Abstract
Background
Genetic variation in the β-2 adrenergic receptor gene (ADRB2) has been implicated in asthma severity and control with conflicting results. Epigenetic variation in the β-2 adrenergic receptor gene (ADRB2) may play an important role in asthma phenotype.
Objective
We aimed to evaluate whether DNA methylation of ADRB2 is associated with asthma phenotypes in inner city school aged children.
Methods
Multiple CpG sites in the promoter region of ADRB2 gene were analyzed in 177 children enrolled in the School Inner-City Asthma Study (SICAS). Blood or saliva-derived DNA was measured by bisulfite-polymerase chain reaction pyrosequencing assay. Average percent DNA methylation across the sites was evaluated for association with asthma severity (report of dyspnea, nighttime symptoms, rescue medication use, and baseline spirometry) and morbidity (school absences and unscheduled healthcare visits). Three clades composed of highly correlated methylation sites within the methylated segment of ADRB2 were further analyzed.
Results
Methylation of individual sites generally ranged from 0 – 6% with average percent methylation across sites of 2.4%. Univariate analyses strongly favored the association of higher percent methylation with lower asthma severity measured by report of dyspnea. Furthermore, there was a non-significant trend toward less rescue medication use, nighttime symptoms, school absences, activity limitation due to asthma, and improved lung function measurements with increased methylation. Multivariate analysis demonstrated methylation of ADRB2 gene significantly associated with less dyspnea (odds ratio (OR) 0.2, 95%confidence interval (CI), 0.1 – 0.6, P = 0.002). Each of the 3 clades of methylation sites showed a strong, but not statistically significant, effect on decreased dyspnea.
Conclusions and Clinical Relevance
DNA methylation in the ADRB2 gene is associated with decreased asthma symptom severity, suggesting a role for methylation in asthma phenotypes.
Introduction
Asthma is a complex disease whose manifestations are influenced by host characteristics and environmental exposures. Genotype – phenotype correlations in asthma have been inconsistent and likely belie the environmental contribution to phenotype, the genetic complexity of the disease and the influence of the myriad biological processes that influence the expression of genomic DNA.
It has long been suggested that the β-2 adrenergic receptor (β2AR) plays an important role in the development of asthma. Szentivanyi proposed in 1968 that bronchial hyperresponsiveness is a result of partial beta-adrenergic blockade[1]. The merits of this hypothesis were supported by studies demonstrating reduced beta-adrenergic response, reduced β2AR density and enhanced β2AR downregulation in lymphocytes and human airway smooth muscle cells derived from asthmatics as compared to controls[2–7]. The β-2 adrenergic receptor has been mapped to chromosome 5q31-33, a region identified by several genome-wide and regional surveys as harboring an atopy or asthma-susceptibility locus[8–11]. Along with additional evidence demonstrating linkage with the 5q31-33 locus and bronchial hyperresponsiveness[12], these studies prompted the investigation of β2AR as a candidate gene for asthma and its related phenotypes. Additionally, β2-agonists are the mainstay of asthma therapy, but significant interindividual clinical differences in response to these medications have been noted[13]. The precise determinants of this variability are unclear, but genetic variation at the receptor level may be of importance. This series of observations have made the β2AR one of the most extensively studied genes in asthma genetics and pharmacogenetics. An overview of this work illustrates the complexity of the β2AR’s biology, as well as the difficulties in elucidating the genetic determinants of complex disease and pharmacogenetics.
The β2AR is a G-protein coupled receptor present in respiratory epithelium, airway smooth muscle and lymphocytes, and is the principle target of beta-agonist bronchodilators. The gene encoding the receptor, ADRB2, has been extensively studied as a candidate gene for asthma susceptibility, asthma phenotypes, and response to medications[14, 15]. While the cumulative body of evidence has not supported ADRB2 as an asthma susceptibility gene[16, 17], results have been mixed in regard to asthma severity[18], nocturnal asthma, airway hyperresponsiveness, lung function and response to medication. Inconsistencies between studies may be due to population differences in genetic background, epistatic gene-gene interactions, or physiologic conditions that alter the receptor expression and function[16]. Additionally, epigenetic variation affecting gene expression may be an important determinant of clinical asthma phenotype.
Epigenetics describes molecular factors and processes that regulate genome activity independent of DNA sequence. One such process is the methylation of cytosine of CpG dinucleotides, frequently resulting in transcriptional silencing of neighboring genes. Programmed methylation of CpG clusters (“CpG islands”) is critical for normal development and cellular differentiation, as well as X-chromosome inactivation and genetic imprinting. CpG methylation also results from environmental stimuli[19], including in utero and environmental tobacco smoke[20, 21]. In complex diseases like asthma, DNA methylation offers a potential mechanism for environmental modification of genetic responses, including those at the ADRB2 locus. Indeed, a 407 base pair variably methylated CpG island overlaps the 5′ untranslated region and leading coding sequence of ADRB2, and may represent an important source of epigenetic regulation of this gene. A single previous study has shown a positive association of methylation of the ADRB2 gene with asthma severity in a cohort of Caucasian children[22]. It is in this context that we herein describe the variation of DNA methylation in the ADRB2 CpG island and its relationship to clinical asthma phenotypes in a cohort of extensively phenotyped school age inner city, ethnically and racially diverse children with asthma. We aimed to determine the effect of methylation at the ADRB2 promoter region to asthma symptoms, morbidity, and lung function. Some of the results of this study have been previously reported in abstract form[23].
Materials and Methods
Study Population
This study was nested in the ongoing School Inner City Asthma Study (SICAS), an epidemiologic study of the effect of environmental exposures in school classrooms and asthma morbidity in inner city school children. Recruitment is summarized elsewhere[24]. Briefly, students with asthma were screened and recruited from entire urban elementary schools. Children who had a physician’s diagnosis of asthma, which has been shown to correlate with indices of disease[25, 26], AND cough, wheezing, dyspnea, or whistling in the chest in the previous 12 months were considered as students with asthma and were eligible for the study[24]. Eligibility requirements were modeled after other National Inner-City asthma studies[27]. Exclusion criteria included significant pulmonary disease other than asthma, significant cardiovascular disease requiring daily medication, use of beta-blocker medication, and an inability to complete study procedures. For the purposes of this study, only children who provided samples for DNA extraction were selected. We obtained 136 spit saliva-derived and 41 blood-derived DNA samples from 177 participants enrolled between July 2008 and July 2010. One subject had both blood and saliva obtained at the same visit. Complete baseline questionnaire data was available for all tested outcomes for each child.
DNA Methylation
Blood and saliva-derived DNA were collected for evaluation of ADRB2 methylation. CpG sites were selected from the ADRB2 CpG island promoter region upstream from the transcriptional start site for high probability of affecting transcription, and thereby gene expression.
DNA methylation assays were performed by staff who were blinded to the study (EpigenDX, Worcester, MA), by bisulfate modification followed by PCR amplification and pyrosequencing. Percent methylation at each site was calculated as: C% = C Peak / (C Peak + T Peak). Correlation of in vitro prepared standards with expected methylation percentages demonstrated an r-squared of 0.8 or higher. In addition to assay quality control measures, duplicate samples for two subjects and one blood and saliva sample from the same subject, were compared. Three CpG sites were excluded due to greater than 50% variability from the mean between duplicate samples. The distributions of methylation at each position for saliva and blood-derived DNA were comparable. Therefore, the methylation profile at each locus is a combination of these sources. For analysis, the average percent methylation across the valid methylation sites was used as the predictor. Subsequently, this was divided into 3 clades of highly correlated sites for more specific investigation of methylation regions.
Measurement of Lung Function
At the baseline clinic visit, pre- and post-bronchodilator spirometry (Koko spirometer, nSpire Health, inc., Longmont, CO, USA) was performed according to ATS guidelines[28] and compared to predicted outcomes from the NHANES III[29]. Sixty two subjects had only pre-bronchodilator spirometry measured and therefore do not have a measure of bronchodilator response. Each spirometry session was reviewed for acceptability and repeatability according to ATS guidelines[28, 30].
Measurement of Asthma Severity and Morbidity
Baseline caregiver questionnaire recorded clinical asthma symptoms and activities of daily life concurrent with DNA sampling. We used 4 week recall of asthma symptoms and rescue medication use to evaluate the severity of asthma symptoms and impairment each subject suffered immediately to the time of sample attainment. Specifically, caregivers were asked to report whether: (1) the child had wheeze or difficulty breathing when exercising, (2) wheezing during the day when not exercising, (3) waking up at night with wheezing or difficulty breathing,(4) missed any days of school and (5) missed any daily activities due to his/her asthma over the previous 4 weeks. In order to quantify asthma symptom severity in the context of the impairment domain of NHLBI criteria[31], several variables were derived from categorical questions regarding the frequency of shortness of breath, nighttime symptoms, and rescue medication use. Since the responses were not identical to characterizations in the guidelines, some categorizations were approximated. The characterization of symptom severity was as follows: (1) A response for shortness of breath (termed ‘dyspnea’ herein) was deemed positive if it occurred greater than twice weekly over the previous 4 weeks, (2) frequent nighttime awakening due to asthma was defined as one or more interrupted nights in the previous 4 weeks, and (3) frequent rescue medication use was defined as use of rescue medication greater than or equal to twice per week over the previous 4 weeks.
The number of unscheduled healthcare visits for asthma, which included emergency department visits and unscheduled clinic or doctor’s office visits, and estimated number of days absent from school due to asthma over the previous 12 months were also obtained by caregiver questionnaire.
Statistical Analysis
Univariate and multivariate regression approaches were used to evaluate the relationships between average % methylation and asthma outcomes. While the inclusion of sample type in the univariate and multivariate analyses had little effect on the outcomes, sample type was included as a covariate to minimize potential differential tissue effect. Relationships from univariate analysis between the primary predictor and outcomes of interest that met a threshold association of alpha ≤ 0.0035 based on the Bonferroni correction for multiple testing (P = 0.05 ÷ 14 comparisons = 0.0035 were carried forward to multivariate regression analysis. Multivariate analysis also evaluated the more specific clades of highly associated (correlation >0.5) methylation sites in order to more specifically understand the methylation site effect on clinical outcome. Age, race and gender were included in all models as potential confounders. Additional potential confounders were evaluated in each statistical model and were retained if associated with outcomes of interest at a level of alpha ≤ 0.05 and had reasonable likelihood to be associated with methylation. In this regard, the following variables were considered: family income, environmental tobacco smoke exposure, family history (parents, siblings, grandparents) of asthma, preterm birth, physician diagnosis of hayfever/allergic rhinitis and physician diagnosis of eczema. Logistic, linear, and Poisson regression was used for respective binomial, Gaussian, and Poisson distributed outcomes. SAS version 9.2 (SAS Institute Inc., Cary, NC) was used for all statistical analysis. We performed post-hoc analyses to assess the possibility of genotype-specific methylation at amino acid position 16 (Arg16Gly).
Several steps were taken to assess the impact of including blood and saliva in a combined analysis. First, a stratified analysis limited to saliva-derived DNA or blood-derived DNA was performed for the significant associations. Second, a separate analysis of the residuals versus outcomes when sample type was included in the regression was undertaken to evaluate the confounding influence of sample type on the relationship of methylation with the asthma phenotypes. In essence, this demonstrates the effect of methylation independent of the sample source (the residual effect after sample type is accounted for) on the outcomes of interest.
Results
Study population
One hundred and seventy seven children enrolled in the School Inner City Asthma Study had blood or saliva-derived DNA analyzed for methylation of the ADRB2 gene. The mean age of the study participants was 7.9 years old and gender was evenly represented. Table I illustrates the demographic and baseline characteristics of the cohort and distribution of outcome measures. The group was racially and ethnically diverse with over one third of participants reporting to be Black and another one third Hispanic. Nearly 20% were of mixed race and/or ethnicity. Based on annual household income, greater than one third of participants were impoverished. Lung function was within the normal predicted range for all subjects.
Table 1.
Descriptive characteristics of the School Inner City Asthma Study participants
| Characteristic | Number of Subjects (%), except where noted |
|---|---|
| Female | 80 (45.2) |
| Race | |
| White | 8 (4.5) |
| Black | 67 (37.9) |
| Hispanic | 61 (34.5) |
| Mixed | 33 (18.6) |
| Other | 8 (4.5) |
| Age, years, mean (SD) | 7.9 (2.0) |
| Poverty | |
| Household income <$25,000 | 67(37.9) |
| Household income ≥$25,000 | 83 (46.9) |
| Family history of asthmaa | 137 (77.4) |
| Diagnosis of eczema | 88 (49.7) |
| Diagnosis of allergic rhinitis | 29 (16.4) |
| Environmental tobacco smoke exposure | |
| Never | 48 (27.1) |
| Occasional or more frequent | 129 (72.9) |
| Preterm birthb | 25 (14.1) |
| Spirometryc | |
| FEV1 precent predicted, mean (SD) | 107.2 (20.1) |
| FEV1/FVC ratio × 100, mean (SD) | 88.2 (6.3) |
| Bronchodilator response, mean % change (SD) | 12.4 (14.1) |
| School absences due to asthma in previous 12 months, Days, median (IQR) | 4 (1, 8) |
| Number of unscheduled healthcare visits for asthma in previous 12 months, median (range) | 0 (0,1) |
| Dyspnead | 26 (14.7) |
| Frequent rescue medication used | 40 (22.6) |
| Frequent nighttime awakening due to asthmad | 82 (46.3) |
| Perceived poor asthma controld | 48 (27.1) |
| Wheeze/dyspnea with exercisee | 78 (44.1) |
| Wheeze/dyspnea without exercisee | 37 (20.9) |
| Awaken with wheeze/dyspneae | 61 (34.5) |
| School absence due to asthmae | 39 (22.0) |
| Missed any daily activities due to asthmae | 40 (22.6) |
SD, standard deviation; FEV1, forced expiratory volume in 1 second; FEV1/FVC, FEV1 per forced vital capacity; Bronchodilator response = ((post-beta agonist FEV1 − pre- beta agonist FEV1)/ post-beta agonist FEV1) × 100.
Family history includes immediate family plus grandparents
less than 37 weeks gestational age
FEV1 percent predicted and FEV1/FVC n=145, Bronchodilator response n=93
Dyspnea: shortness of breath greater than twice weekly over the previous 4 weeks; Frequent rescue medication use: determined as ≥ twice weekly over previous 4 weeks; Frequent nighttime awakening due to asthma: Greater than twice in the previous 4 weeks; perceived poor asthma control: caregiver report regarding control over previous 4 week period.
Reported presence of symptom over previous 4 week period.
ADRB2 methylation patterns
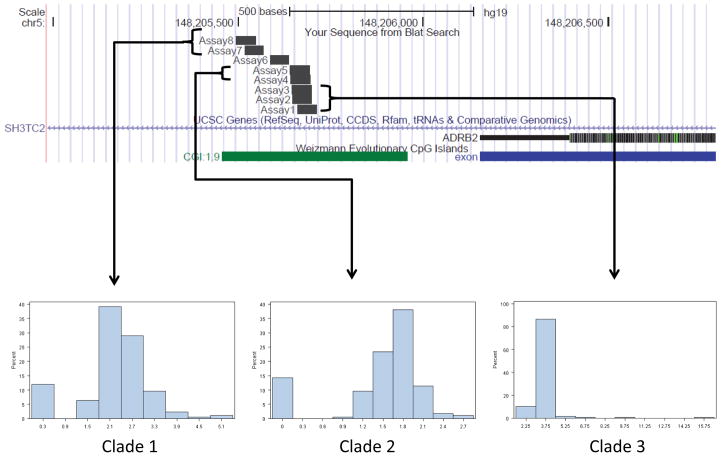
Eight of eleven typed ADRB2 methylation sites met quality control criteria. The average variability from the mean for duplicate samples was 0.34, and between blood and saliva was 0.30. Figure 1 illustrates the location of the CpG sites in relation to the ADRB2 gene as well as representative histograms for several of the methylation sites, grouped by clade. The proportion of methylation generally ranged from 0 – 6% with the average %methylation of 2.4%(Table 2). CpG sites were clustered based on high intra-site correlation of methylation (table E1). For two of the three groups, there was a high correlation of percent methylation for sites within the group (r2 = 0.73 for positions 4 and 5 [clade 2]; r2 = 0.75–0.79 between positions 6, 7 and 8 [clade 3] (Supplementary Table E1)). However, despite their physical proximity to one another and shared primer sets, correlation between sites 1, 2 and 3 (clade 1) were not high (r2 ≤ 0.54 for all comparisons).
Figure 1.
Upper panel: UCSC genome browser map of methylation sites investigated. Lower Panel: selected histograms for methylation Clades. UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly.
Table 2.
Distribution of percent methylation of CpG sites in β-2 Adrenergic Receptor gene
| CpG Sites | Location, Chromosome 5a | Mean, % | Standard Deviation | Interquartile range | |
|---|---|---|---|---|---|
| Clade 1 | Position 1 | 148205686-7 | 3.1 | 0.5 | 2.8, 3.2 |
| Position 2 | 148205673-4 | 3.5 | 1.1 | 3.1, 3.6 | |
| Position 3 | 148205671-2 | 2.7 | 0.5 | 2.5, 3.0 | |
| Clade 2 | Position 4 | 148205669-70 | 1.3 | 0.7 | 1.2, 1.6 |
| Position 5 | 148205667-8 | 1.5 | 0.7 | 1.4, 1.8 | |
| Clade 3 | Position 6 | 148205612-3 | 2.1 | 1.0 | 1.9, 2.7 |
| Position 7 | 148205542-3 | 2.2 | 0.8 | 1.9, 2.6 | |
| Position 8 | 148205520-1 | 2.8 | 0.6 | 2.4, 3.0 | |
| Average %Methylation | 148205520 -148205687 | 2.4 | 0.5 | 2.2, 2.7 |
UCSC Genome Browser on Human Feb. 2009 (GRCh37/hg19) Assembly.
Associations of ADRB2 methylation status with asthma severity phenotypes
We next assessed the relationship of ADRB2 methylation to measures of asthma severity. In univariate analysis (Table 3), average % methylation demonstrated a strong association with dyspnea and notable trends with other markers of improved asthma control, including decreased asthma symptoms, less frequent rescue medication use, fewer school absentee days, and higher measures of lung function. After adjusting for age, race, gender, preterm birth, family history of asthma, diagnosis of eczema and sample source % methylation of the ADRB2 promoter continued to exhibit a strong inverse association with dyspnea (OR = 0.2; 95% CI 0.1 – 0.6, p = 0.002; table 4). Furthermore, sensitivity analysis by DNA sample type and, separately, the residual effect after accounting for sample type was consistent with the findings of full cohort. Each clade showed strong, but not significant, associations with decreased dyspnea (table 4).
Table 3.
Univariate analysis of average %methylation of ADBR2 and asthma outcomes
| Outcome | Mean ADRB2 Methylation | ||
|---|---|---|---|
| Severitya | Odds Ratio | 95% confidence interval | P-value |
| Dyspnea | 0.2 | 0.1 – 0.5 | 0.0006 |
| Frequent nighttime awakening due to asthma | 0.7 | 0.4 – 1.3 | 0.22 |
| Frequent rescue medication use | 0.4 | 0.2 – 0.8 | 0.01 |
| Perceived poor asthma control | 0.6 | 0.3 – 1.2 | 0.14 |
| Wheeze/dyspnea with exercise | 0.6 | 0.3 – 1.1 | 0.08 |
| Wheeze/dyspnea without exercise | 0.6 | 0.3 – 1.3 | 0.21 |
| Awaken with wheeze/dyspnea | 0.7 | 0.4 – 1.3 | 0.22 |
| School absence due to asthma | 0.7 | 0.3 – 1.4 | 0.26 |
| Missed any activities due to asthma | 0.6 | 0.3 – 1.2 | 0.14 |
| Spirometryb | |||
| FEV1% | 1.4 | −5.0 – 7.8 | 0.67 |
| Post bronchodilator change in FEV1 | −2.9 | −8.2 – 2.4 | 0.28 |
| FEV1/FVC Ratio | 1.0 | −1.0 – 2.9 | 0.30 |
| No. of days of school absence due to asthma/12 months | −0.2 | −0.7 – 0.3 | 0.47 |
| No. of unscheduled healthcare visits due to asthma/12 months | −0.1 | −0.6 – 0.4 | 0.76 |
Severity parameters tested using logistic regression adjusting for sample type
Quantitative outcomes tested by linear regression analysis for spirometry outcomes and Wald test for Poisson distribution for number of school absences and number of unscheduled healthcare visits, beta coefficients are reported. All analyses adjusted for sample type.
Table 4.
The association between percent methylation of ADRB2 and Dyspnea
| Dyspnea | |||||
|---|---|---|---|---|---|
| Predictor | Odds Ratioa | 95%Confidence Interval | % Methylation Positive Response mean (SD)b | % Methylation Negative Response mean (SD) b | p-value |
| Average %methylation of ADRB2 | 0.2 | 0.1 – 0.6 | 2.1 (0.5) | 2.4 (0.5) | 0.002 |
|
| |||||
| Clade 1 | 0.2 | 0.04 – 0.7 | 2.9 (0.5) | 3.1 (0.6) | 0.01 |
| Clade 2 | 0.4 | 0.2 – 0.8 | 1.1 (0.7) | 1.5 (0.6) | 0.01 |
| Clade 3 | 0.4 | 0.2 – 0.7 | 2.0 (0.6) | 2.4 (0.7) | 0.004 |
adjusted for age, race, gender, preterm birth, family history of asthma, diagnosis of eczema and sample type
means derived from univariate comparisons
Methylation quantitative trait locus (mQTL) analysis
Recent data suggests that DNA sequence variation (for example, single nucleotide polymorphisms, SNPs) can confer allele-specific methylation changes at adjacent CpG sites. It is therefore possible that our observed associations of ADRB2 CpG methylation with asthma severity phenotypes is an indirect consequence of neighboring functional DNA sequence variants. Given that a common coding variant – a substitution of Arginine for Glycine at amino acid 16 – has been inconsistently associated with asthma severity and pharmacogenetic phenotypes, we genotyped the Arg16Gly variant and tested for association with the average % methylation at the ADRB2 promoter. We found no evidence of allele-specific differences in methylation, suggesting that our observed methylation-phenotype relationships are not due to indirect genetic effects for this SNP.
Discussion
We found that increased DNA methylation at the promoter region of the ADRB2 gene was associated with decreased asthma symptom severity, as measured by report of dyspnea, and trended toward an inverse association with airway obstruction measured by spirometry. These results provide evidence that epigenetic modification of the ADRB2 gene may directly influence the clinical asthma phenotype observed in school-aged children at high risk for asthma morbidity. This is the first evidence that increased methylation at the ADRB2 gene may be associated with improved asthma phenotypes.
While the association demonstrated here is compelling, the interpretation of these findings in the absence of gene expression and functional assays related to ADRB2 methylation is limited. DNA CpG hypermethylation typically leads to gene silencing and overall decreased expression of the gene product. These effects are frequently tissue specific, and understanding the potential mechanisms by which methylation of ADRB2 may promote milder asthma requires consideration of the main asthma-relevant tissues in which ADRB2 is expressed: airway epithelium and smooth muscle, and peripheral blood mononuclear cells (PBMCs).
In the airways, increased B2AR density has been associated with fatal and severe asthma in children and adults[32][33], however these findings are inconsistent[34]. In untreated adults with mild asthma there is an inverse relationship between FEV1 percent predicted and pulmonary receptor density measured by positron emission tomography (PET)[35]. Additionally, gene expression assays have demonstrated increased ADRB2 expression in bronchial mucosa in adults with severe asthma compared to those with mild to moderate disease[36] and maximal expression in those with persistent airflow limitation[37]. Therefore, it is plausible that suppressing beta 2 adrenergic receptor expression by methylation of ADRB2 in the bronchial mucosa and smooth muscle may lead to a milder asthma phenotype.
In peripheral mononuclear cells, the β2AR is known to mediate a proinflammatory pathway favoring the expression of Th2 cytokines[38, 39]. DNA methylation, among other epigenetic modulations, was recently shown decrease ADRB2 gene expression in Th2 driven cells in a mouse model[40]. In regards to the current study in which genetic analysis from blood and spit saliva, both samples representative of mononuclear cells, the effect of increased methylation on improved asthma symptoms and suggestion of better airway caliber may be mediated through decreased expression of Th2 cytokines. Mechanistically, this would favorably affect airways inflammation, thereby mitigating airway obstruction and asthma symptoms.
Despite these associations, results from pharmacologic studies have demonstrated expression of ADRB2 in airway epithelial cells and reduced airway hyperresponsiveness[41] and similarly, drug induced downregulation of leukocyte β2AR has been associated with increased symptoms and worsened lung function[4, 6, 42]. These studies suggest that decreased ADRB2 expression leads to greater airway hyperresponsiveness and a TH2 predominant phenotype, contrary to the paradigm we propose. Fu, et al.[22] recently demonstrated findings consistent with this alternative mechanistic hypothesis. In their cohort of school-aged children they found a positive relationship between whole blood ADRB2 methylation and asthma severity. Both our study and that by Fu et al. demonstrate similar distribution of methylation, age of participants and definition of asthma, though the methylated region of ADRB2, the population, and the timing of assessment differed substantially. The current study enrolled a racially and ethnically diverse population and measured outcomes at the time of genetic sample acquisition, and focused on methylation upstream of the transcriptional start site of ADRB2, while Fu et al. studied methylation of the 5′UTR downstream of the transcriptional start site in Caucasian children and assessed outcomes over the subsequent year as annualized severity[22].
This study has several potential limitations. We studied DNA isolated from either whole blood or saliva (which contains a mix of leukocytes and monocytes[44] as well as buccal epithelial cells), raising the possibility that the heterogeneity of these cell types may influence our results due to tissue specific differences in methylation patterns. Several lines of investigation reassure us that this is not the case. First, saliva and blood methylation demonstrated similar distributions at each site. Second, the relationship of methylation to the outcomes was not altered by the inclusion of sample type as a covariate. Third, completely removing the effect of sample source by analyzing the residuals from the main analysis with the outcomes did not substantially change the findings of this analysis. Finally, we performed a sensitivity analysis stratified by sample type which confirmed the effect size and directionality of the main study results. These analyses demonstrate that despite the lack of adequate sample size to find significant effect, the findings in the selected tissue are consistent with the findings of the overall cohort. Together, these results are reassuring that our findings are not due to the use of two tissue types. We are further limited in the narrow portion of the ADRB2 gene in which we focused for assessment of methylation. This region was chosen for its high probability for affecting gene transcription and thereby, expression, however, we concede that other regions may be important in this regard. Furthermore, expression and function studies are lacking for the ADRB2 gene and the influence of methylation and were beyond the scope of this study.
The findings presented here are an important step in elucidating the relationship between methylation at the ADRB2 gene clinical asthma phenotypes. Mechanistic studies to determine the effect of methylation on gene transcription and B2AR function are needed to fully understand the consequences of methylation of ADRB2. Vigorous investigation of epigenetic DNA modification of ADRB2 may offer the opportunity to discover new pathways in the disease and new pharmacologic targets[45]. Clarifying the role of epigenetic effects on the ADRB2 gene, as well as other candidate genes, allows for further understanding of complex asthma phenotypes by determining functional modifications to genomic expression. Extensive allele specific and haplotype specific methylation has been described across the genome[46–49]. While there was no genotype-specific methylation pattern at the ARG16GLY position in our data, it will be important to investigate the relationship between methylation of the ADRB2 gene and a more exhaustive array of polymorphisms in future studies before concluding that methylation is independent of genotype. We anticipate that integrative studies that explore the joint contributions of both genetic and epigenetic variation on phenotype will advance our understanding of complex diseases, particularly those with both significant genetic and environmental contributions, such as asthma.
Supplementary Material
Acknowledgments
Funding source: Funding for this research was provided by the American Thoracic Society Fellow Career Development Award, which was made possible by an educational grant from GlaxoSmithKline and Boston Children’s Hospital Pipeline Grant, and by grants K23AI106945-01 (PI: Gaffin), K24 AI 106822, R01 AI 073964 and R01 AI 073964-02S1 (PI: Phipatanakul) from the National Institutes of Health. This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (NIH Award #UL1 RR 025758 and financial contributions from Harvard University and its affiliated academic health care centers). The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic health care centers, the National Center for Research Resources, or the National Institutes of Health
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- 1.Szentivanyi A. The β-adrenergic theory of the atopic abnormality in bronchial asthma. J Allergy. 1968;42:203–32. [Google Scholar]
- 2.Shelhamer JH, Marom Z, Kaliner M. Abnormal beta-adrenergic responsiveness in allergic subjects. II. The role of selective beta 2-adrenergic hyporeactivity. J Allergy Clin Immunol. 1983;71:57–61. doi: 10.1016/0091-6749(83)90547-x. [DOI] [PubMed] [Google Scholar]
- 3.Parker CW, Smith JW. Alterations in cyclic adenosine monophosphate metabolism in human bronchial asthma. I. Leukocyte responsiveness to -adrenergic agents. The Journal of clinical investigation. 1973;52:48–59. doi: 10.1172/JCI107173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brooks SM, McGowan K, Altenau P. Relationship between beta-adrenergic binding in lymphocyte and severity of disease in asthma. Chest. 1979;75:232–83. [PubMed] [Google Scholar]
- 5.Kariman K. Beta-adrenergic receptor binding in lymphocytes from patients with asthma. Lung. 1980;158:41–51. doi: 10.1007/BF02713701. [DOI] [PubMed] [Google Scholar]
- 6.Galant SP, Duriseti L, Underwood S, Insel PA. Decreased beta-adrenergic receptors on polymorphonuclear leukocytes after adrenergic therapy. N Engl J Med. 1978;299:933–6. doi: 10.1056/NEJM197810262991707. [DOI] [PubMed] [Google Scholar]
- 7.Tashkin DP, Conolly ME, Deutsch RI, Hui KK, Littner M, Scarpace P, Abrass I. Subsensitization of beta-adrenoceptors in airways and lymphocytes of healthy and asthmatic subjects. Am Rev Respir Dis. 1982;125:185–93. doi: 10.1164/arrd.1982.125.2.185. [DOI] [PubMed] [Google Scholar]
- 8.Marsh DG, Neely JD, Breazeale DR, Ghosh B, Freidhoff LR, Ehrlich-Kautzky E, Schou C, Krishnaswamy G, Beaty TH. Linkage analysis of IL4 and other chromosome 5q31.1 markers and total serum immunoglobulin E concentrations. Science. 1994;264:1152–6. doi: 10.1126/science.8178175. [DOI] [PubMed] [Google Scholar]
- 9.Noguchi E, Shibasaki M, Arinami T, Takeda K, Maki T, Miyamoto T, Kawashima T, Kobayashi K, Hamaguchi H. Evidence for linkage between asthma/atopy in childhood and chromosome 5q31-q33 in a Japanese population. Am J Respir Crit Care Med. 1997;156:1390–3. doi: 10.1164/ajrccm.156.5.9702084. [DOI] [PubMed] [Google Scholar]
- 10.Bleecker ER, Amelung PJ, Levitt RC, Postma DS, Meyers DA. Evidence for linkage of total serum IgE and bronchial hyperresponsiveness to chromosome 5q: a major regulatory locus important in asthma. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 1995;25 (Suppl 2):84–8. doi: 10.1111/j.1365-2222.1995.tb00430.x. discussion 95–6. [DOI] [PubMed] [Google Scholar]
- 11.Ober C, Tsalenko A, Parry R, Cox NJ. A second-generation genomewide screen for asthma-susceptibility alleles in a founder population. Am J Hum Genet. 2000;67:1154–62. doi: 10.1016/s0002-9297(07)62946-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Postma DS, Bleecker ER, Amelung PJ, Holroyd KJ, Xu J, Panhuysen CI, Meyers DA, Levitt RC. Genetic susceptibility to asthma--bronchial hyperresponsiveness coinherited with a major gene for atopy. N Engl J Med. 1995;333:894–900. doi: 10.1056/NEJM199510053331402. [DOI] [PubMed] [Google Scholar]
- 13.Drazen JM, Israel E, Boushey HA, Chinchilli VM, Fahy JV, Fish JE, Lazarus SC, Lemanske RF, Martin RJ, Peters SP, Sorkness C, Szefler SJ. Comparison of regularly scheduled with as-needed use of albuterol in mild asthma. Asthma Clinical Research Network. N Engl J Med. 1996;335:841–7. doi: 10.1056/NEJM199609193351202. [DOI] [PubMed] [Google Scholar]
- 14.Hawkins GA, Weiss ST, Bleecker ER. Clinical consequences of ADRbeta2 polymorphisms. Pharmacogenomics. 2008;9:349–58. doi: 10.2217/14622416.9.3.349. [DOI] [PubMed] [Google Scholar]
- 15.Litonjua AA. The significance of beta2-adrenergic receptor polymorphisms in asthma. Curr Opin Pulm Med. 2006;12:12–7. doi: 10.1097/01.mcp.0000198068.50457.95. [DOI] [PubMed] [Google Scholar]
- 16.Raby BA, Weiss ST. Beta2-adrenergic receptor genetics. Curr Opin Mol Ther. 2001;3:554–66. [PubMed] [Google Scholar]
- 17.Thakkinstian A, McEvoy M, Minelli C, Gibson P, Hancox B, Duffy D, Thompson J, Hall I, Kaufman J, Leung TF, Helms PJ, Hakonarson H, Halpi E, Navon R, Attia J. Systematic review and meta-analysis of the association between {beta}2-adrenoceptor polymorphisms and asthma: a HuGE review. American journal of epidemiology. 2005;162:201–11. doi: 10.1093/aje/kwi184. [DOI] [PubMed] [Google Scholar]
- 18.Contopoulos-Ioannidis DG, Manoli EN, Ioannidis JP. Meta-analysis of the association of beta2-adrenergic receptor polymorphisms with asthma phenotypes. J Allergy Clin Immunol. 2005;115:963–72. doi: 10.1016/j.jaci.2004.12.1119. [DOI] [PubMed] [Google Scholar]
- 19.Breton CV, Byun HM, Wenten M, Pan F, Yang A, Gilliland FD. Prenatal tobacco smoke exposure affects global and gene-specific DNA methylation. Am J Respir Crit Care Med. 2009;180:462–7. doi: 10.1164/rccm.200901-0135OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joubert BR, Haberg SE, Nilsen RM, Wang X, Vollset SE, Murphy SK, Huang Z, Hoyo C, Midttun O, Cupul-Uicab LA, Ueland PM, Wu MC, Nystad W, Bell DA, Peddada SD, London SJ. 450K Epigenome-Wide Scan Identifies Differential DNA Methylation in Newborns Related to Maternal Smoking During Pregnancy. Environmental health perspectives. 2012 doi: 10.1289/ehp.1205412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wan ES, Qiu W, Baccarelli A, Carey VJ, Bacherman H, Rennard SI, Agusti A, Anderson W, Lomas DA, Demeo DL. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Human molecular genetics. 2012;21:3073–82. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu A, Leaderer BP, Gent JF, Leaderer D, Zhu Y. An environmental epigenetic study of ADRB2 5′-UTR methylation and childhood asthma severity. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2012;42:1575–81. doi: 10.1111/j.1365-2222.2012.04055.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaffin JM, Raby BA, Hoffman EB, Gold DR, Phipatanakul W. Methylation Of The Beta-2 Adrenergic Receptor Gene Is Associated With Decreased Asthma Severity In Inner City School Children. American Journal of Respiratory and Critical Care Medicine. 2012;1:A5604. [Google Scholar]
- 24.Phipatanakul W, Bailey A, Hoffman EB, Sheehan WJ, Lane JP, Baxi S, Rao D, Permaul P, Gaffin JM, Rogers CA, Muilenberg ML, Gold DR. The school inner-city asthma study: design, methods, and lessons learned. J Asthma. 48:1007–14. doi: 10.3109/02770903.2011.624235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss ST. Epidemiology and heterogeneity of asthma. Ann Allergy Asthma Immunol. 2001;87:5–8. doi: 10.1016/s1081-1206(10)62188-6. [DOI] [PubMed] [Google Scholar]
- 26.Weiss ST, Tager IB, Speizer FE, Rosner B. Persistent wheeze. Its relation to respiratory illness, cigarette smoking, and level of pulmonary function in a population sample of children. Am Rev Respir Dis. 1980;122:697–707. doi: 10.1164/arrd.1980.122.5.697. [DOI] [PubMed] [Google Scholar]
- 27.Mitchell H, Senturia Y, Gergen P, Baker D, Joseph C, McNiff-Mortimer K, Wedner HJ, Crain E, Eggleston P, Evans R, 3rd, Kattan M, Kercsmar C, Leickly F, Malveaux F, Smartt E, Weiss K. Design and methods of the National Cooperative Inner-City Asthma Study. Pediatr Pulmonol. 1997;24:237–52. doi: 10.1002/(sici)1099-0496(199710)24:4<237::aid-ppul3>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 28.Miller MR, Hankinson J, Brusasco V, Burgos F, Casaburi R, Coates A, Crapo R, Enright P, van der Grinten CP, Gustafsson P, Jensen R, Johnson DC, MacIntyre N, McKay R, Navajas D, Pedersen OF, Pellegrino R, Viegi G, Wanger J. Standardisation of spirometry. Eur Respir J. 2005;26:319–38. doi: 10.1183/09031936.05.00034805. [DOI] [PubMed] [Google Scholar]
- 29.Hankinson JL, Odencrantz JR, Fedan KB. Spirometric reference values from a sample of the general U.S. population. Am J Respir Crit Care Med. 1999;159:179–87. doi: 10.1164/ajrccm.159.1.9712108. [DOI] [PubMed] [Google Scholar]
- 30.Beydon N, Davis SD, Lombardi E, Allen JL, Arets HG, Aurora P, Bisgaard H, Davis GM, Ducharme FM, Eigen H, Gappa M, Gaultier C, Gustafsson PM, Hall GL, Hantos Z, Healy MJ, Jones MH, Klug B, Lodrup Carlsen KC, McKenzie SA, Marchal F, Mayer OH, Merkus PJ, Morris MG, Oostveen E, Pillow JJ, Seddon PC, Silverman M, Sly PD, Stocks J, Tepper RS, Vilozni D, Wilson NM. An official American Thoracic Society/European Respiratory Society statement: pulmonary function testing in preschool children. Am J Respir Crit Care Med. 2007;175:1304–45. doi: 10.1164/rccm.200605-642ST. [DOI] [PubMed] [Google Scholar]
- 31.Expert Panel Report 3 (EPR-3): Guidelines for the Diagnosis and Management of Asthma-Summary Report 2007. J Allergy Clin Immunol. 2007;120:S94–138. doi: 10.1016/j.jaci.2007.09.043. [DOI] [PubMed] [Google Scholar]
- 32.Bai TR, Mak JC, Barnes PJ. A comparison of beta-adrenergic receptors and in vitro relaxant responses to isoproterenol in asthmatic airway smooth muscle. Am J Respir Cell Mol Biol. 1992;6:647–51. doi: 10.1165/ajrcmb/6.6.647. [DOI] [PubMed] [Google Scholar]
- 33.Spina D, Rigby PJ, Paterson JW, Goldie RG. Autoradiographic localization of beta-adrenoceptors in asthmatic human lung. Am Rev Respir Dis. 1989;140:1410–5. doi: 10.1164/ajrccm/140.5.1410. [DOI] [PubMed] [Google Scholar]
- 34.Sharma RK, Jeffery PK. Airway beta-adrenoceptor number in cystic fibrosis and asthma. Clin Sci (Lond) 1990;78:409–17. doi: 10.1042/cs0780409. [DOI] [PubMed] [Google Scholar]
- 35.Qing F, Rahman SU, Rhodes CG, Hayes MJ, Sriskandan S, Ind PW, Jones T, Hughes JM. Pulmonary and cardiac beta-adrenoceptor density in vivo in asthmatic subjects. Am J Respir Crit Care Med. 1997;155:1130–4. doi: 10.1164/ajrccm.155.3.9116998. [DOI] [PubMed] [Google Scholar]
- 36.Selivanova PA, Kulikov ES, Kozina OV, Trofimenko IN, Freidin MB, Chernyak BA, Ogorodova LM. Differential expression of the beta2-adrenoreceptor and M3-cholinoreceptor genes in bronchial mucosa of patients with asthma and chronic obstructive pulmonary disease. Ann Allergy Asthma Immunol. 2012;108:39–43. doi: 10.1016/j.anai.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Selivanova PA, Kulikov ES, Kozina OV, Gereng EA, Freidin MB, Ogorodova LM. Morphological and molecular characteristics of “difficult” asthma. J Asthma. 2010;47:269–75. doi: 10.3109/02770900903584001. [DOI] [PubMed] [Google Scholar]
- 38.Agarwal SK, Marshall GD., Jr Beta-adrenergic modulation of human type-1/type-2 cytokine balance. J Allergy Clin Immunol. 2000;105:91–8. doi: 10.1016/s0091-6749(00)90183-0. [DOI] [PubMed] [Google Scholar]
- 39.Panina-Bordignon P, Mazzeo D, Lucia PD, D’Ambrosio D, Lang R, Fabbri L, Self C, Sinigaglia F. Beta2-agonists prevent Th1 development by selective inhibition of interleukin 12. The Journal of clinical investigation. 1997;100:1513–9. doi: 10.1172/JCI119674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McAlees JW, Smith LT, Erbe RS, Jarjoura D, Ponzio NM, Sanders VM. Epigenetic regulation of beta2-adrenergic receptor expression in T(H)1 and T(H)2 cells. Brain Behav Immun. 2011;25:408–15. doi: 10.1016/j.bbi.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McGraw DW, Forbes SL, Mak JC, Witte DP, Carrigan PE, Leikauf GD, Liggett SB. Transgenic overexpression of beta(2)-adrenergic receptors in airway epithelial cells decreases bronchoconstriction. Am J Physiol Lung Cell Mol Physiol. 2000;279:79–89. doi: 10.1152/ajplung.2000.279.2.L379. [DOI] [PubMed] [Google Scholar]
- 42.Szefler SJ, Ando R, Cicutto LC, Surs W, Hill MR, Martin RJ. Plasma histamine, epinephrine, cortisol, and leukocyte beta-adrenergic receptors in nocturnal asthma. Clin Pharmacol Ther. 1991;49:59–68. doi: 10.1038/clpt.1991.11. [DOI] [PubMed] [Google Scholar]
- 43.Parola AL, Kobilka BK. The peptide product of a 5′ leader cistron in the beta 2 adrenergic receptor mRNA inhibits receptor synthesis. J Biol Chem. 1994;269:4497–505. [PubMed] [Google Scholar]
- 44.Vidovic A, Vidovic Juras D, Vucicevic Boras V, Lukac J, Grubisic-Ilic M, Rak D, Sabioncello A. Determination of leucocyte subsets in human saliva by flow cytometry. Archives of oral biology. 2012;57:577–83. doi: 10.1016/j.archoralbio.2011.10.015. [DOI] [PubMed] [Google Scholar]
- 45.Cosio BG, Mann B, Ito K, Jazrawi E, Barnes PJ, Chung KF, Adcock IM. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. Am J Respir Crit Care Med. 2004;170:141–7. doi: 10.1164/rccm.200305-659OC. [DOI] [PubMed] [Google Scholar]
- 46.Schalkwyk LC, Meaburn EL, Smith R, Dempster EL, Jeffries AR, Davies MN, Plomin R, Mill J. Allelic skewing of DNA methylation is widespread across the genome. Am J Hum Genet. 2010;86:196–212. doi: 10.1016/j.ajhg.2010.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. Am J Respir Crit Care Med. 184:191–7. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morales E, Bustamante M, Vilahur N, Escaramis G, Montfort M, de Cid R, Garcia-Esteban R, Torrent M, Estivill X, Grimalt JO, Sunyer J. DNA hypomethylation at ALOX12 is associated with persistent wheezing in childhood. Am J Respir Crit Care Med. 185:937–43. doi: 10.1164/rccm.201105-0870OC. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.