofilin (green). Scale bar: 10μm. (d) Staining of F-actin (green), PSD95 (red), and βIII-tubulin (blue) in dendritic spines and corresponding dendrites of LRRK2+/+ and LRRK2−/− hippocampal neurons at 15DIV. Scale bar: 5μm. (e) F-actin area staining in the dendritic spine of LRRK2+/+ and LRRK2−/− hippocampal neurons (n=6–8 neurons per genotype; n=85–90 spines per genotype). Data represent mean ± SEM. Unpaired t-test, ***p<0.001. (f, j) Spines and dendritic regions of LRRK2−/− (f) and LRRK2+/+ (j) hippocampal neurons transfected with GFP, GFP-S3A, and GFP-S3D cofilin plasmids, along with mCherry at 15DIV. Scale bar: 5μm. (g, k) Density of dendritic spines and filopodia was quantified from LRRK2−/− and LRRK2+/+ neurons. (h–i, l–m) Bar graphs showing average head spine diameter and dendritic spine length of transfected LRRK2−/− (h, i) and LRRK2+/+ neurons (l, m) (LRRK2−/−, nGFP=73 spines, nS3A-GFP=80 spines; LRRK2+/+ nGFP=101 spines, nS3D-GFP=79 spines; Four to seven neurons per genotype). Data represent mean ± SEM. Unpaired t-test, *p<0.05, **p<0.01, ***p<0.001
Consistently, increased phosphorylation of S3 (pS3) cofilin staining was found in neurons of LRRK2−/− hippocampal sections and in the dendritic spine of cultured LRRK2−/− hippocampal neurons (Supplementary Fig. 2b, Fig. 2c). Notably, the reduction of pS3 cofilin in the LRRK2+/+ mice brain was coincided with the increase of LRRK2 expression during the postnatal development (Fig. 2a), suggesting that LRRK2 may suppress the phosphorylation of cofilin. In agreement with this idea, the levels of pS3 cofilin were reduced in the brain of P2 to P15 LRRK2 transgenic mice (Supplementary Fig. 2c), which overexpress human wild-type (WT) LRRK221. Considering the lack of apparent physical interaction between cofilin and LRRK2 (Supplementary Fig. 2d), LRRK2 may regulate cofilin phosphorylation indirectly through modulating the activity of other protein kinases or phosphatases.
An elevation of pS3 cofilin affects the spine formation
The abnormal increase of pS3 cofilin may impair actin dynamics and spine formation in LRRK2−/− neurons. In support of this notion, a significant decrease of filamentous actin (F-actin) content was observed in the dendritic spine of cultured LRRK2−/− neurons after 15 days in vitro (DIV) (Fig. 2d,e). The actin cytoskeleton protruding from the dendritic shaft of LRRK2−/− neurons appeared thin and elongated (Fig. 2d), which is consistent with a higher incidence of thin filopodia like protrusions found in the P15 LRRK2−/− SPNs (Fig. 1a–c).
To establish a direct connection between cofilin phosphorylation and spine morphology, we introduced green fluorescent protein (GFP)-tagged active or inactive forms of cofilin into cultured LRRK2−/− or LRRK2+/+ hippocampal neurons, and examined the alteration of dendritic spine morphology. We found that LRRK2−/− neurons transfected with the constitutively active cofilin S3A mutation22 developed more and larger spines and rescued the spine morphology defects in LRRK2−/− neurons (Fig. 2f–i). On the contrary, LRRK2+/+ neurons transfected with the inactive cofilin S3D mutation22 displayed longer and smaller spines with a concomitant increase of filopodia as well as spines with abnormal long protrusions, similar to the alterations observed in LRRK2−/− neurons (Fig. 2j–m). These data suggest that the persistent phosphorylation of cofilin in the SPNs of P15 LRRK2−/− mice may alter the actin dynamics, resulting in the abnormal dendritic spine morphology.
LRRK2 regulates pS3 cofilin via a PKA-dependent pathway
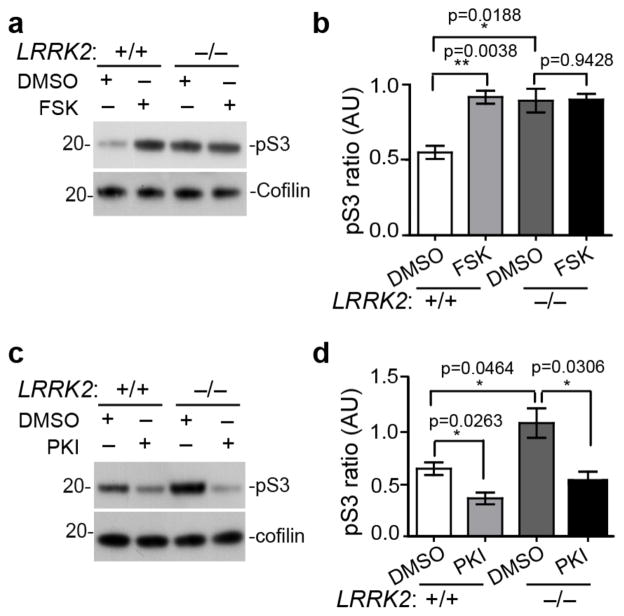
We next examined the signaling pathways by which LRRK2 regulates cofilin phosphorylation. LIM kinase and Slingshot are typically involved in the phosphorylation and de-phosphorylation of cofilin12. However, in P15 LRRK2−/− pups, no obvious alteration of LIMK1 phosphorylation, an indicator of LIMK1 kinase activity, was found between P15 LRRK2+/+ and LRRK2−/− brain extracts (Supplementary Fig. 3a,b). In addition, the phosphorylated and total levels of Slingshot-1 L (SSH1L), a member of the Slingshot family that dephosphorylates cofilin23, were also comparable between genotypes (Supplementary Fig. 3a,c). To identify alternative mechanisms of cofilin activity regulated by LRRK2, we compared the levels of pS3 cofilin by Western blot analysis of cultured LRRK2+/+ and LRRK2−/− cortical neurons treated with the activators and inhibitors of various intracellular signaling pathways directly or indirectly related with the regulation of cofilin phosphorylation24. LRRK2+/+ and LRRK2−/− neurons displayed similar changes of cofilin phosphorylation levels in response to modulators of AKT/protein kinase B, phosphoinositide 3-kinase (PI3K), protein kinase C (PKC), mammalian target of rapamycin (mTOR), pyruvate dehydrogenase kinase (PDK1), and MAPK/ERK kinase (MEK) pathways (Supplementary Fig. 3d–g). In contrast, application of forskolin (FSK) that promotes the production of cAMP and activation of PKA pathway caused a significant increase in cofilin phosphorylation in LRRK2+/+ neurons, but not in LRRK2−/− neurons (Fig. 3a,b). Meanwhile, treatment with a PKA inhibitor PKI resulted in comparable levels of p-cofilin in LRRK2+/+ and LRRK2−/− neurons (Fig. 3c,d). These data suggest an impairment of regulation of PKA signaling in LRRK2−/− neurons.
Figure 3. PKA signaling is involved in LRRK2-dependent phosphorylation of cofilin.
(a, b) Western Blot analysis of cultured LRRK2+/+ and LRRK2−/− cortical neurons treated with DMSO or FSK (50μM for 1 hr) and (c, d) DMSO or PKI (5μM for 24 hrs) for the expression of phosphorylated and total cofilin. Three or more independent cultures were analyzed per genotype and per treatment. Data represent mean ± SEM. Unpaired t-test, t(4)=6.019, 3.820, and 0.0764, respectively in (b). t(4)=3.441, 2.851, and 3.278, respectively in (d). *P<0.05, **p<0.01. The full-length images in Fig. 3a and 3c are shown in Supplementary Fig. 3m and 3n.
To further confirm these findings, we tested the effect of FSK application on a canonical PKA substrate, cAMP response element-binding protein (CREB) in LRRK2+/+ and LRRK2−/− neurons. FSK treatment induced one-fold increase of phosphorylated CREB (pCREB) in LRKK2+/+ neurons, but failed to increase the ratio of pCREB in LRRK2−/− neurons (Supplementary Fig. 3h–j). In addition, we found a significant increase of pCREB in the nuclear extract of P15 LRKK2−/− striatal tissues (Supplementary Fig. 3k,l). All the above suggest an aberrant up-regulation of basal PKA activity in the developing LRKK2−/− neurons.
LRRK2 selectively interacts with PKARIIβ
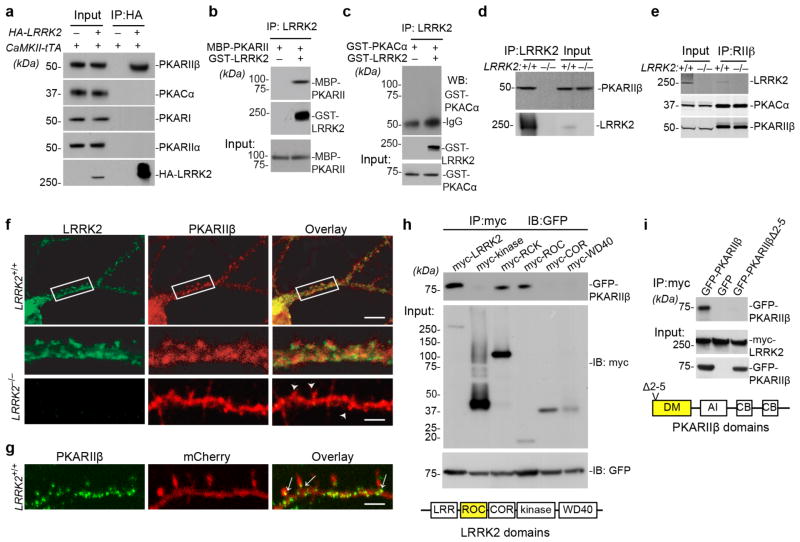
To determine at which level LRRK2 exerts its effect on PKA activation, we first examined the production of cAMP in cultured LRRK2+/+ and LRRK2−/− neurons treated with vehicle or FSK. In both conditions, no significant difference in cAMP production was observed (Supplementary Fig. 4a,b). We next investigated whether LRRK2 directly interacted with PKA. Co-immunoprecipitation (Co-IP) of LRRK2 with PKA subunits from the striatal extract of human WT LRRK2 transgenic mice25 revealed a selective interaction between LRRK2 and PKARIIβ subunit but not with the catalytic subunit (PKACα) or other regulatory subunits (Fig. 4a). The direct interaction of LRRK2 and PKARIIβ was further confirmed by an in vitro binding assay using recombinant LRRK2 and PKARIIβ proteins (Fig. 4b). Although PKA can phosphorylate LRRK226, no stable direct interaction between LRRK2 and PKACα recombinant proteins was observed (Fig. 4c). Moreover, endogenous LRRK2 and PKARIIβ proteins were co-IPed by an LRRK2-specific antibody from the light membrane fraction of LRRK2+/+ brain homogenates (Fig. 4d), of which both LRRK2 and PKARIIβ were highly enriched (Supplementary Fig. 4c). Meanwhile, endogenous PKARIIβ and PKACα proteins were also co-IPed with LRRK2 by a PKARIIβ-specific antibody (Fig. 4e), suggesting that LRRK2 co-exists with PKARIIβ and PKACα in the same protein complex via direct interaction with PKARII β.
Figure 4. LRRK2 interacts with PKARIIβ.
(a) Co-IP of transgenic human wild-type LRRK2 and endogenous mouse PKARIIβ from the forebrain homogenate of P15 CaMKII-tTA/tetO-LRRK2 double and littermate CaMKII-tTA single transgenic mice. The N-terminal of transgenic LRRK2 is HA-tagged. (b,c) Co-IP of GST-tagged recombinant human LRRK2 proteins with maltose-binding protein (MBP)-tagged recombinant mouse PKARIIβ proteins (b) and GST-tagged recombinant mouse PKACα proteins (c). (d) Co-IP of endogenous mouse LRRK2 and PKARIIβ from the light membrane fractions of P15 mouse striatal tissues. (e) Co-IP of endogenous mouse LRRK2, PKACα and PKARIIβ from the light membrane fractions of P15 mouse striatal tissues using a PKARIIβ antibody. (f) Co-staining of endogenous LRRK2 (green) and PKARIIβ (red) in cultured mouse hippocampal neurons at 15DIV. The middle two panels show the boxed areas in the top panel. The bottom panel shows LRRK2 (green) and PKARIIβ (red) staining in 15DIV LRRK2−/− hippocampal neurons (arrowheads point to dendritic spines). Scale bars: 20μm (top panel), 5μm (lower panels). (g) Representative image shows subcellular localization of endogenous PKARIIβ (green) in the dendrite of mCherry-transfected 15DIV LRRK2+/+ hippocampal neurons. Arrowheads indicate the localization of PKARIIβ at the base of dendritic spines. Scale bars: 5μm. (h) Co-IP of GFP-PKARIIβ with myc-tagged human wild-type full-length LRRK2, as well as LRRK2 kinase, RCK (Roc-COR-Kinase), ROC, COR, and WD40 domains from transfected HEK293 cells. (i) Co-IP of myc-tagged human wild-type full-length LRRK2 with GFP, GFP-PKARIIβ, and GFP-PKARIIβΔ2–5 from transfected HEK293 cells. DM: dimerization domain, AI: auto-inhibitory domain, CB: cAMP binding domain. The full-length images in Fig. 4a–e, 4h, and 4j are shown in Supplementary Fig. 4g–k, 4l, and 4m.
Immunostaining revealed co-localization of endogenous LRRK2 and PKARIIβ signals in the dendrites of LRRK2+/+ hippocampal pyramidal neurons (Fig. 4f). While, PKARIIβ staining was mainly distributed along the dendritic shaft and at the base of dendritic spines in LRRK2+/+ neurons (Fig. 4g), PKARII β signal was evenly distributed in the dendritic shaft and spines of LRRK2−/− neurons (Fig. 4f). Meanwhile, a loss of LRRK2 did not affect the expression and phosphorylation of PKARII β, the expression of PKACα, and the association and co-localization of PKARII β with PKACα in LRRK2−/− neurons (Supplementary Fig. 4d–f). Together, these observations suggest that LRRK2 may regulate PKA activity by controlling the subcellular localization of PKARIIβ along the dendritic shaft.
Further co-IP experiments showed that PKARIIβ selectively interacted with the Ras of complex proteins (ROC) domain of LRRK2, while, the deletion of the N-terminal two-to-five residues of PKARIIβ (PKARIIβΔ2–5) prevented the interaction of PKARIIβ with LRRK2 (Fig. 4h,4i)..
LRRK2 confines PKARIIβ to the dendritic shafts
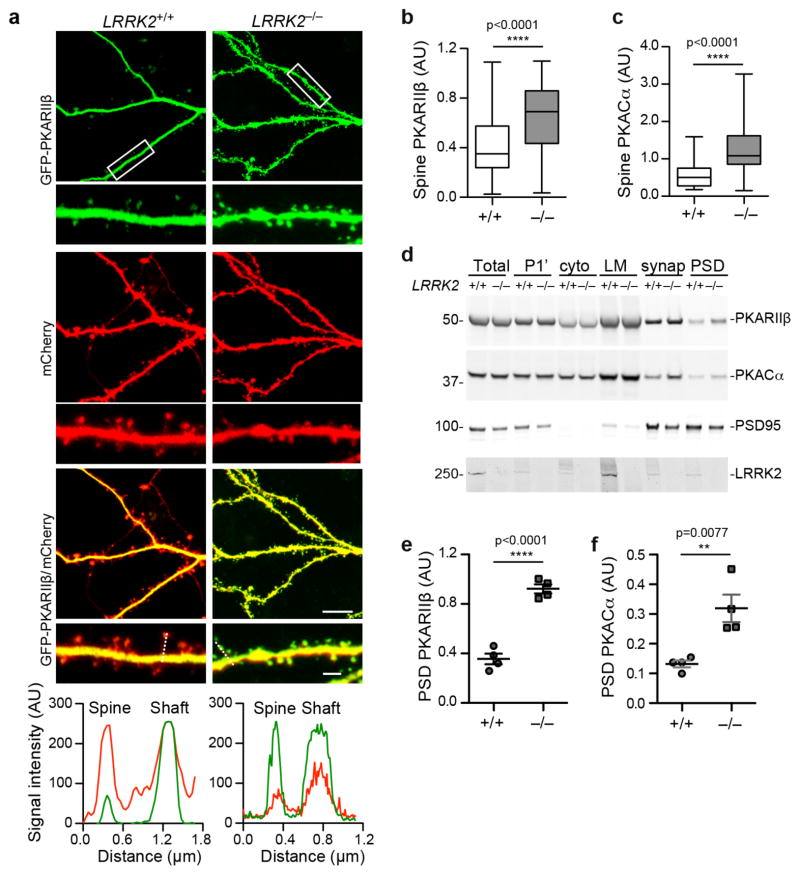
To further investigate the alteration of PKARIIβ subcellular localization in LRRK2−/− neurons (Fig. 4f), we transfected cultured LRRK2+/+ and LRRK2−/− hippocampal neurons with GFP-tagged PKARIIβ and cell volume marker mCherry. Confocal imaging revealed a predominant presence of PKARIIβ in the dendritic shafts of LRRK2+/+ neurons (Fig. 5a). By contrast, a significant increase of dendritic spine PKARIIβ signal was observed in LRRK2−/− neurons (Fig. 5a,b). A similarly increase of GFP-tagged PKACα in the dendritic spines was also observed in LRRK2−/− neurons (Fig. 5c, Supplementary Fig. 5a). Furthermore, subcellular fractionation of P15 mouse forebrain tissues revealed significant increase of PKARIIβ and PKACα levels in the postsynaptic density (PSD) fraction of LRRK2−/− samples compared to the age-matched LRRK2+/+ controls (Fig. 5d–f). Therefore, these studies indicate a critical involvement of LRRK2 in controlling the accessibility of PKARIIβ and PKA holoenzyme into the dendritic spines.
Figure 5. LRRK2 regulates the subcellular localization of PKARIIβ.
(a) Fluorescent images of GFP-PKARIIβ and mCherry in dendrites of transfected LRRK2+/+ and LRRK2−/− hippocampal neurons at 15DIV. Scale bars: 5μm (lower magnification), 1μm (higher magnification). The RGB signal profile shows mCherry (red) and GFP-PKARIIβ (green) expression across the spine and adjacent dendritic shaft section depicted by the discontinuous white line on LRRK2+/+ and LRRK2−/− hippocampal neurons. (b,c) The relative ratio of GFP-PKARIIβ (b) and GFP-PKACα (c) in the dendritic spine versus adjacent dendritic shaft of LRRK2+/+ and LRRK2−/− hippocampal neurons at 15DIV (n=52–95 spines of seven neurons per genotype). Data represent mean ± SEM. Unpaired t-test, t(114)=7.013 in (c). ***p<0.001, ****p<0.0001. (d) Western blot analysis of endogenous PKARIIβ, PKACα, PSD95, and LRRK2 expression in the total, pelleted nuclear and other insoluble (P1′), cytosol, light membrane (LM), synaptosome (synap) and PSD fractions of P15 LRRK2+/+ and LRRK2−/− mouse brain homogenates. The full-length images are shown in Supplementary Fig. 5e. (e,f) Quantified data of PKARIIβ (e) and PKACα (f) protein levels in the PSD fraction of P15 LRRK2+/+ and LRRK2−/− mouse brain homogenates (n=4 per genotype). Data represent mean ± SEM. Unpaired t-test, t(6)=9.929 and 3.935, respectively. **P<0.01, ****p<0.0001.
To verify a direct effect of LRRK2 on the subcellular localization of PKARIIβ, reintroduction of myc-tagged WT LRRK2 into LRRK2−/− neurons prevented the dendritic spine translocation of PKARIIβ (Supplementary Fig. 5b). Moreover, transfection of PKARIIβΔ2–5 that disrupted the interaction of PKARIIβ with LRRK2 allowed for the diffusion of PKARIIβΔ2–5 into the dendritic spines of LRRK2+/+ neurons (Supplementary Fig. 5c). By contrast, linking the MAP2 microtubule-binding domain (MTDB) with PKARIIβΔ2–5, which allows for a direct association of PKARIIβΔ2–5 with the microtubules27, prevented the localization of PKARIIβΔ2–5 into the dendritic spine of LRRK2+/+ neurons (Supplementary Fig. 5c). Interestingly, PKARIIβ-Δ2–5–MTBD was also excluded from the spines of LRRK2−/− neurons (Supplementary Fig. 5d), emphasizing the importance of microtubule association in anchoring PKARIIβ within the dendritic shaft. Considering LRRK2 binds to microtubules and regulates the dynamic assembly of microtubule network28,25, our data further support the importance of the LRRK2 microtubule binding property in providing the structural framework for retaining PKARIIβ in the dendritic shaft.
Alteration of GluR1 phosphorylation in LRRK2−/− neurons
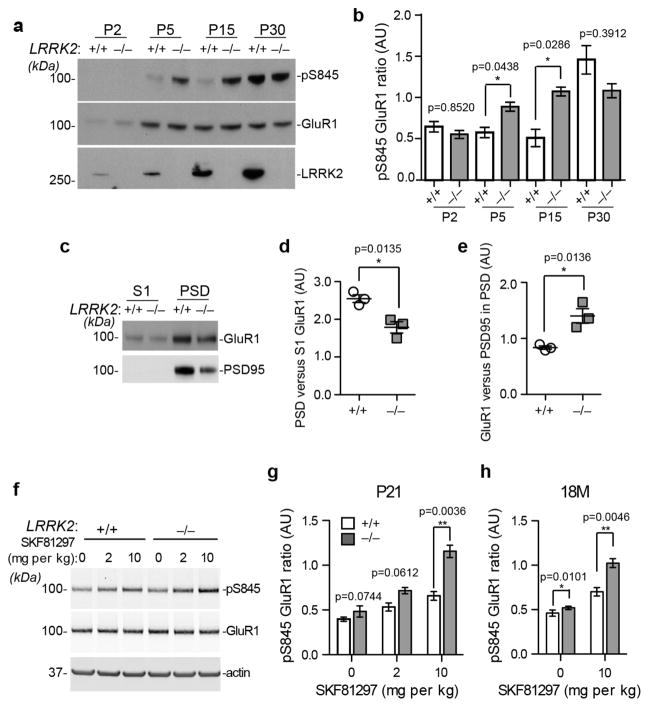
To examine the potential impact of aberrant PKA signaling on the postsynaptic targeting of AMPA-type glutamate receptors, we examined the PKA-dependent phosphorylation of GluR1 at residue Serine 845 (pS845)29. The level of pS845GluR1 was significantly increased in the forebrain homogenate of P5 and P15 LRRK2−/− pups compared to the age-matched littermate controls (Fig. 6a,b). The difference in GluR1 phosphorylation disappeared in P30 LRRK2+/+ and LRRK2−/− mice (Fig. 6a,b). FSK treatment did not further increase the level of pS845 GluR1 in cultured LRRK2−/− neurons (Supplementary Fig. 6a,b). LRRK2 kinase inhibitor LRRK2-IN-130 showed no effect on FSK-induced phosphorylation of GluR1 and cofilin in cultured LRRK2+/+ neurons (Supplementary Fig. 6c–e), suggesting that LRRK2 kinase activity is not involved in regulating PKA activity in neurons.
Figure 6. Loss of LRRK2 causes alteration of PKA-dependent phosphorylation of GluR1 in young and aged mice.
(a) Western blot analysis of pS845 GluR1, total GluR1 and LRRK2 expression in the forebrain homogenates of P2, P5, P15, and P30 LRRK2+/+ and LRRK2−/− mice. (b) Quantification analysis of pS845 GluR1 levels normalized to total GluR1 levels derived of three independent experiments. Data represent mean ± SEM. Unpaired t-test, t(4)=0.5791, 6.282, 3.576, and 1.061, respectively. *p<0.05, **p<0.01. (c) Western blot analysis of GluR1 and PSD95 in the S1 and PSD fractions of P15 LRRK2+/+ and LRRK2−/− mouse brains. (d,e) Quantification analysis of GluR1 levels in the PSD versus S1 fractions of brain extracts (d), as well as GluR1 levels versus PSD95 levels in the PSD fraction of brain extracts (e). n=3 per genotype. Data represent mean ± SEM. Unpaired t-test, t(4)=4.218 (d) and 4.206 (e). *p<0.05. (f) Western blot analysis of pS845 and total GluR1 in the striatum of P21 LRRK2+/+ and LRRK2−/− mice after treated with saline (0) or Drd1 agonist SKF81297 at 2 and 10mg per kg bodyweight. Actin was used as the loading control. (g,h) Quantification analysis of pS845 GluR1 ratio in the striatal tissues of P21 (g) and 18-month-old mice (h) treated with SKF81297. n=4 per genotype per treatment. Data represent mean ± SEM. Unpaired t-test. t(8)=2.501, 2.177, and 4.605 in (g). t(6)=3.703 and 4.401 in (h). *p<0.05, **p<0.01. The full-length images of Fig. 6a,c,f are shown in Supplementary Fig. 6h-j.
The phosphorylation of GluR1 by PKA promotes the trafficking of GluR1 to synapses31. Although the overall level of GluR1 in the PSD fraction was significantly decreased in the brain homogenate of P15 LRRK2−/−−/− pups compared to littermate controls (Fig. 6c,d), when normalized against PSD95 protein the level of GluR1 was actually higher in LRRK2−/− neurons (Fig. 6e). These observations suggest that LRRK2−/− neurons may have less mature dendritic spines, but these mature spines may contain more GluR1 for synaptic transmission.
Dopamine activates the PKA signaling via Drd1 family dopamine receptors19. Intraperitoneal injection of a selective Drd1 agonist SKF8129732 caused significant increase of pS845 GluR1 in the striatal tissues of P21 LRRK2+/+ and LRRK2−/− mice, whereas significantly more pS845 GluR1 was detected in the LRRK2−/− samples (Fig. 6f,g). Furthermore, SKF81297 stimulation also induced more pS845 GluR1 in the striatum of 18-month-old LRRK2−/− mice compared to the age-matched controls (Fig. 6h). In contrast, SKF81297 did not affect protein kinase C (PKC)-mediated phosphorylation of GluR1 at S83133 in the striatum of P21 LRRK2+/+ and LRRK2−/− mice (Supplementary Fig. 6f,g). Taken collectively, these data reveal an aberrant elevation of PKA activity induced by Drd1 activation in both young and aged LRRK2−/− mice, suggesting that LRRK2 continues to function as a negative regulator of PKA after the postnatal developmental period.
LRRK2 R1441C mutation impairs its regulatory role on PKA
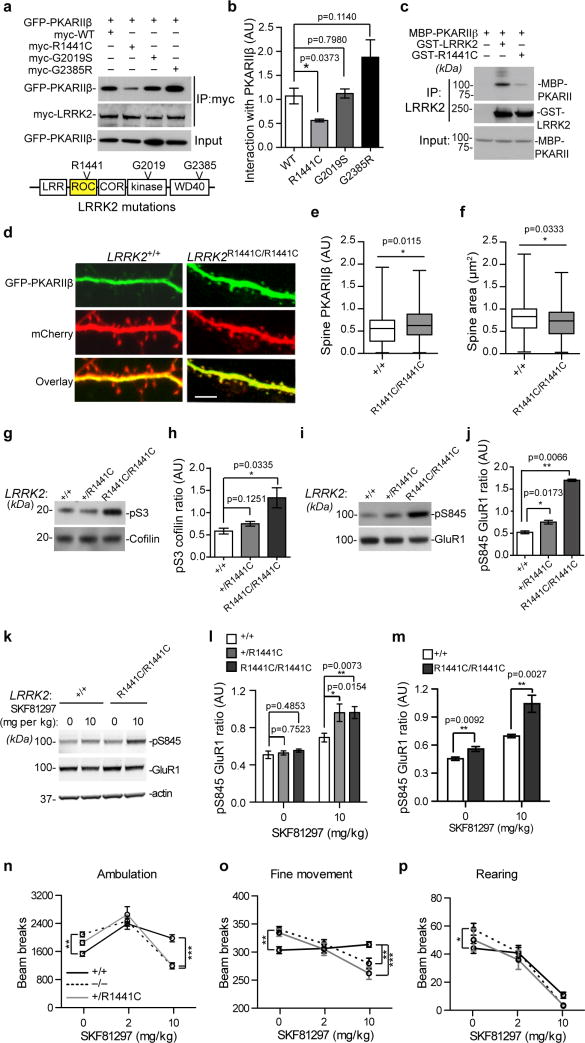
To examine the impact of Parkinson’s disease-related LRRK2 missense mutations on the interaction with PKARIIβ, we found that the R1441C mutation at the ROC domain of LRRK2 significantly compromised the interaction between LRRK2 and PKARIIβ (Fig. 7a,b). In vitro recombinant protein binding assays further confirmed the reduced interaction (43.33 7.53%, unpaired t-test, t4=7.52, p=0.0017) between R1441C LRRK2 and PKARIIβ (Fig. 7c). We next examined the effect of R1441C LRRK2 on the subcellular localization of PKARIIβ in hippocampal neurons cultured from homozygous LRRK2 R1441C knock-in (LRRK2R1441C/R1441C) mice34. A significant increase of PKARIIβ signals was found in the dendritic spine of LRRK2KI/KI neurons compared to the littermate control LRRK2+/+ neurons (Fig. 7d,e). In addition, LRRK2R1441C/R1441C neurons displayed decreased spine area compared to the controls (Fig. 7f). Meanwhile, we also observed a dose-dependent increase of pS3 cofilin and pS845 GluR1 in the brains of P15 LRRK2+/RC and LRRK2R1441C/R1441C mice (Fig. 7g–j). Together, these data suggest that the R1441C mutation may compromise the regulatory function of LRRK2 on PKA during synaptogenesis.
Figure 7. The R1441C mutation impairs the regulatory function of LRRK2 on PKA.
(a) Co-IP of GFP-PKARIIβ with wild-type, R1441C, G2019S, and G2835 LRRK2 from transfected cells (a). (b) The relative binding affinity of GFP-PKARIIβ with LRRK2 proteins (n=3). t(4)=3.070, 0.2735, and 2.016. (c) Co-IP of GST-LRRK2 and GST-R1441C with MBP-PKARIIβ recombinant proteins. (d) GFP-PKARIIβ and mCherry signals in the spines and dendrites of 15DIV cultured hippocampal neurons. Scale bar: 5μm. (e,f) Quantification of GFP-PKARIIβ signal in the spines (f, n=70–75 spines from six neurons per genotype), and the spine size (g, n=60 spines from four neurons per genotype)..t=2.552 and 2,145, respectively. (h–k) The pS3 versus total cofilin (h,i) and pS845 versus total GluR1 (j,k) in the forebrain homogenate of P15 LRRK2+/+, LRRK2+/R1441C and LRRK2R1441C/R1441C mice. n=3 per genotype. t(4)=1.935 and 3.182 in (h), 3.918 and 5.191 in (j). (l–n) The pS845 versus total GluR1 in the striatum of P21 LRRK2+/+ (n=8), LRRK2+/R1441C (n=4), and LRRK2R1441C/R1441C (n=4) mice (l,m), and 18-month-old LRRK2+/+ (n=4) and LRRK2RC/RC (n=4) mice (n). t(10)=0.3237, 0.7222, 2.865, and 3.283 in (m). t(6)=3.303 and 4.083 in (m). (o–q) The ambulatory (o), fine (p), and rearing movement (q) of P21 LRRK2+/+, LRRK2−/−, and LRRK2+/RC mice after treated with saline (n=16, 37, and 10) or SKF81297 at 2 (n=21, 28, and 11) and 10mg per kg bodyweight (n=27, 28, and 10). Data represent mean ± SEM. Unpaired test (b,e,f,h,j,l,m) and two-way ANOVA plus Bonferroni post-tests (m–q) were used.
To test the response of LRRK2+/R1441C and LRRK2R1441C/R1441C mice to dopamine stimulation, we found that SKF81297 treatment also induced an abnormal increase of pS845 GluR1 in the striatum of P21 and 18-month-old knock-in mice compare to the age-matched control mice (Fig. 7k–m). Notably, both LRRK2+/RC and LRRK2R1441C/R1441C mice showed similar alterations of GluR1 phosphorylation after treated with SKF81297 (Fig. 7l,m).
To examine the functional consequence of Drd1 activation, we monitored the movement of saline- or SKF81297-treated P21 LRRK2+/+, LRRK2−/−, and LRRK2+/R1441C mice in the Open-field tests. Under the saline treatment, LRRK2−/− mice showed significantly increased ambulatory, fine (e.g. grooming), and rearing movements compared to LRRK2+/+ mice (Fig. 7n–p). Although LRRK2+/R1441C mice displayed a similar increase of motor activity in the saline-treated condition, the difference is not statistically significant (Fig. 7n–p). The administration of SKF81297 generally promoted the ambulatory and fine movements, but suppressed the rearing of control LRRK2+/+ mice (Fig. 7n–p). Notably, both LRRK2−/− and LRRK2+/R1441C mice were significantly less responsive to higher dosage of SKF81297 stimulation compared to the control mice (Fig. 7n–p). These behavioral data suggest that the LRRK2 R1441C missense mutation may compromise the normal function of LRRK2 in the SPNs in response to dopamine stimulation.
DISCUSSION
In this study we provide substantial evidence that LRRK2 regulates synaptogenesis and dopamine receptor activation in the SPNs through modulation of PKA activity. Abnormal dendritic spine morphology and synaptic transmission were observed in the SPNs of P15 LRRK2−/− pups, which may be attributable to the aberrant increase of PKA-mediated phosphorylation of S3 cofilin, S845 GluR1 and other substrates in the LRRK2-deficient neurons. LRRK2 continues to regulate PKA activity in the SPNs of young and aged mice as evident by the abnormal phosphorylation of S845 GluR1 in response to dopamine receptor Drd1 activation, which may contribute to the reduced responsiveness of LRRK2−/− mice to the Drd1 agonist-induced motor activity. LRRK2 may regulate the specificity and efficacy of PKA signaling in the SPNs by selectively interacting with PKARIIβ subunits and controlling the predominantly dendritic shaft localization of PKA holoenzyme. Thereby, a loss of LRRK2 allowed more translocation of PKA holoenzyme into the dendritic spines where PKA is in closer proximity to the source of cAMP production and the synaptic substrates35, resulting in excessive phosphorylation of key synaptic proteins involving in synaptic formation and transmission. Perhaps more notably, the Parkinson’s disease-related LRRK2 R1441C mutation significantly impaired the interaction between LRRK2 and PKARIIβ, and caused similar alterations of PKA signaling as the LRRK2-null counterparts. Therefore, our findings provide new molecular insights into the pathophysiological mechanism of LRRK2 in Parkinson’s disease.
In an attempt to identify the molecular players and signaling pathways underlying the dendritic spine morphological changes in LRRK2−/− neurons, we observed a significant increase of cofilin phosphorylation in these neurons. The phosphorylation of cofilin is generally accepted to decrease its binding with F-actin and prevent its disassembly of F-actin36. Consistent with an earlier study37, the decrease of cofilin activity in LRRK2−/− neurons impaired the dynamic assembly of F-actin as evident by the accumulation of F-actin aggregates in the soma and the decrease of F-actin content in the dendritic spines. Accordingly, the introduction of constitutively active S3A-cofilin in LRRK2−/− neurons prevented F-actin aggregation. The inhibition of cofilin activity by siRNA knockdown has also been shown to cause longer, thinner, and more complex dendritic protrusions in cultured neurons38, as we have observed in the LRRK2−/− neurons. Collaboratively, the introduction of S3A-cofilin enabled to rescue the dendritic spine abnormalities in the LRRK2−/− neurons. It is worth to point out that cofilin not only depolymerizes or severs F-actin according to the classical model, it also promotes the net assembly and polymerization of the newly generated barbed ends under specific conditions39, which may explain how the inactivation of cofilin led to aggregation of F-actin at the soma and reduced the F-actin content in the dendritic spine of LRRK2−/− neurons. Together, our observations reveal an active involvement of LRRK2 in the formation of dendritic spines through modulating cofilin-mediated actin dynamics.
LRRK2 modulates the activity of PKACα by anchoring both PKACα and PKARIIβ along the dendritic shaft. The spatial restriction of PKARIIβ in the dendritic shaft plays an important role in regulating PKACα activity during the excitatory synaptic transmission27. For instance, dendritic MAP2 is considered as a neuronal AKAP that anchors PKARIIβ along with PKACα in the dendritic shaft40. After cAMP binds to PKARIIβ, the catalytic subunits begin to dissociate from the complex and enter the dendritic spine27. Neurons derived from MAP2Δ1-158 mutant mice that also display increased translocation of PKARIIβ into dendritic spines show increase of PKA-dependent phosphorylation of GluR127. Based on these previous observations27,40 we propose LRRK2 as an additional dendritic AKAP that interacts with PKARIIβ, acting synergistically with, or independently of MAP2, particularly at the time of active dendritic spine morphogenesis. Similarly to what happened in the MAP2Δ1-158 mutant mice40, the absence of LRRK2 allowed significantly more PKACα and PKARIIβ subunits in the dendritic spines of LRRK2−/− neurons, which is likely to bring PKA holoenzyme closer to the upstream components of cAMP-mediated signaling transduction pathways, such as the dopamine receptors and adenylate cyclase, as well as the downstream substrates, such as the synaptic protein GluR1, resulting in the abnormally increased PKA activity. Therefore, there may exist competition between dendritic shaft AKAPs, such as MAP2 and LRRK2, and synaptic AKAPs such as AKAP79/15041, in anchoring the PKA holoenzyme in the dendritic shaft and spines. The loss of LRRK2 may shift the balance to more PKA binding with the synaptic AKAPs, leading to more PKA activity. This hypothesis is further supported by a recent computational analysis of the importance of subcellular location of PKA holoenzyme in dopamine receptor Drd1-induced activation of PKA35. The key revelation of this study is that cAMP exhibits a strong spatial gradient within the dendritic spines and shaft upon dopamine activation, of which the distance between PKA and the source of cAMP is more important in controlling the PKA activity. Given the presence of Drd1 receptor, G protein (Golf), and adenylate cyclase 5 (AC5) mainly in the dendritic spines of SPNs35, the increase translocation of PKA holoenzyme in the spines of LRRK2−/− SPNs thereby renders these neurons more sensitive to PKA activation in response to dopamine stimulation.
Considering the predominant effects of LRRK2 occurs between P5 and P15, LRRK2 may be a key negative regulator of PKA signaling at this critical time period for synaptogenesis. In line with this notion, PKARIIβ is important for the induction of LTP in young mice (P10–P14) but not in older ones (P21–P28), of which another protein kinase CaMKII becomes predominant42. Having shown morphological changes in dendritic spines as a result of LRRK2’s absence, we examined excitatory synaptic transmission in LRRK2−/− neurons. We demonstrate a decrease in the frequency of mEPSCs in the SPNs of P15 pups that may reflect the improper maturation of dendritic spines and synapses in these neurons. This observation is further supported by the decrease in the expression of synaptic markers such as the postsynaptic adaptor proteins PSD95 in the PSD fraction of P15 LRRK2−/− brains. On the other hand, the amplitude of the mEPSCs was slightly but significantly increased in P15 LRRK2−/− neurons, despite the overall decrease in GluR1 level in the PSD fraction. However, provided that LRRK2−/− neurons had less PSD proteins, further normalization of GluR1 against PSD95 protein showed an actual increase of GluR1 in the PSD fraction of LRRK2−/− mice. Therefore, LRRK2−/− neurons had overall less mature dendritic spines and synapses, but their mature spines contained more GluR1, resulting in reduced mEPSC frequency but increased mEPSC amplitude. These data are also of interest considering that increased GluR1 phosphorylation is observed in the P5 and P15 LRRK2−/− pups. Enhanced GluR1 phosphorylation may facilitate its trafficking to the perisynaptic sites followed by lateral diffusion into the dendritic spines43, which may lead to more GluR1 targeted to the postsynaptic site of mature dendritic spines in LRRK2−/− neurons. The increased synaptic targeting of GluR1 as well as the related increase of mEPSC amplitude may serve as a compensatory mechanism to salvage the overall impaired synaptic transmission due to the delay of spine maturation in LRRK2−/− neurons.
The impairment of synaptic transmission underlies the key pathophysiological phenotypes prior to neurodegeneration in Alzheimer’s disease, Parkinson’s disease, and other degenerative neurological disorders44. Parkinson’s disease-related dopamine deficiency has been shown to cause substantial changes of the spine morphology and function of SPNs3. Golgi staining of postmortem Parkinson’s disease brains has revealed 20–30% reduction in striatal spine density and a reduction of the size of the dendritic trees of SPNs45,46. Together, these early studies demonstrate a critical involvement of SPNs in the pathogenic processes of Parkinson’s disease. In light of the function of LRRK2 in regulating PKA activity, Drd1 in the SPNs also utilizes the PKA pathway as a means to relay extracellular dopamine stimulation to intracellular signaling transduction1. It also should be noted that phenotypes of PKARIIβ knockout mice are mainly related to the striatal functions, emphasizing the requirement of PKARIIβ in motor learning and related dopaminergic signaling18. Therefore, we suspect that LRRK2-deficiency may interfere with the PKA signaling in the striatal SPNs and compromise their responses to dopaminergic transmission. Indeed, Drd1 agonist caused aberrant increase of PKA-mediated phosphorylation of GluR1 in the striatum of LRRK2−/− mice, which may be attributable to the less responsiveness of LRRK2−/− mice to Drd1 agonist-induced motor activity. Interestingly, Parkinson’s disease-related LRRK2 R1441C knock-in mice showed similar abnormal GluR1 phosphorylation and motor behavioral phenotypes as the LRRK2−/− mice in responding to Drd1 stimulation. These observations are also consistent with an early report that LRRK2 R1441C knock-in mice are less responsive to amphetamine-induced locomotor activity34, as the main effect of amphetamine in the striatum is to causes an increase of dopamine release47. Considering LRRK2 functions as a dimer48, our data suggest the R1441C missense mutation may exert a dominant-negative effect on the regulatory function of LRRK2 in modulating PKA activity by dimerization with the wild-type proteins.
The activation of Drd1 appears to exert different effects on different types of motor activities, which increased the ambulatory and fine movements, but suppressed the rearing, suggesting the existence of different Drd1-invovled neural circuits in regulating specific motor outputs. The loss of LRRK2 mainly compromised the ambulatory and fine motor responses to Drd1 activation. Given LRRK2 is more abundant in a subset of SPNs at the striosomes6, it is of interest to determine the exact neural circuits in the striatum and basal ganglion affected by LRRK2. Notably, a recent neural tracing study reveals that SPNs in the striosomes can directly project to the nigrostriatal dopaminergic neurons49. Although the functionality of this newly defined circuit remains to establish, SPNs in the striosomes may directly modulate the excitability and dopamine release of nigrostriatal dopaminergic neurons. Considering the increase of pS845 GluR1 in the LRRK2−/− and LRRK2+/R1441C SPNs may enhance the postsynaptic transmission and activation of these inhibitory GABAnergic neurons, we suspect that LRRK2−/− and LRRK2+/R1441C SPNs may further inhibit the activity of nigrostriatal dopaminergic neurons, resulting in suppression of dopamine release. Certainly, more physiological and behavioral studies will need to fully understand the function of this circuit in both normal and Parkinson’s disease conditions.
ONLINE METHODS
Animals
LRRK2−/− mice, CaMKII-tTA/tetO-WT-LRRK2 transgenic mice, LRRK2 wild-type BAC transgenic mice and LRRK2 R1441C knock-in mice were created as described previously8,21,25,34, and maintained in C57BL6 strain background. Two to four mice were housed in each cages and in a 12h light/dark cycle and fed regular diet ad libitum. All mouse work follows the guidelines approved by the Institutional Animal Care and Use Committees of the National Institute of Child Health and Human Development, NIH.
Golgi–Cox impregnation of brain tissue and spine measurements
Mice were perfused intracardially with 0.9% saline. The brains were dissected, and impregnated using a standard Golgi–Cox solution (1% potassium dichromate, 1% mercuric chloride, and 0.8% potassium chromate), according to a previously described method50. The brains immersed in the Golgi–Cox solution were stored at room temperature for 6d, immersed in a sucrose solution (30%) for 2d, and sectioned coronally (150μm) using a vibratome. Sections were mounted on gelatinized slides and stained according to a previously described method51. Dorsolateral striatal Golgi-stained sections were imaged under brightfield illumination on a Zeiss microscope system and a 100× oil immersion lens. Healthy SPNs were chosen randomly for quantification from three mice per group, and spines on tertiary dendrites were considered from an individual blind to the genotype of neurons. The density of spines and filopodia was analyzed by Neurolucida 10 software (MBF Inc., Williston, VT, USA). For spine size, images were acquired and the full length and head diameter were measured manually with ImageViewer52, a custom-made software for spine analysis written in Matlab (The MathWorks). For frequency distributions, spines were separated into thin mushroom and stubby spines based on length to head width ratio. Each spine was categorized as stubby if without a neck, and with a total length to head width ratio less than 0.3. Spines with total length to head width ratio more than 1 were categorized as thin, and those between 0.31 and 0.99 were categorized as mushroom. Filopodia were protrusions greater than 2.5μm in length without a neck. The comparison of cumulative frequency distributions was evaluated by the Kolmogorov-Smirnov test (KS-test), and average comparison of length, width and densities were performed with Student’s t test or U of Mann-Whitney test. Distributions in frequencies of different category of spine were compared with Chi-Squared test.
Electrophysiology
Brain slices containing striatum were prepared from P14–15 or P21–28 LRRK2−/− and LRRK2+/+ littermates. The animals were anesthetized deeply with isoflurane and decapitated. The brains were placed in ice-cold modified artificial CSF (aCSF) containing the following: 194mM sucrose, 30mM NaCl, 4.5mM KCl, 1mM MgCl2, 26mM NaHCO3, 1.2mM NaH2PO4, and 10mM dextrose, saturated with 95% O2/5% CO2. Coronal sections (280μm) through striatum were prepared using a vibratome (Leica Microsystems). Sections were equilibrated for at least 1h at 33°C in 95% O2/5% CO2 continuously bubbled aCSF containing the following: 124mM NaCl, 4.5mM KCl, 2mM CaCl2, 1mM MgCl2, 26mM NaHCO3, 1.2mM NaH2PO4, and 10mM D-glucose. Sections were subsequently incubated in the same solution at room temperature. Electrodes (2–5MΩ resistance) were pulled from borosilicate glass on a Flaming/Brown micropipette puller (Sutter Instruments). Pipettes were filled with solution (290–300mOsm) containing the following: 120mM CsMeSO3, 5mM NaCl, 10mM tetraethylammonium-Cl, 10mM HEPES, 5mM QX-314, 1.1mM EGTA, 0.3mM Na-GTP, and 4mM Mg-ATP, with pH set at 7.2 with CsOH53. Miniature EPSCs (mEPSCs) were performed between 29°C and 31°C and in the presence of picrotoxin (50μM) and TTX (1μm). Currents were measured in conventional ruptured-patch whole-cell mode in striatal medium spiny neurons (SPNs) voltage-clamped at – 60mV using a Multiclamp 700A amplifier (Molecular Devices). mEPSCs were recorded in gap-free mode for 5–10min and analyzed by using the Mini Analysis Program (Synaptosoft Inc)54 [Amplitude threshold (15), Area threshold (20), Period to search local maximum (15000μs), Time before peak for baseline (10000μs), Period to average baseline (10000μs), Period to search decay time (5000μs), Fraction to find decay (0.001)]. To measure paired pulse facilitation (PPF), recordings were performed in the presence of picrotoxin (100μm). Paired stimuli at an interpulse interval of 50ms were delivered and the ratio of peak amplitudes of the EPSCs was calculated.
Western blot and co-IP
Tissue or neurons were lysed in 50mM Tris-HCl pH7.6, 150mM NaCl, 2mM EDTA, 2% SDS or RIPA buffer (Sigma Aldrich) supplemented with complete protease inhibitor mixture (Roche Applied Biosciences) and phosphatase inhibitor mixture (Pierce Biotechnology). Proteins were separated by 4–12% NuPage Bis-Tris PAGE (Invitrogen) and transferred to nitrocellulose membranes. Antibodies specific for LRRK2 [Epitomics 3514-1, IP 1:1000; UC Davis/NIH NeuroMab Facility 73–188, Western blot (WB) 1:1000, immunocytochemistry (ICC) 1:200; and Abcam ab133475, WB 1:1000, ICC 1:200], pLRRK2 (S935, Abcam ab133450, WB 1:1000), PKACα (BD Bioscience 610981, WB 1:1000 and Cell Signaling 47825, IP 1:1000, WB 1:1000, ICC 1:1000), PKARIα (BD Biosciences 51-9002044, WB 1:1000), PKARIIα (BD Biosciences 51-9002051, WB 1:1000), PKARIIβ (BD Biosciences 610625, WB 1:10000, ICC 1:1000, IP: 1:1000), pPKARIIβ (S114, BD Biosciences 612550, WB 1:1000), pCofilin [S3, Cell Signaling 3313S, WB 1:1000, ICC 1:200, immunohistochemistry (IHC), 1:200], Cofilin (cell Signaling 3318, WB 1:1000), pLIMK1 (T508, Cell Signaling 3841, WB 1:1000), LIMK1 (Cell Signaling 3842, WB 1:1000), Hsp90 (BD Biosciences 610418, WB 1:1000), pCREB (S133, Cell Signaling 9198, WB 1:1000), CREB (Cell Signaling 9104, WB 1:1000), pGluR1 (S845, Sigma A4477, WB 1:1000; S831, Sigma A4352, WB 1:1000), GluR1 (Abcam ab31232, WB 1:1000), β-actin (Sigma A2228, WB 1:1000), PSD95 (Sigma P246, WB 1:1000; Millipore MAB1598, ICC 1:200 ), PP1 (Sigma P7607, WB 1:1000), 14-3-3ζ (Cell Signaling 7413, WB 1:1000), HA (Roche Applied Science 11867423001, WB 1:1000), myc (Sigma M4439, WB 1:1000, IP 1:1000), pSSH1L (S978, ECM Biosciences SP3901, WB 1:1000), SSH1L (ECM Biosciences SP1711, WB 1:1000), β-tubulin III (Millipore AB9345, WB 1:1000), Diap1 (Cell Signaling 5486P, WB 1:1000), Diap2 (Cell Signaling 5474P, WB 1:1000), Arp2 (Cell Signaling 3128P, WB 1:1000), Arp3 (Cell Signaling 4738P, WB 1:1000), CAP1 (Sigma, SAB1406999, WB 1:1000), and cdc42/Rac (Cell Signaling 2416, WB 1:1000) were used. Protein bands were detected using the enhanced chemiluminescence (ECL) system (Pierce Biotechnology) and quantified using the Scion Image System. For the co-immunoprecipitation experiments, brain extracts or cellular extracts of transfected cells (500–1500μg of total protein) were lysed with IP buffer (20mM Hepes, 300mM NaCl, 5% (v/v) glycerol, 0.2mM EDTA and 1% NP40. The lysates were incubated with appropriate dilutions of the various antibodies overnight at 4°C. Antibody-bound protein complexes were collected with either protein G or protein A (Pierce Rockford, IL, USA) for 1h at 4 °C, and then the pellets were washed five times with IP buffer at 4°C. The beads were resuspended in SDS sample buffer (Invitrogen) and the recovered proteins were separated by SDS-PAGE gel electrophoresis, and detected by immunoblot analysis.
Primary neuronal cell cultures
Primary neuronal cultures from cortex, striatum and hippocampus from pups at P0 were prepared as described previously55. In brief, neurons were dissociated by papain buffer (Sigma Aldrich), and were then placed in poly-D-lysine slides (BD) or plates in Basal Eagle Medium (Sigma-Aldrich). Arabinosylcytosine (Sigma-Aldrich) was used to inhibit glial cell growth. Forskolin (FSK), PKI (PKA inhibitor) and LRRK2 kinase inhibitor LRRK2-IN-1 (Tocris Bioscience, Bristol, UK) as well as all the other compounds were added directly to the medium of cortical neurons from stock solution and unless otherwise stated these compounds were from Calbiochem.
Transfection of primary neurons
Primary hippocampal or striatal neurons were transfected between P15-P21 DIV using the calcium phosphate method as described previously56. Neuronal cultures were incubated with the DNA-calcium phosphate precipitate for approximately 1.5hr. The precipitate was then dissolved by the incubation of the cells in a medium that had been pre-equilibrated in a 10% CO2 incubator. The cells were in turn transferred to their original conditioned medium and fixed at about 24–48hr after transfection. GFP-tagged PKARIIβ, PKACα, PKARIIβΔ2–5, and PKARIIβΔ2–5 expression vector and mCherry expression construct were obtained from Dr. Haining Zhong. Myc-tagged human wild-type LRRK2 and LRRK2 ROC domain expression constructs were obtained from Dr. Mark Cookson (NIA, NIH). GFP-tagged cofilin expression vector was purchased from Origene Technologies (Rockville, MD). GFP-S3A and GFP-S3D cofilin mutant constructs were generated using QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA).
Immunofluorescence staining and image acquisition
Neurons were fixed in 4% paraformaldehyde (PFA) as described previously8. Briefly, they were permeabilized with Triton-X-100 and incubated with 10% goat serum (Sigma-Aldrich) for 1hr to block unspecific binding and incubated overnight with the primary antibody. Fluorescent images were captured using a Zeiss confocal microscope (LSM510META). Spinal intensities and areas quantification was performed using the Image J Software. Regions of interest (ROIs) were set up automatically and the green (PKARIIβ) and red (mCherry) signals within the ROIs were divided to adjacent dendritic green and red intensities. The ratio of the normalized green to red signals in each genotype represents the relative intensity of the PKARIIβ inside the spines. The data presented involve about 80–150 spines sampled from three randomly selected microscopic fields of at least three independent experiments from a person blind to the genotype of neurons.
cAMP ELISA determination
cAMP levels of cortical neurons of each genotype (LRRK2+/+, LRRK2−/−) treated or untreated with FSK were quantified by using a cAMP Elisa kit (R&D Systems) according to the manufacturer’s protocol. Briefly, three independent experiments with each sample assaying in duplicates were used for the analysis. The goat anti-mouse antibody that is coated on the microplate binds to a monoclonal antibody specific for cAMP. The cAMP present in the samples compete with an amount of HRP-labeled cAMP for binding on the monoclonal antibody. A substrate solution is incubated in the microplate to determine the bound enzyme activity. The intensity of the color is proportional to the cAMP inversebly concentration to the concentration of cAMP in the sample.
Subcellular fractionation of brains
PSD fractions were prepared from mouse brains as previously described57. All procedures performed at 4°C. Mouse brains were rapidly decapitated and homogenized in four volumes of ice-cold Buffer A (0.32M sucrose, 5mM HEPES, pH7.4, 1mM MgCl2, 0.5mM CaCl2, protease and phosphatase inhibitor mixture) with Teflon homogenizer (12 strokes). Homogenized brain extract was spun at 1400 × g for 10min. Supernatant (S1) was saved and pellet (P1) was homogenized again with Teflon homogenizer with buffer A (5 strokes). After centrifugation at 700 × g, the supernatant (S1′) was saved and pooled with S1. Pooled S1 and S1′ were centrifuged at 13,800 × g for 10min to collect the crude synaptosomal pellet (P2) and the supernatant (S2), which contains the cytosol and light membranes. S2 was centrifuged at 100,000 × g for 15min to produce the cytosolic fraction (S2′) and the light membrane (LM) fraction. P2 was resuspended in Buffer B (0.32M sucrose, 6mM Tris, pH 8.0 supplemented with protease and phosphatase inhibitors cocktail) with Teflon homogenizer (five strokes). The P2 suspension was loaded onto a discontinuous sucrose gradient (0.85M/1M/1.15M sucrose solution in 6mM Tris, pH8.0), followed by centrifugation for 2h at 82,500 × g. The synaptosome fraction between 1M and 1.15M sucrose was collected and resuspended with 6mM Tris with 0.5% Triton. The PSD fraction was purified by centrifugation at 18,900 rpm with a Ti70.1 rotor.
Intraperitoneal injection of Drd1 agonist SKF-81297, Open-field test, and Western blot analysis of striatal proteins
Healthy mice removed from the light cycle were placed in the dark room for 30min before behavioral test. 5min after the intraperitoneal injection of 0.9% (w/v) saline, 2mg/kg or 10mg/kg Drd1 agonist, R(+)-SKF-81297 hydrobromide (Sigma), mice were placed in the chamber for Open-field test using Flex-Field activity system (San Diego Instruments, CA) as described previously32,58. Immediately after the behavioral test, mice were decapitated and the striatal tissues were dissected out. Striatal tissues were homogenized and sonicated in 1% SDS lysis buffer and denatured at 70°C for 10min, as described previously59. 20ug protein was resolved in 4–12% Tris-Bis NuPage gel and transferred to PVDF membrane. Blots were incubated with antibodies against total GluR1 and pS845 GluR1. Expression level and the ratios between phosphorylated and total proteins were determined using a LI-COR system and Odyssey software and expressed relative to saline vehicle controls.
Statistics
Graph Pad Prism 5 was used for the Statistical Analysis. Statistical significances were determined by comparing means of different groups using t test or two-way ANOVA followed by post-tests. Distributions of mEPSC amplitudes and frequencies as well as morphological analysis distributions were compared using Chi-Squared test or Kolmogorov-Smirnov (K-S) tests. Band intensities of Western blots were compared with unpaired Student t-tests (two-sided). Summary data is presented as mean SEM. *P<0.05; **p<0.01; ***p<0.001, ****p<0.0001.
Supplementary Material
Acknowledgments
This work was supported by the intramural research programs of National Institute on Aging (AG000944, AG000928, HC) and National Institute of Alcohol Abuse and Alcoholism (DL). We thank Dr. Haining Zhong of Vollum Institute for providing PKA expression vectors, Dr. Mark Cookson of NIA for providing human LRRK2 expression vector, Drs. Jie Shen and Youren Tong of Harvard University for providing LRRK2 R1441C knock-in mice, Dr. Patrick Lewis of University College London for providing LRRK2 wild-type and R1441C recombinant proteins, Dr. Veronica Alvarez from NIAAA, Dr. Bo Ma from NIA and Drs. Zheng Li, Jie-Min Jia, and Song Jiao of National Institute of Mental Health for their advice and technical support. We also thank Ms. Laura Donahue and Ms. Namratha Sastry for their assistance in editing the manuscript.
Footnotes
AUTHOR CONTRIBUTIONS
L.P and H.C conceived the project, designed the experiments and wrote the manuscript. L.P. and J. Y. generated and analyzed biochemical and cell biology data in Fig. 2–7 and Supplementary Fig. 1–6. J. Y. generated and analyzed data in Fig. 4–7 and Supplementary Fig. S4–6 and performed the Open-field tests in Fig. 7. S.C. and D.L. performed Golgi-COX staining and electrophysiology experiments and data analysis in Fig. 1. G.L and J. Y. performed immunofluorescence experiment of brain section in Supplementary Fig. 2. C.X. conducted primary neuronal cultures, immunofluorescence staining, and transfection experiments. L.S. generated the cofilin constructs shown in Supplemental Fig. 2. X.G. performed the PPF experiment in Fig 1. X.L. was actively involved in the mice generation. N.C. helped with electrophysiology experiments.
References
- 1.Gerfen CR, Surmeier DJ. Modulation of striatal projection systems by dopamine. Annu Rev Neurosci. 2011;34:441–466. doi: 10.1146/annurev-neuro-061010-113641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith Y, Villalba RM, Raju DV. Striatal spine plasticity in Parkinson’s disease: pathological or not? Parkinsonism Relat Disord. 2009;15(Suppl 3):S156–161. doi: 10.1016/S1353-8020(09)70805-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calabresi P, Picconi B, Tozzi A, Di Filippo M. Dopamine-mediated regulation of corticostriatal synaptic plasticity. Trends Neurosci. 2007;30:211–219. doi: 10.1016/j.tins.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 4.Graybiel AM, Ragsdale CW, Jr, Moon Edley S. Compartments in the striatum of the cat observed by retrograde cell labeling. Exp Brain Res. 1979;34:189–195. doi: 10.1007/BF00238352. [DOI] [PubMed] [Google Scholar]
- 5.Singleton AB, Farrer MJ, Bonifati V. The genetics of Parkinson’s disease: progress and therapeutic implications. Movement disorders : official journal of the Movement Disorder Society. 2013;28:14–23. doi: 10.1002/mds.25249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mandemakers W, Snellinx A, O’Neill MJ, de Strooper B. LRRK2 expression is enriched in the striosomal compartment of mouse striatum. Neurobiol Dis. 2012;48:582–593. doi: 10.1016/j.nbd.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 7.Westerlund M, et al. Developmental regulation of leucine-rich repeat kinase 1 and 2 expression in the brain and other rodent and human organs: Implications for Parkinson’s disease. Neuroscience. 2008;152:429–436. doi: 10.1016/j.neuroscience.2007.10.062. [DOI] [PubMed] [Google Scholar]
- 8.Parisiadou L, et al. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tada T, Sheng M. Molecular mechanisms of dendritic spine morphogenesis. Curr Opin Neurobiol. 2006;16:95–101. doi: 10.1016/j.conb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Yoshihara Y, De Roo M, Muller D. Dendritic spine formation and stabilization. Curr Opin Neurobiol. 2009;19:146–153. doi: 10.1016/j.conb.2009.05.013. [DOI] [PubMed] [Google Scholar]
- 11.Meng Y, et al. Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron. 2002;35:121–133. doi: 10.1016/s0896-6273(02)00758-4. [DOI] [PubMed] [Google Scholar]
- 12.Bernstein BW, Bamburg JR. ADF/cofilin: a functional node in cell biology. Trends Cell Biol. 2010;20:187–195. doi: 10.1016/j.tcb.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nadella KS, et al. Regulation of actin function by protein kinase A-mediated phosphorylation of Limk1. EMBO Rep. 2009;10:599–605. doi: 10.1038/embor.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frey U, Huang YY, Kandel ER. Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- 15.Francis SH, Corbin JD. Structure and function of cyclic nucleotide-dependent protein kinases. Annu Rev Physiol. 1994;56:237–272. doi: 10.1146/annurev.ph.56.030194.001321. [DOI] [PubMed] [Google Scholar]
- 16.Brandon EP, Idzerda RL, McKnight GS. PKA isoforms, neural pathways, and behaviour: making the connection. Curr Opin Neurobiol. 1997;7:397–403. doi: 10.1016/s0959-4388(97)80069-4. [DOI] [PubMed] [Google Scholar]
- 17.Ventra C, et al. The differential response of protein kinase A to cyclic AMP in discrete brain areas correlates with the abundance of regulatory subunit II. J Neurochem. 1996;66:1752–1761. doi: 10.1046/j.1471-4159.1996.66041752.x. [DOI] [PubMed] [Google Scholar]
- 18.Brandon EP, et al. Defective motor behavior and neural gene expression in RIIbeta-protein kinase A mutant mice. J Neurosci. 1998;18:3639–3649. doi: 10.1523/JNEUROSCI.18-10-03639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder GL, Fienberg AA, Huganir RL, Greengard P. A dopamine/D1 receptor/protein kinase A/dopamine- and cAMP-regulated phosphoprotein (Mr 32 kDa)/protein phosphatase-1 pathway regulates dephosphorylation of the NMDA receptor. J Neurosci. 1998;18:10297–10303. doi: 10.1523/JNEUROSCI.18-24-10297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong W, Scott JD. AKAP signalling complexes: focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 21.Li Y, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson’s disease. Nat Neurosci. 2009;12:826–828. doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagaoka R, Abe H, Obinata T. Site-directed mutagenesis of the phosphorylation site of cofilin: its role in cofilin-actin interaction and cytoplasmic localization. Cell Motil Cytoskeleton. 1996;35:200–209. doi: 10.1002/(SICI)1097-0169(1996)35:3<200::AID-CM3>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 23.Niwa R, Nagata-Ohashi K, Takeichi M, Mizuno K, Uemura T. Control of actin reorganization by Slingshot, a family of phosphatases that dephosphorylate ADF/cofilin. Cell. 2002;108:233–246. doi: 10.1016/s0092-8674(01)00638-9. [DOI] [PubMed] [Google Scholar]
- 24.Gungabissoon RA, Bamburg JR. Regulation of growth cone actin dynamics by ADF/cofilin. J Histochem Cytochem. 2003;51:411–420. doi: 10.1177/002215540305100402. [DOI] [PubMed] [Google Scholar]
- 25.Lin X, et al. Leucine-rich repeat kinase 2 regulates the progression of neuropathology induced by Parkinson’s-disease-related mutant alpha-synuclein. Neuron. 2009;64:807–827. doi: 10.1016/j.neuron.2009.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li X, et al. Phosphorylation-dependent 14-3-3 binding to LRRK2 is impaired by common mutations of familial Parkinson’s disease. PLoS One. 2011;6:e17153. doi: 10.1371/journal.pone.0017153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong H, et al. Subcellular dynamics of type II PKA in neurons. Neuron. 2009;62:363–374. doi: 10.1016/j.neuron.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gandhi PN, Wang X, Zhu X, Chen SG, Wilson-Delfosse AL. The Roc domain of leucine-rich repeat kinase 2 is sufficient for interaction with microtubules. J Neurosci Res. 2008;86:1711–1720. doi: 10.1002/jnr.21622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blackstone C, Murphy TH, Moss SJ, Baraban JM, Huganir RL. Cyclic AMP and synaptic activity-dependent phosphorylation of AMPA-preferring glutamate receptors. J Neurosci. 1994;14:7585–7593. doi: 10.1523/JNEUROSCI.14-12-07585.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deng X, et al. Characterization of a selective inhibitor of the Parkinson’s disease kinase LRRK2. Nat Chem Biol. 2011;7:203–205. doi: 10.1038/nchembio.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Esteban JA, et al. PKA phosphorylation of AMPA receptor subunits controls synaptic trafficking underlying plasticity. Nat Neurosci. 2003;6:136–143. doi: 10.1038/nn997. [DOI] [PubMed] [Google Scholar]
- 32.Kim DS, Szczypka MS, Palmiter RD. Dopamine-deficient mice are hypersensitive to dopamine receptor agonists. J Neurosci. 2000;20:4405–4413. doi: 10.1523/JNEUROSCI.20-12-04405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roche KW, O’Brien RJ, Mammen AL, Bernhardt J, Huganir RL. Characterization of multiple phosphorylation sites on the AMPA receptor GluR1 subunit. Neuron. 1996;16:1179–1188. doi: 10.1016/s0896-6273(00)80144-0. [DOI] [PubMed] [Google Scholar]
- 34.Tong Y, et al. R1441C mutation in LRRK2 impairs dopaminergic neurotransmission in mice. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:14622–14627. doi: 10.1073/pnas.0906334106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveira RF, Kim M, Blackwell KT. Subcellular location of PKA controls striatal plasticity: stochastic simulations in spiny dendrites. PLoS Comput Biol. 2012;8:e1002383. doi: 10.1371/journal.pcbi.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bamburg JR, Bernstein BW. ADF/cofilin. Curr Biol. 2008;18:R273–275. doi: 10.1016/j.cub.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Bellenchi GC, et al. N-cofilin is associated with neuronal migration disorders and cell cycle control in the cerebral cortex. Genes Dev. 2007;21:2347–2357. doi: 10.1101/gad.434307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hotulainen P, et al. Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J Cell Biol. 2009;185:323–339. doi: 10.1083/jcb.200809046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrianantoandro E, Pollard TD. Mechanism of actin filament turnover by severing and nucleation at different concentrations of ADF/cofilin. Mol Cell. 2006;24:13–23. doi: 10.1016/j.molcel.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 40.Harada A, Teng J, Takei Y, Oguchi K, Hirokawa N. MAP2 is required for dendrite elongation, PKA anchoring in dendrites, and proper PKA signal transduction. J Cell Biol. 2002;158:541–549. doi: 10.1083/jcb.200110134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bauman AL, Goehring AS, Scott JD. Orchestration of synaptic plasticity through AKAP signaling complexes. Neuropharmacology. 2004;46:299–310. doi: 10.1016/j.neuropharm.2003.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Yasuda H, Barth AL, Stellwagen D, Malenka RC. A developmental switch in the signaling cascades for LTP induction. Nat Neurosci. 2003;6:15–16. doi: 10.1038/nn985. [DOI] [PubMed] [Google Scholar]
- 43.He K, et al. Stabilization of Ca2+-permeable AMPA receptors at perisynaptic sites by GluR1-S845 phosphorylation. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20033–20038. doi: 10.1073/pnas.0910338106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen J. Impaired neurotransmitter release in Alzheimer’s and Parkinson’s diseases. Neurodegener Dis. 2010;7:80–83. doi: 10.1159/000285511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stephens B, et al. Evidence of a breakdown of corticostriatal connections in Parkinson’s disease. Neuroscience. 2005;132:741–754. doi: 10.1016/j.neuroscience.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 46.Zaja-Milatovic S, et al. Dendritic degeneration in neostriatal medium spiny neurons in Parkinson disease. Neurology. 2005;64:545–547. doi: 10.1212/01.WNL.0000150591.33787.A4. [DOI] [PubMed] [Google Scholar]
- 47.Sulzer D. How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron. 2011;69:628–649. doi: 10.1016/j.neuron.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng J, et al. Structure of the ROC domain from the Parkinson’s disease-associated leucine-rich repeat kinase 2 reveals a dimeric GTPase. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:1499–1504. doi: 10.1073/pnas.0709098105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- 50.Glaser EM, Van der Loos H. Analysis of thick brain sections by obverse-reverse computer microscopy: application of a new, high clarity Golgi-Nissl stain. J Neurosci Methods. 1981;4:117–125. doi: 10.1016/0165-0270(81)90045-5. [DOI] [PubMed] [Google Scholar]
- 51.Gibb R, Kolb B. A method for vibratome sectioning of Golgi-Cox stained whole rat brain. J Neurosci Methods. 1998;79:1–4. doi: 10.1016/s0165-0270(97)00163-5. [DOI] [PubMed] [Google Scholar]
- 52.Steiner P, et al. Destabilization of the postsynaptic density by PSD-95 serine 73 phosphorylation inhibits spine growth and synaptic plasticity. Neuron. 2008;60:788–802. doi: 10.1016/j.neuron.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mathur BN, Capik NA, Alvarez VA, Lovinger DM. Serotonin induces long-term depression at corticostriatal synapses. J Neurosci. 2011;31:7402–7411. doi: 10.1523/JNEUROSCI.6250-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tang K, Low MJ, Grandy DK, Lovinger DM. Dopamine-dependent synaptic plasticity in striatum during in vivo development. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:1255–1260. doi: 10.1073/pnas.031374698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cai H, et al. Loss of ALS2 function is insufficient to trigger motor neuron degeneration in knock-out mice but predisposes neurons to oxidative stress. J Neurosci. 2005;25:7567–7574. doi: 10.1523/JNEUROSCI.1645-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jiang M, Chen G. High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat Protoc. 2006;1:695–700. doi: 10.1038/nprot.2006.86. [DOI] [PubMed] [Google Scholar]
- 57.Peng J, et al. Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J Biol Chem. 2004;279:21003–21011. doi: 10.1074/jbc.M400103200. [DOI] [PubMed] [Google Scholar]
- 58.Chandran JS, et al. Progressive behavioral deficits in DJ-1-deficient mice are associated with normal nigrostriatal function. Neurobiol Dis. 2008;29:505–514. doi: 10.1016/j.nbd.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Napolitano F, et al. Role of aberrant striatal dopamine D1 receptor/cAMP/protein kinase A/DARPP32 signaling in the paradoxical calming effect of amphetamine. J Neurosci. 2010;30:11043–11056. doi: 10.1523/JNEUROSCI.1682-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.