To the Editor
Angermeyer et al. (2013) claim that “Merkel Cell Polyomavirus-Positive Merkel Cell Carcinoma Cells Do Not Require Expression of the Viral Small T Antigen” (Angermeyer et al., 2013). This controversial conclusion is based on their inability to detect Merkel cell polyomavirus (MCV) small T (sT) protein expression and to inhibit cell growth by putative sT knockdown in MCV-MCC cells.
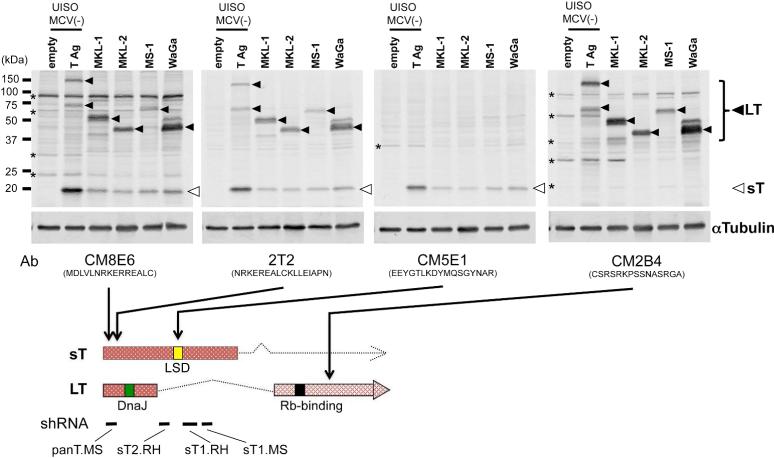
These findings contradict existing evidence showing MCV sT protein expression in MCV-MCC cancer tissues (Shuda et al., 2011) and cell lines (see Figure. 2, Houben et al., 2010) (Guastafierro et al., 2013). To investigate this discrepancy, we tried replicating Angermeyer et al's results using the same antibodies to detect MCV sT (CM8E6 (Kwunet al., 2009), CM5E1 (Shuda et al., 2011) and 2T2 ((Wang et al., 2012), kindly provided by C. Buck) on a panel of MCV-MCC cell lines (Figure 1). MCV sT and large T (LT) are alternatively-spliced viral oncoproteins sharing a common N-terminus but having different C-termini, thus CM8E6 and 2T2 detects all isoforms of T antigens, while CM5E1 detects only sT and CM2B4 detects only LT and related isoforms. Differences in protein expression levels between MCV LT and sT are likely dependent on either premRNA or post-transcriptional protein processing. For positive and negative controls, we used UISO cells transiently transfected with the MCV T antigen locus (JN038578) or with corresponding empty vector. UISO, commonly described as being from MCC origin (Houben et al., 2007), is negative for MCV and miRNA ontology studies show it clusters with cell lines of breast cancer origin (Renwick et al., 2013). In contrast to Angermeyer et al., the 19 kD MCV sT band is readily detected in all MCV-MCC cell lines (open arrows) but not in UISO cells.
Figure 2.
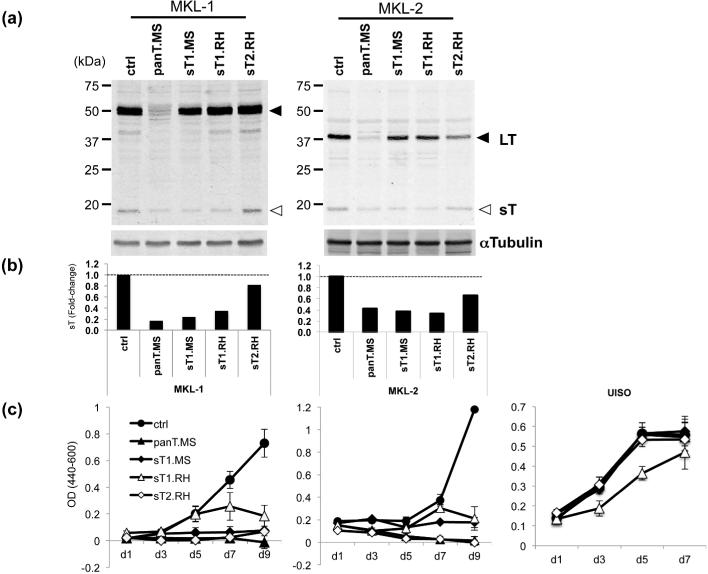
Merkel cell polyomavirus (MCV) small T (sT) antigen knockdown inhibits MCV-positive Merkel cell carcinoma (MCC) cell proliferation.
(a) MCV-positive MCC cell lines, MKL-1 and MKL-2, were transduced with pLKO.1-based lentiviral shRNAs targeting both LT and sT (panT.MS) or sT alone (sT1.MS, sT1.RH, and sT2.RH) as described (Houben et al., 2010). Both LT (closed arrows) and sT (open arrows) proteins are detected by 2T2. (b) Expression of LT and sT was quantitated by LI-COR IR immunoblotting system using αTubulin for normalization. Relative sT expression to sh ctrl is shown. (c) shRNA-transduced MCV-positive (MKL-1 and MKL-2) cells and MCV-negative (UISO) cells were subjected to Wst-1 cell proliferation assay. Error bars indicate standard deviation.
Figure 1.
Detection of Merkel cell polyomavirus (MCV) small T (sT) antigen expression by multiple MCV T antigen antibodies.
MCV-positive MCC cells (MKL-1, MKL-2, MS-1 and WaGa) and MCV-negative UISO cells transfected with MCV genomic T antigen gene or empty vector as positive and negative controls, were immunoblotted with multiple MCV T antigen antibodies. αTubulin was used as a loading control. Both large T (LT, closed arrows) and small T (sT, open arrows) were detected by CM8E6 and 2T2, sT by CM5E1, and LT by CM2B4. Asterisks indicate non-specific bands. Peptide sequences used for monoclonal antibody production and shRNA targeting sites are shown in the bottom diagram of T antigen transcripts with a DnaJ (green box), an Rb-binding (black box) as well as large T stabilization (LSD, yellow box (Kwun et al., 2013)) domains.
Is MCV sT required for MCC cell proliferation?
The knockdown by Angermeyer et al. used different shRNAs (Angermeyer et al., 2013), and directly contradict our findings that sT knockdown inhibits cell replication in MCV-MCC (Shuda et al). Since Angermeyer et al were not able to measure sT protein by immunoblotting, efficacy of knockdown could not be determined. To assess cell proliferation, Angermeyer et al used a competition assay containing mixtures of shRNA-transduced and nontransduced cells that compete with each other for growth. We instead directly measured cell proliferation using standard Wst-1 assays. To resolve this, we generated the same two sT-specific shRNAs cloned in pLKO.1-based lentiviral vector (named here sT1.RH for Roland Houben laboratory and sT2.RH) used in their study and compared them to an shRNA previously described to target sT alone (designated here as sh sT1.MS for Masahiro Shuda laboratory), an shRNA targeting both LT and sT (sh panT.MS) and a scrambled negative control shRNA (sh ctrl) (Shuda et al., 2011). Both sh sT1.MS and sh panT.MS inhibit sT protein expression measured by quantitative LICOR immunoblotting (Figure 2A and B) and cell growth (Figure 2C) as previously described (Shuda et al., 2011). One of Angermeyer et al.'s shRNA (sh sT1.RH) also inhibits sT expression and significantly inhibits MCV-MCC cell growth. Proliferation of UISO cells, however, was also reduced consistent with an off-target effect that precludes evaluating it as a targeting agent for MCV sT. The other shRNA (sh sT2.RH) has minimal (MKL-2) or no (MKL-1) sT knockdown activity (Figure 2B). It nonetheless inhibits MCV-MCC cell growth. Given the inability to monitor sT knockdown and off-target effects for the sT.RH shRNAs used in Angermeyer et al's knockdown studies, attempts to rescue MCC cell proliferation using combinations of LT and sT expression during sT knockdown are not interpretable.
Using the same shRNA constructs described by Angermeyer et al., we show that their conclusion that MCV sT plays no role in MCV is not correct. We recommend using sh sT1.MS, which is efficacious in sT knockdown and we are unaware of any off-target activity. Mixed cell competition assays to measure proliferation are fraught with uncertainty since paracrine effects can distort proliferation measurements and more traditional cell counting or Wst-1 measurements are preferred. Finally, as co-equal authors that independently developed T antigen shRNA knockdowns for the report describing T antigen knockdown in MCC (Houben et al., 2010), we disagree with these authors’ assertion that pan-T knockdown induces apoptosis in MCC. Weak PARP cleavage (Figure. 5B, Houben et al., 2010) can be seen in some cell lines during knockdown, but it is not universally present and Casp3 or Casp9 cleavage is completely absent. As confirmed by Angermeyer et al., MCV sT is the only known transforming oncoprotein of MCV in rodent cells while MCV LT alone is not sufficient to transform rodent fibroblast cells (Angermeyer et al., 2013; Shuda et al., 2011). In the SV40 T antigen model of human cell transformation, expression of both LT and sT is required (Hahn et al., 1999). Taking into consideration the higher tumorigenic barrier in human cells as compared to rodent cells and that the spliced sT isoform is expressed together with LT in most MCC (Shuda et al., 2011), it is likely that MCV sT co-contributes with MCV LT to MCC carcinogenesis.
Acknowledgements
This study was funded by the National Institutes of Health grants CA136363 and CA120726 to PSM and YC, who are also supported as American Cancer Society Research Professors. MS was supported in part by the University of Pittsburgh Skin Cancer SPORE CA12197305.
Footnotes
Conflict of interests
Yuan Chang and Patrick Moore are on patents for Merkel cell polyomavirus diagnostic reagents, which have been assigned to the University of Pittsburgh.
References
- Angermeyer S, Hesbacher S, Becker JC, et al. Merkel cell polyomavirus-positive merkel cell carcinoma cells do not require expression of the viral small T antigen. The Journal of investigative dermatology. 2013;133:2059–64. doi: 10.1038/jid.2013.82. [DOI] [PubMed] [Google Scholar]
- Guastafierro A, Feng H, Thant M, et al. Characterization of an early passage Merkel cell polyomavirus-positive Merkel cell carcinoma cell line, MS-1, and its growth in NOD scid gamma mice. Journal of virological methods. 2013;187:6–14. doi: 10.1016/j.jviromet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, et al. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–8. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Houben R, Ortmann S, Schrama D, et al. Activation of the MAP kinase pathway induces apoptosis in the Merkel cell carcinoma cell line UISO. The Journal of investigative dermatology. 2007;127:2116–22. doi: 10.1038/sj.jid.5700857. [DOI] [PubMed] [Google Scholar]
- Houben R, Shuda M, Weinkam R, et al. Merkel cell polyomavirus-infected Merkel cell carcinoma cells require expression of viral T antigens. J Virol. 2010;84:7064–72. doi: 10.1128/JVI.02400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwun HJ, Guastafierro A, Shuda M, et al. The minimum replication origin of merkel cell polyomavirus has a unique large T-antigen loading architecture and requires small T-antigen expression for optimal replication. J Virol. 2009;83:12118–28. doi: 10.1128/JVI.01336-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwun HJ, Shuda M, Feng H, et al. Merkel Cell Polyomavirus Small T Antigen Controls Viral Replication and Oncoprotein Expression by Targeting the Cellular Ubiquitin Ligase SCF(Fbw7.). Cell Host Microbe. 2013;14:125–35. doi: 10.1016/j.chom.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renwick N, Cekan P, Masry PA, et al. Multicolor microRNA FISH effectively differentiates tumor types. J Clin Invest. 2013;123:2694–702. doi: 10.1172/JCI68760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuda M, Kwun HJ, Feng H, et al. Human Merkel cell polyomavirus small T antigen is an oncoprotein targeting the 4E-BP1 translation regulator. J Clin Invest. 2011;121:3623–34. doi: 10.1172/JCI46323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Li J, Schowalter RM, et al. Bromodomain protein Brd4 plays a key role in Merkel cell polyomavirus DNA replication. PLoS pathogens. 2012;8:e1003021. doi: 10.1371/journal.ppat.1003021. [DOI] [PMC free article] [PubMed] [Google Scholar]