Abstract
Evidence suggests that the catabolic process of macroautophagy (autophagy hereafter) can either suppress or promote cancer. The essential autophagy gene ATG6/BECN1 encoding the Beclinl protein has been implicated as a haploinsufficient tumor suppressor in breast, ovarian and prostate cancers. The proximity of BECN1 to the known breast and ovarian tumor suppressor breast cancer 1, early onset, BRCA1, on chromosome 17q21, has made this determination equivocal. Here the mutational status of BECN1 was assessed in human tumor sequencing data from The Cancer Genome Atlas (TCGA) and other databases. Large deletions encompassing both BRCA1 and BECN1, and deletions of only BRCA1 but not BECN1, were found in breast and ovarian cancers, consistent with BRCA1 loss being a primary driver mutation in these cancers. Furthermore, there was no evidence for BECN1 mutation or loss in any other cancer, casting doubt on whether BECN1 is a tumor suppressor in most human cancers.
Keywords: autophagy, cancer, BECN1, BRCA1, TCGA, CNV, mutation
Introduction
Autophagy captures and degrades intracellular proteins and organelles in lysosomes to preserve protein and organelle quality and to recycle building blocks to sustain metabolism and survival in starvation (1, 2). Autophagy promotes the health, function, and survival of cells and tissues, and generally, the loss of autophagy is destructive. In mammals, autophagy deficiency is linked to tissue degeneration, chronic inflammation, susceptibility to metabolic stress, and premature lethality.
There is evidence that autophagy both promotes and suppresses cancer, however, this has not been rigorously addressed in humans (3). Monoallelic disruption of BECN1 on chromosome 17q21 has been reported in 40 to 75% of human breast, ovarian, and prostate tumors, suggesting that autophagy is a tumor suppression mechanism (4-6). BECN1 allelic loss was also found in 9 out of 22 breast cancer cell lines by fluorescence in situ hybridization (FISH) analysis, although no coding or splice site mutations were found (4). The small sample sizes and poorly matched comparisons of cell lines and normal tissues and the modest frequencies of loss of heterozygosity used for these investigations is, however, insufficient to support the claim that BECN1 is a haploinsufficient tumor suppressor.
BECN1 is located on chromosome 17q21 next to BRCA1, a known tumor suppressor gene and whose loss is a driver of breast and ovarian cancer. BRCA1 is a critical regulator of DNA repair by homologous recombination (HR) and its loss causes DNA repair defects and cancer predisposition (7). The close proximity of BECN1 and BRCA1 complicates determination if allelic loss of BECN1 is a driver or passenger mutation in breast and ovarian cancers. Further, the mutational status of BECN1 in other cancers has not been rigorously assessed.
In support of the concept that autophagy is a tumor suppression mechanism and that allelic loss of BECN1 promotes cancer, Beclin1+/− mice are prone to mammary hyperplasia, liver and lung carcinomas and lymphomas (8, 9). However, mosaic whole body knock out of the essential autophagy gene Atg5, or liver-specific knock out of the essential autophagy gene Atg7, produces only benign liver hepatomas and no other neoplasms (10). Thus autophagy defects promote development of benign liver tumors in mice but may also block their progression. Autophagy-independent functions of Becnl may also contribute to the suppression of non-liver neoplasms or these events may be limited to genetically manipulated mice and not relevant to human cancers. The vast majority of germline mutations in BRCA1 are loss-of-function mutations (frameshift, indels, nonsense mutations, or missense), or focal deletions, not gross deletions in the BRCA1 locus at 17q21 that extend to encompass BECN1. Thus Beclin1+/− mice do not reflect a human condition.
In contrast, autophagy promotes the survival of tumor cells in hypoxic tumor regions (11) as well as the growth, survival and malignancy of RAS- and BRAF-driven cancers (3, 12-15). Autophagy promotes tumorigenesis by suppressing p53 activation and maintaining mitochondrial function essential for cellular metabolism and survival (16). Without autophagy tumors accumulate defective mitochondria, have growth and metabolic defects, and progresses to a more benign fate. This is consistent with a large body of literature indicating that autophagy is required for survival in starvation and stress, functions that are conserved from yeast to mammals that are also important for growth of cancer (2, 3, 17).
Germline mutations in BRCA1, BRCA2, and PALB2, predispose to hereditary breast cancer and the three proteins function together to maintain genome stability by promoting faithful repair of double strand breaks by HR (18). Mammary epithelial cell-specific knockout of Palb2 causes mammary tumorigenesis with long latency that is suppressed by allelic loss of Becnl, suggesting that autophagy is tumor-promoting (19). Deletion of Trp53 abrogates tumorigenesis impairment upon allelic loss of Becnl in Pa/fr2-deficient mammary tumors, thus the combination of autophagy defect and loss of a critical DNA repair mechanism augments the p53 anti-tumor response (19). Since loss of both Palb2 and autophagy promote DNA damage and p53 activation, (18, 20, 21), this explains enhanced p53 activity and why autophagy suppresses the p53 response and mammary tumorigenesis.
The important unanswered question here is whether mutations in essential autophagy genes are found in human cancers using current genomic information, and if they are found, are they loss- or gain-of-function mutations? Note that recent assessment of oncogenes and tumor suppressor genes assembled from the current human tumor sequencing data does not include any autophagy genes (22), but this was not examined specifically. To begin to resolve the potential conflicting role of autophagy in human cancer, we examined the publicly available human tumor sequencing and gene expression databases (TCGA) to determine the mutational and expression status of BECN1 in a broad array of human cancers. We first assessed BECN1 for single nucleotide variations (SNVs) and copy number variations (CNVs) in human breast, ovarian and prostate cancer genome sequences. Since BECN1 is adjacent to BRCA1, we specifically looked for deletions of BECN1 that do not encompass BRCA1. We found enrichment for truncating mutations of BRCA1, deletion of the chromosomal region that included BRCA1 only, and deletions affecting both BRCA1 and BECN1, but not truncating mutations of BECN1 or deletions of only BECN1. Analysis of all other cancers that lack BRCA1 deletion indicated no significant recurrence of SNVs or CNVs in BECN1. Thus BECN1 is not mutated or specifically deleted in human cancer, indicating that it is not a tumor suppressor gene.
Materials and Methods
Copy number Variations (CNVs)
To study the copy number status of BECN1 and BRCA1 in different cancers we downloaded over 10,000 processed copy number data from The Cancer Genome Atlas portal (TCGA, https://tcga-data.nci.nih.gov/tcga/). The TCGA Consortium collected tumor and matched normal samples from 24 different cancers and performed SNP and CGH microarray on genomic DNA to find CNV (Table S1). The cancers for which we obtained CNV data include acute myeloid leukemia (LAML), bladder urothelial carcinoma (BLCA), brain lower grade glioma (LGG), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), colon adenocarcinoma (COAD), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), kidney renal clear cell carcinoma (KIRC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC), ovarian serous cyst adenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), sarcoma (SARC), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), thyroid carcinoma (THCA), and uterine corpus endometrioid carcinoma (UCEC). Tumor samples represent primary as well as metastatic tumors.
The TCGA consortium performed CNV calling and provided Level 3 data for CNVs including segment mean values and number of markers for all CNV detected. Briefly, the segment mean is the average of log2 ratio of probes in the segment (log2 (observed intensity/reference intensity)) and represents the extent of copy number changes for that particular genomic segment while the number of markers is the number of probes present in that segmental region. To extract a set of high confidence CNVs, we use threshold of 0.2 in segment mean value for amplifications and −0.2 for deletions. We derived these thresholds by examining the distribution of segment mean values from tumor and normal samples. In addition, we require the number of markers spanning a CNV to be at least ten to decrease false positives in calling CNV. We test the sensitivity of our method by finding the previously reported CNV (amplifications in PIK3CA, EGFR, FOXA1 and HER2; de/etions in MLL3, PTEN, RB1 and MAP2K4) in breast invasive cancer (23). We use the CNV that pass these criteria for further analysis and identify all CNVs that overlap BECN1 or BRCA1.
We used the matched tumor and normal samples to determine the somatic CNVs. We identify CNVs as germline in the tumor if there was an overlapping CNV in the matched normal. The ratio of deletions to amplifications of somatic CNVs found across the genome provide a background ratio for comparison with ratio found at a particular locus. If the ratio of deletion to amplifications is different at a locus than the genome average, then there may be selection for deletions or amplifications at that locus. We use the two-tailed Fisher exact test for determining statistical significance using the average number of deletions and duplications per sample for the background and the number of samples with deletion and amplifications for the locus.
Somatic Mutations
The TCGA provides somatic mutations detected from whole genome and whole exome sequencing from matched tumor and normal samples as level 2 data (Table S2). We extracted the somatic mutations for BECN1 and BRCA1 and indicated their type as missense, nonsense, silent, splice site, and insertion or deletion resulting in frame shift or in frame (Table S3 and Table S4).
Gene Expression
TCGA RNA-seq level 3 data for all cancers, tumor and normal, was processed and normalized and we used the RSEM normalized values for gene expression. Fold change in BECN1 gene expression between tumor and normal tissue were calculated using median expression of tumors and normal. Significance of differential gene expression change in BECN1 is calculated using 2-tailed Wilcoxon test and Bonferroni corrected for multiple hypothesis testing.
Results
Copy Number Variations (CNVs) in BECN1
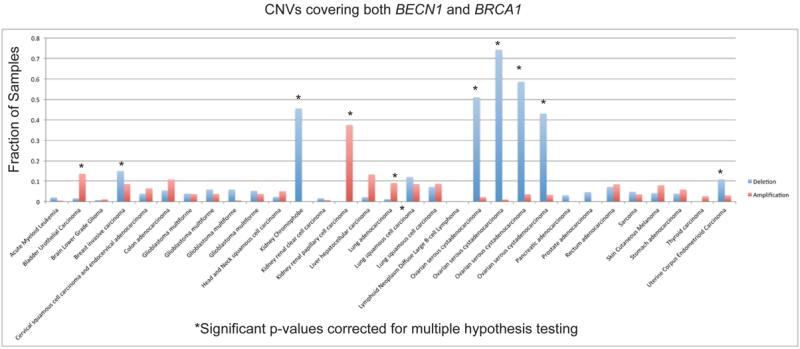
CNVs in BECN1 were assessed in the databases indicated in Table S1 from approximately 10,000 normal/tumor pairs. CNVs were classified into three groups defined by whether the CNV overlapped with BECN1 but not BRCA1, overlapped with BRCA1 but not BECN1, or overlapped with both BECN1 and BRCA1 (Table 1). Most of the CNVs detected are large and overlap both BECN1 and BRCA1. Each CNV was further identified as a deletion, amplification or interrupting amplification if only a part of a gene was amplified (not included in the count for amplifications). As expected, breast and ovarian tumors were significantly enriched for having deletions in the locus containing both BECN1 and BRCA1 (Table 1).
Table 1.
Enrichment for deletions and amplifications for BRCA1 and not BECN1 in human cancers. Only statistically significant enrichments have their Bonferroni corrected p-values shown.
| Cancer | Number of tumor/normal pairs |
Average CNA per tumor | BECN1 alone | BRCA1 alone | Both BECN1 and BRCA1 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Deletion | Amplification | Deletion | Amplification | p-value | Deletion | Amplification | p-value | Deletion | Amplification | p-value | ||
| Acute Myeloid Leukemia | 196 | 146 | 146 | 1 | 1 | 0 | 0 | 4 | 1 | |||
| Bladder Urothelial Carcinoma | 184 | 99 | 106 | 0 | 0 | 0 | 0 | 3 | 25 | 0.01442 | ||
| Brain Lower Grade Glioma | 269 | 34 | 32 | 1 | 0 | 0 | 0 | 2 | 3 | |||
| Breast invasive carcinoma | 999 | 85 | 111 | 3 | 7 | 24 | 4 | 2.42E-03 | 149 | 86 | 4.00E-03 | |
| Cervical squamous cell carcinoma and endocervical adenocarcinoma |
155 | 52 | 61 | 1 | 0 | 2 | 0 | 6 | 10 | |||
| Colon adenocarcinoma | 460 | 70 | 71 | 3 | 2 | 7 | 1 | 25 | 50 | |||
| Glioblastoma multiforme 1 | 434 | 29 | 25 | 1 | 0 | 1 | 1 | 17 | 16 | |||
| Glioblastoma multiforme 2 | 237 | 30 | 27 | 2 | 1 | 1 | 2 | 14 | 9 | |||
| Glioblastoma multiforme 3 | 170 | 35 | 29 | 0 | 0 | 1 | 0 | 10 | 1 | |||
| Glioblastoma multiforme 4 | 534 | 106 | 109 | 0 | 1 | 3 | 1 | 28 | 20 | |||
| Head and Neck squamous cell carcinoma |
390 | 49 | 55 | 0 | 0 | 1 | 1 | 9 | 20 | |||
| Kidney Chromophobe | 66 | 97 | 75 | 1 | 0 | 1 | 0 | 30 | 0 | 0.00021 | ||
| Kidney renal clear cell carcinoma | 550 | 35 | 44 | 0 | 1 | 2 | 0 | 9 | 4 | |||
| Kidney renal papillary cell carcinoma | 168 | 30 | 49 | 0 | 0 | 0 | 0 | 0 | 63 | 8.36E-07 | ||
| Liver hepatocellular carcinoma | 136 | 52 | 88 | 0 | 2 | 1 | 2 | 3 | 18 | |||
| Lung adenocarcinoma | 505 | 50 | 66 | 0 | 2 | 0 | 2 | 6 | 46 | 0.00698 | ||
| Lung squamous cell carcinoma 1 | 117 | 51 | 50 | 1 | 0 | 0 | 0 | 14 | 10 | |||
| Lung squamous cell carcinoma 2 | 505 | 100 | 109 | 2 | 3 | 7 | 0 | 36 | 44 | |||
| Lymphoid Neoplasm Diffuse Large B-cell Lymphoma |
18 | 49 | 48 | 0 | 0 | 0 | 0 | 0 | 0 | |||
| Ovarian serous cystadenocarcinoma 1 | 587 | 105 | 109 | 7 | 2 | 10 | 3 | 299 | 13 | 2.03E-35 | ||
| Ovarian serous cystadenocarcinoma 2 | 97 | 76 | 92 | 2 | 0 | 3 | 0 | 72 | 1 | 1.17E-13 | ||
| Ovarian serous cystadenocarcinoma 3 | 355 | 93 | 121 | 2 | 1 | 3 | 2 | 208 | 13 | 4.05E-31 | ||
| Ovarian serous cystadenocarcinoma 4 | 586 | 253 | 363 | 16 | 4 | 47 | 3 | 2.42E-12 | 252 | 19 | 8.09E-47 | |
| Pancreatic adenocarcinoma | 66 | 29 | 30 | 0 | 0 | 0 | 0 | 2 | 0 | |||
| Prostate adenocarcinoma | 195 | 46 | 29 | 1 | 2 | 1 | 0 | 9 | 0 | |||
| Rectum adenocarcinoma | 167 | 81 | 83 | 0 | 1 | 0 | 0 | 12 | 14 | |||
| Sarcoma | 84 | 71 | 122 | 0 | 1 | 0 | 1 | 4 | 3 | |||
| Skin Cutaneous Melanoma | 338 | 54 | 78 | 0 | 0 | 2 | 2 | 14 | 27 | |||
| Stomach adenocarcinoma | 306 | 44 | 59 | 1 | 1 | 6 | 2 | 12 | 18 | |||
| Thyroid carcinoma | 505 | 15 | 18 | 0 | 0 | 1 | 0 | 0 | 13 | |||
| Uterine Corpus Endometrioid Carcinoma | 505 | 62 | 91 | 3 | 3 | 12 | 2 | 55 | 15 | 1.15E-05 | ||
Other tumor types that exhibited significant enrichment for deletions in both BECN1 and BRCA1 include kidney chromophobe and uterine corpus endometrioid carcinoma (Table 1). Tumor types found having enrichment for amplifications include bladder urothelial carcinoma, kidney renal papillary cell carcinoma and lung adenocarcinoma. Closer examination found that the CNVs in kidney chromophobe and kidney renal papillary cell carcinoma are whole chromosome deletions and amplifications, respectively, which are consistent with known loss and gain of chromosome 17 for these two types of tumors (24).
CNVs that overlap BRCA1 but not BECN1 were enriched for deletions in breast and ovarian tumors, while CNVs that overlap BECN1 but not BRCA1 were not enriched for deletions in any tumor (Table 1). These results are consistent with the loss of BRCA1 being the driver mutation in breast and ovarian tumors. No significant CNVs in BECN1 were detected in any other cancers (Table 1). Loss of chromosome 17q21 and BRCA1 has been reported in prostate cancer only very infrequently (0.45%) (25). For prostate adenocarcinoma, we found 9 deletions (covering both BECN1 and BRCA1) and no amplifications (Table 1). The p-value for enrichment of deletions is 0.024, however, after correcting for multiple hypothesis testing, it is not significant. Prostate adenocarcinoma is a heterogeneous disease and the fraction of this disease where loss of 17q21 is a driver mutation is small compared to breast or ovarian cancer. It is clear, however, that in contrast to previous reports, BECN1 deletions do not significantly occur in the absence of BRCA1 deletion.
Somatic mutations
There are 169 and 32 (ratio of 5.28) mutations found in BRCA1 and BECN1 respectively across all tumor samples (6632) and the numbers are 137 and 31 (ratio of 4.42) if we exclude breast and ovarian tumors where BRCA1 is known to be a tumor suppressor (Table S2). The difference in mutation number is mostly explained by the size of the coding region of the two genes (ratio of protein coding length of BRCA1 to BECN1 is 4.14).
None of the mutations found in BECN1 were nonsense or splice site mutations (Table S3) with the potential to alter function and that are frequently found tumor suppressor mutations. If we restrict analysis to breast and ovarian cancer, there is only one mutation found in BECN1 and it is a missense mutation in an ovarian tumor. In contrast, there are 32 mutations in BRCA1 of which 23 are nonsense, splice site or frame shift mutations all of which lead to truncation of BRCA1 (Table S4).
Across all cancer data from TCGA, there are 30 missense, 0 nonsense, 0 splice site and 11 silent mutations for BECN1 and 135 missense, 20 nonsense, 12 splice site and 39 silent mutations for BRCA1. To find statistical enrichment of missense, nonsense or splice site mutations compared to silent mutations, we use as null model the aggregate of mutations across all samples in breast cancer (778 tumors) yielding 31861 missense, 2339 nonsense, 1075 splice site and 11677 silent mutations. Since the vast majority of mutations detected in tumors are passenger mutations with little or no selective advantage to the tumors, the ratio of missense to silent mutations (2.73), nonsense to silent mutations (0.20), and splice site to silent mutations (0.09) are good approximations for little or no selection of missense, nonsense or splice site over silent mutations. Indeed these ratios are very similar when looking at other cancer types from TCGA. There is statistically significant enrichment for ratio of nonsense to silent and splice site to silent mutations for BRCA1 (2.6 and 3.4 fold enrichment with p-value of 0.0008 and 0.0003 using two-tailed Chi-square test with Yate’s correction). There is no significant enrichment for missense over silent mutations for BRCA1 and BECN1, and no enrichment of nonsense and splice site over silent mutations in BECN1. The proportion of missense, nonsense and splice site mutations for BECN1 is statistically consistent with the occurrence of passenger mutations.
Gene expression changes
The differential expression of BECN1 between tumor and normal tissue for 17 cancer types from TCGA show no significant fold change greater than 2 (Table S5). The greatest decrease in expression of BECN1 occurs in kidney chromophobe where the fold change of tumor to normal is 0.65 which is consistent with loss of chromosome 17 being common in this cancer.
Discussion
Using the genomic information collected on a broad array of human cancers, we assessed the mutational status of the essential autophagy gene BECN1. Despite reports indicating allelic loss of BECN1 in some human cancers, this appears to be explained solely by the proximity of BECN1 to BRCA1. We find no evidence of mutation or focal loss of BECN1 from the analysis of currently available cancer genomic information. Monoallelic loss of the chromosome 17q21 region that encompasses both BECN1 and BRCA1 is found in both breast and ovarian cancer. However as the region is large, this finding does not support a role for BECN1 as the driver. Furthermore, there is no finding of nonsense or splice site mutations in BECN1 in any other cancers.
Germline missense mutations in BRCA1 followed by somatic deletion of the remaining allele in tumors are responsible for inherited cancers. In these cancers, the majority of the deletions are large and take out both genes and a hundred others. While focal deletions and somatic, predicted loss of function mutations (missense, nonsense, frame shift and splice site mutations) are found in BRCA1, they are not found in BECN1. Furthermore, there are no significant germline mutation or allelic loss of BECN1 in breast and ovarian cancer patients, nor are there inactivating mutations in the absence of BRCA1 mutation or loss. This is in agreement with BRCA1 deficiency being a driver mutation in breast and ovarian cancer. Indeed, allelic loss of Becnl suppresses rather than promotes mammary tumorgenesis mediated by Palb2 deficiency (19). As PALB2 is a regulator of BRCA1 and BRCA2 and a known tumor suppressor (18), this suggests that Becnl suppresses tumorigenesis of HR-deficient cancers rather than promoting it.
One interesting tumor type where autophagy may promote tumor progression not included in the analysis here is hepatomas. Mice with allelic loss of Becnl, or bi-allelic deletion of Atg5 or Atg7 in liver are prone to liver tumors. Autophagy deficiency may promote initiation of benign liver tumors by inducing chronic tissue damage, but also autophagy may be needed for progression to more aggressive disease. Indeed, deletion of Atg7 diverts progression of lung adenocarcinomas to benign oncocytomas (13, 14). It will be of interest to examine the mutational status of autophagy genes in human hepatomas and oncocytomas once the sequencing data becomes available. This will test if autophagy defects both promote the genesis of hepatomas while they limit tumor progression to benign disease (hepatomas and oncocytomas).
Supplementary Material
Implications: Contrary to previous reports, BECN1 is not significantly mutated in human cancer and not a tumor suppressor gene as originally thought.
Figure 1.
CNVs covering both BRCA1 and BECN1.
Acknowledgements
The White laboratory acknowledges support from the NIH grants R01 CA163591 and R01 CA130893, the Department of Defense (W81XWH-09-01-0394), the Val Skinner Foundation, the New Jersey Commission for Cancer Research and the Rutgers Cancer Institute of New Jersey. The Chan laboratory acknowledges support from the Rutgers Cancer Institute of New Jersey. The Ganesan laboratory acknowledges support from the Triple Negative Breast Cancer Foundation, the Hugs for Brady Foundation, and the NIH (R01 CA169182 and R01 CA130893).
Footnotes
Disclosure: The authors have nothing to disclose
References
- 1.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–41. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 2.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–8. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nature reviews Cancer. 2012;12:401–10. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aita VM, Liang XH, Murty VV, Pincus DL, Yu W, Cayanis E, et al. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics. 1999;59:59–65. doi: 10.1006/geno.1999.5851. [DOI] [PubMed] [Google Scholar]
- 5.Choi AM, Ryter SW, Levine B. Autophagy in human health and disease. The New England journal of medicine. 2013;368:651–62. doi: 10.1056/NEJMra1205406. [DOI] [PubMed] [Google Scholar]
- 6.Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, et al. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature. 1999;402:672–6. doi: 10.1038/45257. [DOI] [PubMed] [Google Scholar]
- 7.Drost R, Jonkers J. Opportunities and hurdles in the treatment of BRCA1-related breast cancer. Oncogene. 2013 doi: 10.1038/onc.2013.329. [DOI] [PubMed] [Google Scholar]
- 8.Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A, et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest. 2003;112:1809–20. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yue Z, Jin S, Yang C, Levine AJ, Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc Natl Acad Sci U S A. 2003;100:15077–82. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takamura A, Komatsu M, Hara T, Sakamoto A, Kishi C, Waguri S, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Degenhardt K, Mathew R, Beaudoin B, Bray K, Anderson D, Chen G, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25:460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27:1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strohecker AM, Guo JY, Karsli-Uzunbas G, Price SM, Chen GJ, Mathew R, et al. Autophagy Sustains Mitochondrial Glutamine Metabolism and Growth of BRAFV600E-Driven Lung Tumors. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-13-0397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25:717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155:1216–9. doi: 10.1016/j.cell.2013.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki SW, Onodera J, Ohsumi Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PloS one. 2011;6:e17412. doi: 10.1371/journal.pone.0017412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews Molecular cell biology. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huo Y, Cai H, Teplova I, Bowman-Colin C, Chen G, Price S, et al. Autophagy opposes p53-mediated tumor barrier to facilitate tumorigenesis in a model of PALB2-associated hereditary breast cancer. Cancer discovery. 2013 doi: 10.1158/2159-8290.CD-13-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karantza-Wadsworth V, Patel S, Kravchuk O, Chen G, Mathew R, Jin S, et al. Autophagy mitigates metabolic stress and genome damage in mammary tumorigenesis. Genes Dev. 2007;21:1621–35. doi: 10.1101/gad.1565707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathew R, Kongara S, Beaudoin B, Karp CM, Bray K, Degenhardt K, et al. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007;21:1367–81. doi: 10.1101/gad.1545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr., Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Network TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yusenko MV. Molecular pathology of renal oncocytoma: a review. International journal of urology : official journal of the Japanese Urological Association. 2010;17:602–12. doi: 10.1111/j.1442-2042.2010.02574.x. [DOI] [PubMed] [Google Scholar]
- 25.Leongamornlert D, Mahmud N, Tymrakiewicz M, Saunders E, Dadaev T, Castro E, et al. Germline BRCA1 mutations increase prostate cancer risk. British journal of cancer. 2012;106:1697–701. doi: 10.1038/bjc.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.