Abstract
Background
Abnormal calcium handling plays a crucial role in arrhythmias, sudden cardiac arrest (SCA), and congestive heart failure (CHF). Calsequestrin 2 (CASQ2) mutations affect calcium release and initiate malignant ventricular arrhythmias (VA) and SCA syndromes. Common single nucleotide polymorphisms (SNPs) in CASQ2 may be associated with SCA in patients with coronary artery disease (CAD).
Objective
We examined the association of common CASQ2 SNPs with the risk of SCA in patients with CAD.
Methods
CASQ2 SNPs (n=14) were genotyped and analyzed in a case-control study comparing 114 patients with CAD and SCA due to ventricular arrhythmias (VA) to 311 CAD controls without VA or SCA.
Results
Multivariate logistic regression adjusting for age and CHF status identified an association between rs7521023 with SCA (odds ratio [OR] 2.72, 95% confidence interval (CI): 1.44–5.13, p=0.002). The substantial impact of CHF on SCA in the model (OR 26.6, 95% CI: 13.40–52.70, p<0.001) led us to further examine the relationship between CHF, SCA, and CASQ2 SNPs. We identified 2 CASQ2 variants (rs7521023 [OR 0.4, 95% CI 0.25–0.76, p=0.003] and rs6684209 [OR 19.8, 95% CI 3.63–108.2, p<0.001]) associated with CHF after adjusting for SCA, age, gender, and hypertension.
Conclusions
We observed association between a CASQ2 polymorphism and SCA due to VA in patients with CAD adjusting for CHF, and independent associations between CASQ2 SNPs and CHF adjusting for SCA. Further investigation in independent cohorts is needed to confirm these findings.
Keywords: Sudden cardiac arrest, ventricular arrhythmias, ventricular tachycardia, coronary artery disease, congestive heart failure, calsequestrin 2, genetics
Introduction
Sudden cardiac arrest (SCA) remains a major public health problem, accounting for up to 450,000 deaths annually in the United States and up to 15% of overall mortality. 1 Up to 80% of sudden cardiac deaths (SCDs) are attributable to coronary artery disease (CAD).2 Identifying those at increased risk for SCA and likely to benefit most from implantable cardioverter defibrillators (ICDs) remains a challenge due to the low sensitivity and specificity of current risk stratification criteria.3 Genetic susceptibility for SCA in the setting of coronary artery disease (CAD) was suggested by several epidemiologic studies4–6 and confirmed by recent candidate gene association studies 7–12 and genome wide association studies13. Since SCA due to CAD is a common phenotype, common genetic variants (e.g., single nucleotide polymorphisms [SNPs]) are plausible risk factors.
Abnormal calcium handling plays a crucial role in arrhythmias, SCA, and congestive heart failure (CHF).14 Calsequestrin 2 (CASQ2) is an important component of calcium handling as a calcium storage protein in the sarcoplasmic reticulum (SR), playing an important role in the regulation of the cardiac ryanodine receptor (RyR2)-mediated calcium release and SR calcium release refractoriness.14 A missense mutation in CASQ2 affects calcium release and is associated with catecholaminergic polymorphic ventricular tachycardia (CPVT) type 2.15–17 Recently, a candidate gene study reported association of SCD in the setting of CAD with SNPs in GPD1L, NOS1AP, and in the CASQ2 region.18 We tested the hypothesis that specific common polymorphisms in the CASQ2 gene may affect risk for SCA in patients with CAD.
Methods
The University of California at San Francisco (UCSF) Committee on Human Research approved the protocol. Informed consent was obtained from all participants for genetic study.
Study Design: UCSF Cases and Controls
Using a case-control design, we compared a cohort of patients with CAD who had SCA due to documented ventricular arrhythmias (VA) to a control group with CAD but no SCA or VA. We only included cases with both documented SCA and CAD – the most common phenotype of SCA. One hundred and fourteen consecutive out of hospital SCA cases with CAD presenting to UCSF Medical Center between January 2000 and June 2011 were included. We defined SCA as a cardiac arrest with documented sustained monomorphic ventricular tachycardia (VT) or ventricular fibrillation (VF) requiring resuscitation with either cardioversion or defibrillation, exclusive of torsades de pointes.8,11 Forty-four cases had VF and seventy cases had rapid VT (>200 bpm).
To specifically address the risk for SCA and VA rather than CAD as an intermediate phenotype, a CAD control group was identified from the UCSF Genomic Resource in Arteriosclerosis, a population-based study of atherosclerotic heart disease.19 Patients in the study had presented to the UCSF Cardiology Service and included individuals with prior MI and documented CAD by coronary angiography (≥70% stenosis in ≥1 major arteries) or revascularization procedures (coronary artery bypass graft surgery [CABG] or percutaneous coronary intervention [PCI]. From a cohort of over 16,000 patients, 1,054 potential CAD controls without history of VA or SCA were matched to the SCA cases with respect to age at index MI ± 5 years, sex, ethnicity, and degree of CAD as defined by revascularization procedure and duration of follow up since index MI. Because SCA events in cases occurred in the setting of CAD, in our matching procedure age was defined as age at first MI or first diagnosis of CAD in both cases and controls. Of available controls we then chose older controls to ensure a long enough follow-up time (mean 15 years) to be certain that they did not develop VT/VF or SCD. The age of CAD controls is reported as current age of controls, i.e., age at first MI or first diagnosis of CAD + mean 15 years follow-up time without VT/VF or SCD. From the 1,054 patients who met matching criteria to the SCA cases, we randomly selected 311 patients as CAD controls to achieve a 3-to-1 control to case ratio.
Blood collection and genotyping
Blood samples were obtained by venipuncture, and genomic DNA was extracted from peripheral blood lymphocytes (Invitrogen, Carlsbad, California) for the SCA cases. Genotyping was performed blinded to clinical status; positive and negative control samples were included. DNA samples were quantitated with a Nanodrop spectrophotometer (ND-1000) and normalized to a concentration of 50 ng/μl (diluted in 10 mM Tris/1 mM ethylene-diamine-tetra-acetic acid). Cases and controls were genotyped using the TaqMan Assay (Applied Biosystems) and read on the ABI PRISM 7900HT Sequence Detection System (PerkinElmer). When indeterminate readings or discrepancies appeared, plates were repeated or genotyping results were verified by sequencing.
SNP Selection
SNP selection was performed using the dbSNP b126; candidate SNPs were required to be common, defined as those with an estimated minor allele frequency (MAF) ≥ 0.05. Thirteen tagging SNPs spanning the 70 kb chromosomal region containing the CASQ2 gene were selected. One of two nonsynonymous polymorphisms in CASQ2 which were recently reported to be associated with catecholaminergic polymorphic ventricular tachycardia (rs4074536) was also included; the other nonsynonymous polymorphism in CASQ2 (rs10801999) was not included since the MAF was less than 0.05.14,20
To ensure robust genetic association analyses, we performed rigorous quality control (QC) filtering SNPs. None of the SNPs (n=14) deviated from Hardy-Weinberg expectations (p-value>0.05). Five percent of the samples were re-genotyped, with one SNP (rs10754345) excluded because it exhibited an insufficient concordance (i.e., 75%). Thirteen of 14 SNPs in CASQ2 passed all QC filters and were included in the genetic association analyses. Potential functional roles of the exonic non-synonymous missense SNP associated with SCA was examined using PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), a comprehensive search engine that tests a series of functional effects.21
Statistical analyses
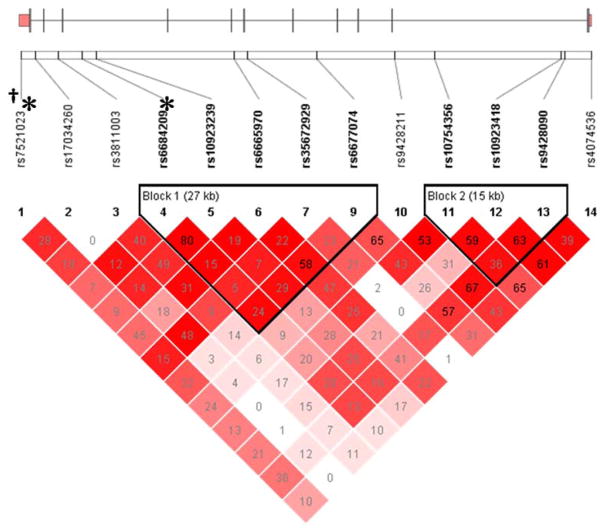
Data are displayed using frequencies for categorical and means ± standard deviations (SD) for continuous data. Analyses were carried out using STATA SE 8.0. Data significance was determined using Student’s t-test for unpaired variables, and using parametric ANOVA to determine differences in continuous variables between more than one group or Wilcoxon rank sum as appropriate based on normality of the distributions. Allele and genotype frequencies were determined by gene counting. Hardy-Weinberg equilibrium was assessed by the chi-square exact test. Measures of linkage disequilibrium, D′ and r2, were computed from the case and control genotypes with Haploview 4.1 (http://www.broad.mit.edu/mpg/haploview/) and heat maps of pairwise D′ values are presented in Figure 1.
Figure 1.
Calsequestrin 2 Gene Structure and Linkage Disequilibrium. An ideogram of Calsequestrin 2 (CASQ2) is presented above the white bar that represents the physical distance along human chromosome 1 (position 31, chr1:116242626-116311426; GRCh37/hg19 assembly, NM_001232). Exons are represented as tick marks. Gray lines connecting the exons represent introns. Reference sequence identifiers (rsID) for each single nucleotide polymorphism (SNP) are plotted both in terms of their physical distance (i.e., the white bar at the top of the figure) and also equidistantly to render the pairwise linkage disequilibrium (LD) estimates that were calculated and visualized with Haploview 4.2. The gene structure for CASQ2 was rendered with FancyGene 1.4. The correlation statistics (r2 and D′) are provided in the heatmap. LD-based haplotype block definition was based on the D′ confidence interval method. The haploblock is indicated in a bolded triangle and its component SNPs are rendered in bold font. Pairwise D′ values (range: 0–1, inclusive) were rendered in color, with darker red diamonds representing D′ values approaching 1.0. When the r2 values (range of 0–100, inclusive) are not equal to 0 or 100, they are provided in a given diamond. Asterisk and dagger for SNPs associated with CHF and SCA respectively.
For SNPs that were members of the same haploblock, haplotype analyses were conducted to localize the association signal within each gene and to determine if haplotypes improved the strength of the association with the phenotype. Haplotypes were constructed using the program PHASE version 2.1.22 To improve the stability of haplotype inference, the haplotype construction procedure was repeated five times using different seed numbers with each cycle.
A series of null SNPs were used to minimize confounding due to population stratification.23–25 The possibility of substructure was verified by principal component analysis,26 using Helix Tree (Golden Helix, Bozeman, MT). Briefly, the number of principal components (PCs) was sought which distinguished any population subgroups (e.g., cases and controls, subpopulations) in the sample by visual inspection of scatter plots of orthogonal PCs (i.e., PC 1 versus PC2, PC2 versus PC3). This procedure was repeated until no discernible clustering of patients was possible (data not shown). One hundred and eight ancestry informative markers (AIMs) were included in the analysis.
For association tests, three genetic models were assessed for each SNP (i.e., additive, dominant, recessive) and haplotype analysis was performed. The genetic model that best fit the data, maximizing the significance of the p-value, was selected for each SNP. Both unadjusted and adjusted associations were calculated. A multivariate logistic regression analysis was employed to test genetic associations and adjust for relevant covariates (age, gender, diabetes, hypertension, CHF status, CAD severity, and smoking status); a backwards stepwise approach was used. Except for PCs, only predictors with a p-value of <0.05 were retained in the final model. Given the high degree of shared variance observed among the SNPs examined, no correction for multiple testing was pursued. Genetic model fit and both unadjusted and covariate-adjusted odds ratios were estimated using STATA version 9.
Results
Baseline Characteristics
Compared with controls (Table 1), SCA cases were younger (63.1 ± 12.5 vs. 72.4 ± 8.6 years, p<0.001) and CHF was more common (74/114 [64.9%] vs. 37/311 [11.9%], p<0.001). For the minority of controls (121/311 [39%] compared to cases 106/114 [93%]) with echocardiographic data available, controls had a higher ejection fraction (EF) (56.0% ± 14.3% vs. 38.6% ± 5.2%; p<0.001). The matching procedure between SCA cases and CAD controls resulted in similar baseline characteristics such as gender, race, diabetes mellitus, HTN, and age at first MI.
Table 1.
Demographic Characteristics of SCA Cases and CAD Controls
| Baseline Characteristics | SCA Cases (n=114) | CAD Controls (n=311) | P-value | CHF (n=111) | No CHF (n=314) | P-value |
|---|---|---|---|---|---|---|
| Mean (SD) | Mean (SD) | Mean (SD) | Mean (SD) | |||
| Age* (years) | 63.1 ± 12.5 | 72.4 ± 8.6 | <0.001 | 68.2 ± 12.0 | 70.5 ± 10.0 | 0.028 |
| Age at 1st MI (years) | 55.9 ± 13.2 | 55.7 ± 10.7 | 0.574 | 55.1 ± 12.9 | 56.0 ± 10.8 | 0.259 |
| EF† (%) | 38.6 ± 15.2 | 56.0 ± 14.3 | <0.001 | 36.1 ± 14.1 | 58.3 ± 11.9 | <0.0001 |
| n (%) | n (%) | n (%) | n (%) | |||
| Male | 100 (87.7) | 251 (80.7) | 0.091 | 101 (91.0) | 250 (79.6) | 0.006 |
| Race (% Caucasians) | 110 (96.5) | 309 (99.4) | 0.061 | 109 (98.2) | 310 (98.7) | 0.231 |
| Prior MI | 106 (93.0) | 304 (98.4) | 0.033 | 106 (95.5) | 304 (97.4) | 0.309 |
| Diabetes mellitus | 24 (21.1) | 72 (23.3) | 0.624 | 28 (25.2) | 68 (21.8) | 0.459 |
| Hypertension | 79 (69.3) | 194 (62.8) | 0.214 | 86 (77.5) | 187 (59.9) | 0.001 |
| CHF | 74 (64.9) | 37 (11.9) | <0.001 | 111 (100) | 0 (0) | |
| SCA | 111 (100.0) | 0 (0.0) | 74 (66.7) | 40 (12.7) | <0.001 |
Abbreviations: SD = standard deviation
Age of cases at SCA and current age of CAD controls (i.e., age at first MI/CAD + mean 15 years follow-up time without VT/VF or SCD)
EF was available in 121 of the CAD controls (39%) and in 106 of the SCA cases (95.5%)
Genetic Association of CASQ2 SNPs with SCA
To account for non-genetic factors that might affect association with SCA, multivariable logistic regression analyses to identify variables that independently predicted SCA were fitted. Age (OR 0.89, 95% CI 0.86–0.92, p<0.001) and CHF (OR 21.0, 95% CI 11.1–39.7, p<0.001) were independently associated with SCA, therefore both were included in subsequent models that evaluated genotypic predictors. Multivariate logistic regression adjusting for age and CHF identified 4 SNPs (rs7521023, OR 2.7, 95% CI 1.44–5.13, p=0.002; rs17034260, OR 0.5, 95% CI 0.29–0.94, p=0.030; rs6677074, OR 0.5, 95% CI 0.23–0.98; rs9428090, OR 2.3, 95% CI 1.15–4.40, p=0.018) significantly associated with SCA (Table 2). In a model fitted with all 4 genetic predictors (i.e., rs7521023, rs17034260, rs6677074, rs9428090), age, and CHF, only rs7521023 remained significant (p<0.002).
Table 2.
Genetic Association of CASQ2 SNPs with SCA and with CHF status
| rsID | MAF | Allele* | Call Rate | SCA | CHF | ||
|---|---|---|---|---|---|---|---|
| ORadj (95% CI)S | P | ORadj (95% CI)C | P | ||||
| rs7521023 | 0.363 | A>G | 98.6 | 2.7 (1.44–5.13)D | 0.002 | 0.4 (0.25–0.76)D | 0.003 |
| rs17034260 | 0.382 | C>T | 99.8 | 0.5 (0.29–0.94)D | 0.030 | 1.3 (0.85–1.86)A | 0.245 |
| rs3811003 | 0.311 | G>A | 99.8 | 0.8 (0.52–1.27)A | 0.360 | 2.7 (1.16–6.34)R | 0.022 |
| rs66842091 | 0.181 | C>T | 100.0 | 0.8 (0.44–1.28)A | 0.299 | 19.8 (3.63–108.2)R,C | <0.001 |
| rs109232391 | 0.215 | A>T | 99.5 | 0.8 (0.49–1.37)A | 0.458 | 11.1 (2.39–50.9)R | 0.002 |
| rs66659701 | 0.419 | G>A | 98.8 | 1.4 (0.92–2.13)A | 0.112 | 0.53 (0.30–0.95)D | 0.031 |
| rs356729291 | 0.232 | T>G | 99.3 | 0.7 (0.41–1.15)A | 0.155 | 1.13 (0.73–4.41)A | 0.577 |
| rs66770741 | 0.465 | C>A | 100.0 | 0.5 (0.23–0.98)R | 0.045 | 1.90 (1.01–4.47)R | 0.045 |
| rs9428211 | 0.460 | C>T | 97.7 | 0.5 (0.23–1.01)R | 0.052 | 1.23 (0.84–1.85)A | 0.288 |
| rs10754356 | 0.313 | A>G | 99.1 | 0.7 (0.47–1.14)A | 0.163 | 1.23 (0.82–1.86)A | 0.315 |
| rs109234182 | 0.428 | G>A | 100.0 | 0.5 (0.22–1.06)R | 0.071 | 1.08 (0.74–1.59)A | 0.684 |
| rs94280902 | 0.459 | G>A | 99.5 | 2.3 (1.15–4.40)D | 0.018 | 0.88 (0.60–1.30)A | 0.515 |
| rs4074536 | 0.317 | T>C | 99.5 | 0.9 (0.59–1.35)A | 0.587 | 1.10 (0.74–1.62)A | 0.642 |
Abbreviations: 95% CI = 95% confidence interval; MAF = minor allele frequency; ORadj = covariate-adjusted odds ratio; P = p-value; rsID = reference sequence IDentifier.
The alleles for each SNP are listed as follows: common > rare.
SNPs in haploblock 1
SNPs in haploblock 2
An additive genetic model fit the data best.
A dominant genetic model fit the data best.
A recessive genetic model fit the data best.
SNP is associated with SCA after adjustment for age, CHF status.
SNP is associated with CHF status after adjustment for age, gender, hypertension SCA.
Haploblock analysis revealed two haploblocks in the CASQ2 gene region (Figure 1). However, multivariable regression analysis of common haplotypes in each haploblock did not identify any haplotypes associated with SCA.
Genetic Association of CASQ2 SNPs with CHF
It is well established that SCD risk is increased by 6-to-9-fold in the setting of CHF;27 CASQ2 mutation has also previously been reported to affect ventricular function in a mouse model.28 We observed an approximate 20-fold increased risk for SCA in patients with CHF (OR 20.5, 95% CI: 11.0–38.2, p<0.001). Accordingly, we tested for associations between CASQ2 gene variations and CHF, adjusting for SCA. In the initial multivariable logistic regression analysis, age (OR 1.04, 95%CI 1.01–1.06, p=0.012), female gender (OR 0.38, 95% CI 0.17–0.87, p=0.021), hypertension (OR 2.24, 95% CI 1.23–4.06, p=0.008), and SCA (OR 20.4, 95% CI 10.7–38.6, p< 0.01) were independently associated with CHF. Six SNPs were significantly associated with CHF in adjusting for age, gender, HTN, and SCA status (Table 2): rs7521023 (OR 0.4, 95% CI 0.25–0.76, p=0.003), rs3811003 (OR 2.7, 95% CI 1.16–6.34, p=0.022), rs6684209 (OR 19.8, 95% CI 3.63–108.2, p<0.001), rs10923239 (OR 11.1, 95% CI 2.39–50.9, p=0.002), rs6665970 (OR 0.53, 95% CI 0.30–0.95, p=0.031), and rs6677074 (OR 1.90, 95% CI 1.01–4.47, p=0.045).
Analysis of common haplotypes in the two haploblocks identified in CASQ2 (Figure 1), revealed an association between haplotype A6 in haploblock 1 with CHF status (OR 0.65, 95% CI 0.44–0.98, p=0.041). Haplotype A6 is composed of the common alleles at 4 (i.e., rs6684209, rs10923239, rs35672929, rs6677074) of the 5 SNPs (i.e., rs6684209, rs10923239, rs6665970, rs35672929, rs6677074) from which the haplotype was inferred (i.e., C-A-A-T-C). In a model fitted with the seven genetic predictors (i.e., rs7521023, rs3811003, rs66842091, rs109232391, rs66659701, rs66770741, haplotype A6), age, gender, hypertension, and SCA, only rs7521023 (OR 0.49, 95% CI 0.27–0.87, p=0.014) and rs6684209 (OR 14.9, 95% CI 2.67–83.0, p=0.002) were retained after adjusting for age, gender, hypertension, and SCA (p<0.0001).
The functional impact of the 4 SNPs associated with SCA (rs7521023, rs17034260, rs6677074, and rs9428090) and the 6 SNPs associated with CHF (rs7521023, rs3811003, rs6684209, rs10923239, rs6665970, and rs6677074) were assessed using PUPASUITE with no known effect predicted for the minor alleles.29
Discussion
In this candidate gene study, we demonstrate a novel association between a common CASQ2 variant (rs7521023) and risk of SCA due to VA in the setting of CAD, adjusting for age and CHF, both significant non-genetic predictors independently associated with SCA. We also identified 2 CASQ2 variants (rs7521023 and rs6684209) associated with CHF, adjusting for the significant non-genetic predictors of SCA, age, gender, and hypertension.
The CASQ2 gene residing in chromosome 1p13.3 is 68,803 base pairs and encodes for a protein with 399 amino acids. CASQ2 acts as a major Ca2+ storage protein in cardiomyocyte sarcoplasmic reticulum (SR) and participates actively in muscle contraction by localizing Ca2+ at the release site and regulating the amount of Ca2+ released through the ryanodine receptor, RyR2.14 In addition, CASQ2 modulates the luminal Ca2+-dependent closure of the RyR2; thus, CASQ2 determines the functional size and stability of SR Ca2+ stores.30
CASQ2 mutations with functional effect alter calcium handling and contribute to both heart failure and SCA phenotypes.14,28 Thus, it is not surprising that we found associations of common CASQ2 SNPs with these two common and interrelated phenotypes: SCA and CHF. With the exception of one exonic CASQ2 SNP, all of the tagging CASQ2 SNPs in this study were intronic. One variant located in the exonic splicing enhancer region (rs7521023) was associated with both SCA and CHF. Since CASQ2 affects calcium flux, this protein may have a critical interconnected role for contractile function and thus heart failure status as well as ventricular arrhythmogenicity and SCA. CASQ2 rs7521023 may be associated with an increased risk for SCA yet protective for CHF if this SNP is related to decreased CASQ2 function, which would lead to increased cytoplasmic calcium levels. This would in turn improve myocardial contractile function but also lead to increased triggered activity and ventricular arrhythmias and thus increased risk of SCA. Further functional studies are needed to confirm this hypothesized mechanism.
The CASQ2 SNP (rs7521023) associated with SCA adjusted for CHF is a common polymorphism with MAF> 0.35. A report from the Oregon Sudden Unexpected Death Study (Ore-SUDS) examined 1424 SNPs in 18 candidate genes reported association of 3 common variants within or near CASQ2 with risk of out-of-hospital, paramedic collected sudden deaths in patients with CAD. One of the three reported SNPs (intronic rs3010396) was within the coding region of the CASQ2 gene, while the other two were either distant from CASQ2 (over 100,000 bp upstream) or in an intron of an adjoining gene (VANGL1).18 The VANGL1 rs3010396 SNP was not included in the present study and is not in LD with the CASQ2 SNP (rs7521023) significantly associated with SCA herein. CHF was not included in multivariate analysis in the Ore-SUDS study. Notably, the SCA phenotypic criteria implemented in the present study differs from that used in the Ore-SUDS study (i.e., community based SCD victims without requirement for documented arrhythmia in Ore-SUDS and SCA survivors in present study due to documented VT/VF in the setting of CAD), which might partially explain these differing results.
Reduced EF is a well-established risk factor for SCA. 31 Individuals with significant LV systolic dysfunction are at particularly high risk of ventricular arrhythmias and SCA. 31 EF was not included in the multivariate models because EF assessment was only available in a minority of CAD controls as compared to the majority of SCA cases. Since CHF status was available in all participants, CHF was used as a surrogate for low EF.
While prior studies have reported genetic associations with CHF (i.e., the cardiac isoform of myosin-binding protein C MYBPC3, endothelial nitric oxide synthase, Signal transducer and activator of transcription 3 STAT3, interleukin 1 beta, presenilin-1, BCL2-associated athanogene 3 BAG3, peroxisome proliferator activated receptor alpha, FERM domain containing 4B FRMD4B), none have evaluated genetic variation in critical components of the calcium handling pathway. 32 We report the association of 2 CASQ2 SNPs (i.e., rs7521023, rs6684209) with CHF following adjustment for relevant covariates (i.e., age, gender, hypertension, SCA). Whereas a mouse model of a CASQ2 mutation affected ventricular function, 28 the findings presented herein are the first report of an association between common genetic variation in CASQ2 and CHF in humans. If validated in independent cohorts, these CASQ2 SNPs may have clinical utility in testing and risk stratification of CHF patients at risk for SCA.
These CASQ2 SNPs associated with SCA and CHF are intronic, thus they may be in LD with the functional variant that impacts gene splicing, transcription, or gene regulation. Since limited data are available on their functional effects, further study is warranted to identify the causal underlying gene association signals with sequencing, identify causative functional variants, and elucidate their impact on CASQ2 function or levels.
Mechanisms of the sudden arrhythmic death vary with sustained VT usually occurring in the setting of a scar while polymorphic VT/VF occurs typically in the setting of ischemia. To improve power, we analyzed SCA as a group regardless if due to VT or polymorphic VT/VF. Future study in larger cohorts may untangle whether these SNP associations may differ based on type of ventricular tachyarrhythmia.
The strengths of the study include the rigorously phenotyped and adjudicated SCA cases, and the inclusion of matched CAD controls without SCA or VT/VF. A recent report from the National Heart, Lung and Blood Institute and Heart Rhythm Society workshop highlighted the critical need to study well-phenotyped SCD and control populations for future prediction and prevention of SCD.33 The inclusion of a CAD control group was used to avoid confounding association with CAD as an intermediate phenotype. Because of the requirement for documented VT/VF in all SCA cases, the possibility of association with other non-arrhythmic causes of sudden death (e.g. tamponade, aortic aneurysm, or pulmonary embolus) is minimized. It is worth noting that the demographic and morbidity characteristics of the cases and controls in our study are similar to recent genetic association studies of SCA and heart failure trials 13,34.
Limitations to the present study warrant consideration. The identified genetic associations require replication in an independent cohort. Due to the sample size requirements for the interrogation of rare polymorphisms, only common SNPs (i.e., MAF>0.05) were examined; thus rarer polymorphisms such as rs10801999 with functional effects in vitro (i.e., reducing calcium-binding and altering CASQ2 polymerization) were not assessed in this study.14 Because the majority of our study population were Caucasian, further studies are needed to determine whether the reported associations can be generalized in other ethnicities. Although correction for multiple testing was not pursued due to the high degree of correlation among the SNPs, it is noteworthy that the SNP associations retained in the final regression models still exceeded a corrected significance threshold (i.e., p=0.05/13 SNPs = 0.00385). Finally, because our cases were defined by SCA status, validation of the association of CASQ2 polymorphisms with CHF in an independent CHF population is warranted.
Conclusions
We demonstrate a novel association between a common CASQ2 variant and SCA due to malignant VA in the setting of CAD, adjusted for age and CHF. We also report association between common CASQ2 polymorphisms and CHF, adjusted for SCA, thus these CASQ2 polymorphisms may impact the interrelated phenotypes of SCA and CHF. Since cases and controls were selected by SCA status, further investigation of the influence of CASQ2 variants with CHF requires a larger CHF cohort to confirm the findings of this study.
Acknowledgments
Acknowledgement of Financial Support
This work was supported in part by the 2011–2012 Heart Rhythm Society Kenneth M. Rosen Fellowship Award in Cardiac Pacing and Electrophysiology to MMR and grant 5R01 HL102090 from the National Heart, Lung, and Blood Institute to ZHT.
We would like to thank Benjamin Colburn for technical assistance.
Abbreviations
- SNP
single nucleotide polymorphism
- LD
linkage disequilibrium
- CASQ2
Calsequestrin 2
- VA
ventricular arrhythmias
- VT
ventricular tachycardia
- VF
ventricular fibrillation
- SCA
sudden cardiac arrest
- SCD
sudden cardiac death
- ICD
implantable cardioverter-defibrillator
- CAD
coronary artery disease
- MI
myocardial infarction
- CHF
congestive heart failure
- EF
Ejection Fraction
- PTCA
percutaneous transluminal coronary angioplasty
- CABG
coronary artery bypass graft
- OR
odds ratio
- CI
confidence interval
Footnotes
Authors’ conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zheng ZJ, Croft JB, Giles WH, Mensah GA. Sudden cardiac death in the United States, 1989 to 1998. Circulation. 2001;104:2158–2163. doi: 10.1161/hc4301.098254. [DOI] [PubMed] [Google Scholar]
- 2.Zipes DP, Wellens HJ. Sudden cardiac death. Circulation. 1998;98:2334–2351. doi: 10.1161/01.cir.98.21.2334. [DOI] [PubMed] [Google Scholar]
- 3.Tseng ZH. Genetic association studies of sudden cardiac death/arrest: the importance of context. Heart Rhythm. 2009;6(9):1315–7. doi: 10.1016/j.hrthm.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Friedlander Y, Siscovick DS, Weinmann S, et al. Family history as a risk factor for primary cardiac arrest. Circulation. 1998;97(2):155–60. doi: 10.1161/01.cir.97.2.155. [DOI] [PubMed] [Google Scholar]
- 5.Jouven X, Desnos M, Guerot C, Ducimetiere P. Predicting sudden death in the population: the Paris Prospective Study I. Circulation. 1999;99:1978–1983. doi: 10.1161/01.cir.99.15.1978. [DOI] [PubMed] [Google Scholar]
- 6.Dekker LR, Bezzina CR, Henriques JP, et al. Familial sudden death is an important risk factor for primary ventricular fibrillation: a case-control study in acute myocardial infarction patients. Circulation. 2006;114 (11):1140–5. doi: 10.1161/CIRCULATIONAHA.105.606145. [DOI] [PubMed] [Google Scholar]
- 7.Tseng ZH, Vittinghoff E, Musone SL, et al. Association of TGFBR2 polymorphism with risk of sudden cardiac arrest in patients with coronary artery disease. Heart Rhythm. 2009;6(12):1745–50. doi: 10.1016/j.hrthm.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sotoodehnia N, Li G, Johnson CO, et al. Genetic variation in angiotensin-converting enzyme-related pathways associated with sudden cardiac arrest risk. Heart Rhythm. 2009;6(9):1306–14. doi: 10.1016/j.hrthm.2009.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sotoodehnia N, Siscovick DS, Vatta M, et al. Beta2-adrenergic receptor genetic variants and risk of sudden cardiac death. Circulation. 2006;113(15):1842–8. doi: 10.1161/CIRCULATIONAHA.105.582833. [DOI] [PubMed] [Google Scholar]
- 10.Tseng ZH, Aouizerat BE, Pawlikowska L, et al. Common beta-adrenergic receptor polymorphisms are not associated with risk of sudden cardiac death in patients with coronary artery disease. Heart Rhythm. 2008;5(6):814–21. doi: 10.1016/j.hrthm.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Refaat M, Frangiskakis JM, Grimley S, et al. The β2-Adrenergic Receptor Gln27 Polymorphism is Associated With Increased Ventricular Arrhythmias in Patients With Severe Heart Failure. Heart Rhythm. 2009;6(5S):S456. [Google Scholar]
- 12.Gavin MC, Newton-Cheh C, Gaziano JM, Cook NR, VanDenburgh M, Albert CM. A Common Variant in the Beta-2 Adrenergic Receptor and Risk of Sudden Cardiac Death. Heart Rhythm. 2011;8(5):704–710. doi: 10.1016/j.hrthm.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aouizerat BE, Vittinghoff E, Musone SL, et al. GWAS for discovery and replication of genetic loci associated with sudden cardiac arrest in patients with coronary artery disease. BMC Cardiovasc Disord. 2011:11–29. doi: 10.1186/1471-2261-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faggioni M, Knollmann BC. Calsequestrin 2 and arrhythmias. Am J Physiol Heart Circ Physiol. 2012;302(6):H1250–60. doi: 10.1152/ajpheart.00779.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.di Barletta MR, Viatchenko-Karpinski S, Nori A, et al. Clinical phenotype and functional characterization of CASQ2 mutations associated with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2006;114(10):1012–9. doi: 10.1161/CIRCULATIONAHA.106.623793. [DOI] [PubMed] [Google Scholar]
- 16.Knollmann BC, Chopra N, Hlaing T, et al. CASQ2 deletion causes sarcoplasmic reticulum volume increase, premature Ca2+ release, and catecholaminergic polymorphic ventricular tachycardia. J Clin Invest. 2006;116(9):2510–20. doi: 10.1172/JCI29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Spooner PM. Sudden cardiac death: The larger problem.. The larger genome. J Cardiovasc Electrophysiol. 2009;20(5):585–96. doi: 10.1111/j.1540-8167.2008.01419.x. [DOI] [PubMed] [Google Scholar]
- 18.Westaway SK, Reinier K, Huertas-Vazquez A, et al. Common variants in CASQ2, GPD1L, and NOS1AP are significantly associated with risk of sudden death in patients with coronary artery disease. Circ Cardiovasc Genet. 2011;4(4):397–402. doi: 10.1161/CIRCGENETICS.111.959916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pullinger CR, Hennessy LK, Chatterton JE, et al. Familial ligand defective apolipoprotein B. Identification of a new mutation that decreases LDL receptor binding affinity. J Clin Invest. 1995;95(3):1225–1234. doi: 10.1172/JCI117772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wong CH, Koo SH, She GQ, Chui P, Lee EJ. Genetic variability of RyR2 and CASQ2 genes in an Asian population. Forensic Sci Int. 2009;192(1–3):53–5. doi: 10.1016/j.forsciint.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 21.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoggart CJ, Parra EJ, Shriver MD, et al. Control of confounding of genetic associations in stratified populations. Am J Hum Genet. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Halder I, Shriver M, Thomas M, Fernandez JR, Frudakis T. A panel of ancestry informative markers for estimating individual biogeographical ancestry and admixture from four continents: utility and applications. Human Mutation. 2008;29:648–58. doi: 10.1002/humu.20695. [DOI] [PubMed] [Google Scholar]
- 25.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum Mol Genet. 2008;17:R143–50. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 27.Tomaselli GF, Zipes DP. What causes sudden death in heart failure? Circ Res. 2004;95(8):754–63. doi: 10.1161/01.RES.0000145047.14691.db. [DOI] [PubMed] [Google Scholar]
- 28.Sato Y, Ferguson DG, Sako H, et al. Cardiac-specific overexpression of mouse cardiac calsequestrin is associated with depressed cardiovascular function and hypertrophy in transgenic mice. J Biol Chem. 1998;273(43):28470–7. doi: 10.1074/jbc.273.43.28470. [DOI] [PubMed] [Google Scholar]
- 29.Conde L, Vaquerizas JM, Dopazo H, et al. PupaSuite: finding functional single nucleotide polymorphisms for large-scale genotyping purposes. Nucleic Acids Res. 2006;34:W621–5. doi: 10.1093/nar/gkl071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terentyev D, Nori A, Santoro M, et al. Abnormal interactions of calsequestrin with the ryanodine receptor calcium release channel complex linked to exercise-induced sudden cardiac death. Circ Res. 2006;98:1151–1158. doi: 10.1161/01.RES.0000220647.93982.08. [DOI] [PubMed] [Google Scholar]
- 31.Watanabe J, Shinozaki T, Shiba N, et al. Accumulation of risk markers predicts the incidence of sudden death in patients with chronic heart failure. Eur J Heart Fail. 2006;8(3):237–42. doi: 10.1016/j.ejheart.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 32.Villard E, Perret C, Gary F, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32(9):1065–76. doi: 10.1093/eurheartj/ehr105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fishman GI, Chugh SS, DiMarco JP, et al. Sudden Cardiac Death Prediction and Prevention: Report From a National Heart, Lung, and Blood Institute and Heart Rhythm Society Workshop. Circulation. 2010;122:2335–2348. doi: 10.1161/CIRCULATIONAHA.110.976092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrison AC, Felix JF, Cupples LA, et al. Genomic variation associated with mortality among adults of European and African ancestry with heart failure: the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2010;3(3):248–55. doi: 10.1161/CIRCGENETICS.109.895995. [DOI] [PMC free article] [PubMed] [Google Scholar]