Abstract
Intracellular Ca2+ activated calmodulin (CaM) inhibits gap junction channels in the low nM to high μM range of [Ca2+]i. This regulation plays an essential role in numerous cellular processes that include hearing, lens transparency, and synchronized contractions of the heart. Previous studies have indicated that gap junction mediated cell-to-cell communication was inhibited by CaM antagonists. More recent evidence indicates a direct role of CaM in regulating several members of the connexin family. Since the intracellular loop and carboxyl termini of connexins are largely “invisible” in electron microscopy and X-ray crystallographic structures due to disorder in these domains, peptide models encompassing the putative CaM binding sites of several intracellular domains of connexins have been used to identify the Ca2+-dependent CaM binding sites of these proteins. This approach has been used to determine the CaM binding affinities of peptides derived from a number of different connexin-subfamilies.
Keywords: Ca2+, connexin, gap junction regulation, Calmodulin binding
1. Introduction
Modulation of the cytoplasmic Ca2+ concentration ([Ca2+]i) is a ubiquitous mechanism by which cells transduce external signals into biological responses. The signaling cascade initiated by the rise in [Ca2+]i is often mediated via Ca2+-binding proteins such as calmodulin (CaM) [1, 2]. CaM, a key multifunctional transducer of Ca2+ signals in eukaryotes, has four EF-hand Ca2+ binding motifs in two globular N- and C-domains that are separated by a flexible linker. Upon Ca2+ binding, CaM undergoes a large conformational change, exposing hydrophobic patches that are important in its binding to more than 300 target proteins in multiple cellular processes [3–5]. CaM uses either its N- or C-domains with different Ca2+ binding affinities to differentiate between local and global [Ca2+]i changes, and to regulate a diverse group of membrane channels/pumps that include cyclic nucleotide-gated (CNG) channels [6], N-methyl-D-aspartate receptor (NMDA receptor) [7], ryanodine receptors (RyR) [8], Ca2+-activated K+ channels of small or intermediate conductance (SK or IK) [9, 10], Trp family channels [11], Ca2+ channels, and gap junction channels [12–16].
Gap junctions are formed from the docking of paired hexameric connexons, which are also called hemichannels with each connexon comprised of six monomeric connexins. Approximately 21 connexin isoforms have been identified in a multitude of human cell types [17]. According to their gene structure, homology and specific sequence motifs, connexins have been grouped into three major categories, termed the α, β and γ connexin subfamilies [18]. The different connexin subfamily members all share the same structural topology with four α-helical transmembrane domains connected by two extracellular loops and a cytoplasmic loop; both N- and C-terminal face the cytosol. Three highly conserved Cys residues residing in each extracellular loop form intraconnexin disulfide bonds. The transmembrane domains and the extracellular loops are highly conserved among different connexin families. Although the N-terminus is also relatively conserved, the cytoplasmic loop and C-terminus exhibit significant variation among different connexins.
Connexins are almost universally expressed in vertebrate tissues with the exception of several highly differentiated cell types that include skeletal muscle, erythrocytes, and mature sperm cells [19]. Gap junctions facilitate the direct intercellular cytoplasmic connections that allow the exchange of ionic current, secondary messengers, and small metabolites up to 1 kDa between the cytoplasms of adjacent cells. Gap junctions play important roles in many biological processes [20–22], including development, differentiation, cell synchronization, neuronal activity, and the immune response. The functional loss of gap junction may result in embryonic lethality (e.g. Cx45) [23, 24], perinatal death due to congenital malformations (e.g. Cx43−/−) [25], and plays a major role in the pathogenesis of several other diseases (e.g. Charcot-Marie-Tooth disease, Cx32; Oculodentodigital dysplasia, Cx43; Non-syndromic deafness, Cx26, Cx30, Cx43; lens cataracts, C46, Cx50) [26, 27]. Connexin mutations may interfere with gene expression, affect protein or DNA stability, impede protein trafficking, translocation and assembly, or alter the regulation of gap junction.
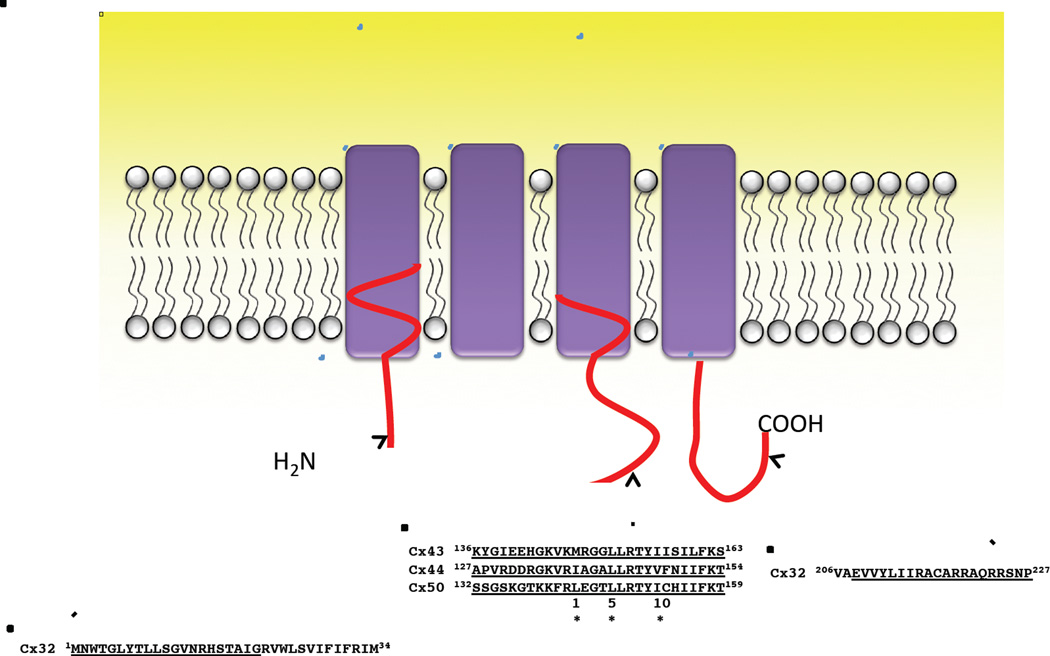
Intercellular communication mediated by gap junctions can be regulated by multiple factors that include [Ca2+]i, H+, CaM, voltage, and phosphorylation of the connexin subunits [28, 29]. The role of CaM in the regulation of gap junction channels has been previously reviewed by Peracchia [30]. In this paper, we review current understanding of the Ca2+-CaM regulation of gap junctions comprised of different connexins that is effected by the direct binding of Ca2+-CaM to specific sequences of these connexins (Fig. 1). We will first discuss the functional regulation of gap junctions by CaM, we will then discuss the molecular basis of such regulation including the CaM binding locations, relative binding affinities, and binding modes for the different connexins, and finally, a gating model for the regulation of the α-subfamily of connexins by Ca2+-CaM is proposed.
Figure 1. Reported CaM-binding sites in rat Cx32, sheep Cx44, human Cx43 and 50.
Two CaM-binding sites are located at the N- and C-terminus of Cx32. The identified CaM-binding sites in α-subfamily connexins are in the second half of the intracellular loop and fit the 1-5-10 subclass binding mode, where each number represents the presence of a hydrophobic residue. (*, Hydrophobic residues).
2. Functional Regulation of Gap Junction by CaM
Over the years, a significant body of data supports the involvement of CaM in the regulation of gap junctions comprised of the three connexin subfamilies. Délèze [31] first showed that Ca2+ is essential for the healing process in mammalian heart muscle by preventing longitudinal diffusion of molecules in cardiac fibers. Subsequently, Rose et al. [32] showed that in salivary gland cells, inhibition of cell-to-cell coupling occurs when the intracellular concentration of Ca2+ was increased from 0.1 μM to 50 μM.
Recognizing that most Ca2+-induced phenomena are mediated by CaM [33], Peracchia and colleagues [34] first tested the hypothesis that gap junctions were regulated by CaM by examining the effect of the CaM inhibitor trifluoperazine (TFP) on the electrical coupling of amphibian embryonic cells exposed to CO2 to lower intracellular pH. While the role of CaM in mediating the action of Ca2+ on enzyme activation was well known in the 1980s, gap junctions represented the first membrane channels shown to be modulated by CaM; only a decade later was the next CaM regulated channel identified [6]. Peracchia and colleagues demonstrated that TFP reversibly inhibits the CO2-induced electrical uncoupling in amphibian embryo cells by interfering with the mechanism which closes the cell-to-cell channels. Subsequently, more specific CaM blockers (calmidazolium and W7) were shown to prevent uncoupling of Xenopus embryonic cells [35] and crayfish axons [36, 37], indicating the generalized nature of this role for CaM in regulating gap junctions. This hypothesis was strengthened by evidence that the gap junction protein Cx32 bound CaM in gel overlays [38–40]. The suppression of CaM expression in oocytes can also inhibit CO2 induced electrical uncoupling and injection of CaM into oocytes can recover it [41]. Cx32 was also shown to colocalize with CaM using immunofluorescence microscopy [42]. Later, Blodow et al. reported that CaM antagonists suppress gap junction coupling of Cx26 in isolated Hensen cells of the guinea pig cochlea [43].
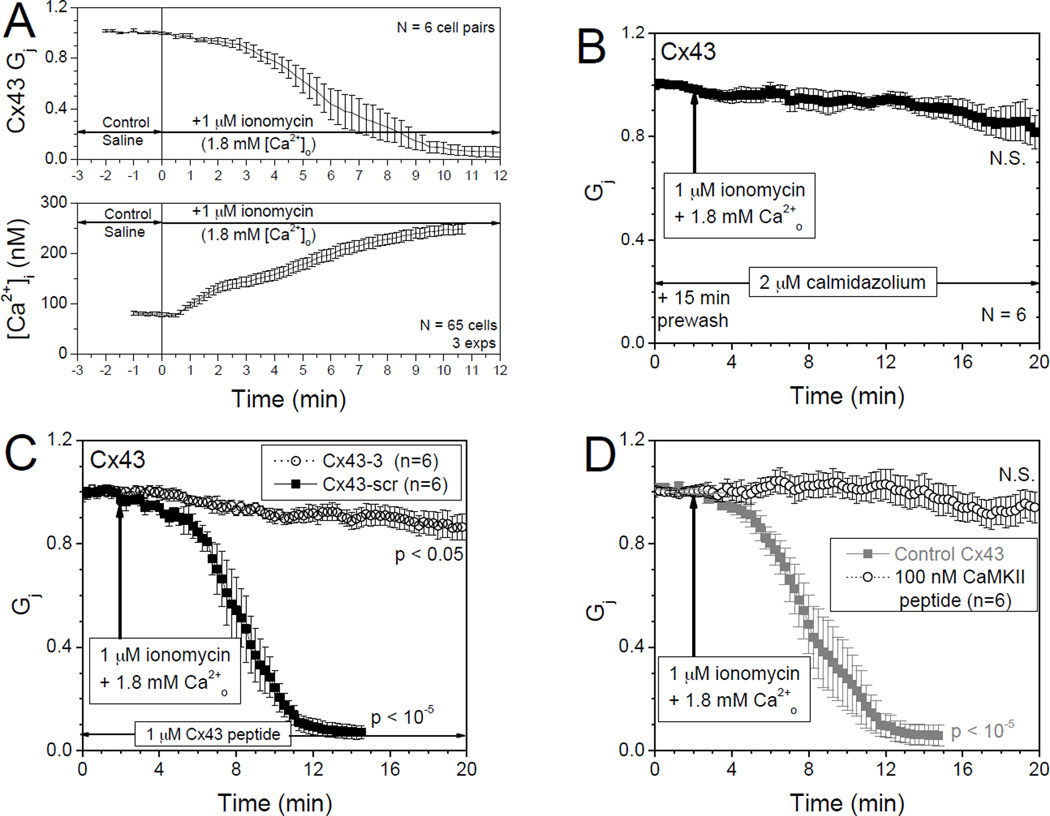
Louis and Lurtz first showed that the gap junction mediated cell-to-cell transfer of dye between lens epithelial cells was inhibited by Ca2+-CaM [44]. The rapid onset of this inhibition (within seconds) suggested that this inhibition was mediated by the direct interaction of CaM with one or more of the lens connexins rather than by the action of a CaM-dependent protein kinase. They subsequently demonstrated in Cx43-transfected HeLa cells that cell-to-cell dye transfer was inhibited by Ca2+-CaM [45]. Cell-to-cell communication was half-maximally inhibited at ~ 300 nM [Ca2+]i [46], and this inhibition was prevented by pre-incubation of lens cultures with CaM antagonists [47]. In HeLa cells transiently expressing the CaM-binding-deficient mutants (Cx43K146E,R148E-EYFP and Cx43M147Q,L151E,I156E-EYFP), elevated [Ca2+]i was unable to inhibit cell-cell dye transfer, confirming that residues 136–158 in the intracellular loop of Cx43 contain the CaM-binding site that mediates the Ca2+-dependent regulation of Cx43 gap junctions [48]. Direct gap junction conductance (Gj) measurements confirmed that increases in [Ca2+ ]i and decreases in Cx43 Gj were temporally correlated and inhibitable by CaM antagonists like calmidazolium, CaMKII CaMi peptide, or the Cx43p136–158 peptide (Fig.2) [49]. We have further shown that inhibition of gap junction conductance by intracellular Ca2+ and CaM can be reversed by 90% with the addition of 10 mM EGTA and removal of external CaCl2 from the bath saline solution, but only if Ca2+-chelation commenced prior to complete uncoupling [49]. On the other hand, Cx40-transfected N2a neuroblastoma cell pairs were not uncoupled by intracellular Ca2+, such lack of intracellular Ca2+ regulation of Cx40 was consistent with the observation that Cx40 does not contain the putative CaM binding site in the same cytosolic loop region [49].
Figure 2. Ca2+/CaM regulation of Cx43 gap junctions.
A. Separate fura-2 and two whole cell patch clamp experimental measurements of [Ca2+]i and normalized gap junction conductance (Gj) in Cx43-N2a cells illustrate the temporal decrease in Gj (upper panel) and increase in [Ca2+]i (lower panel) during 1 µM ionomycin perfusion (1 ml/min). B. Pretreatment with 2 µM calmidazolium prevented the Ca2+-ionomycin induced reduction in Cx43 Gj. Inclusion of inhibitory CaM binding peptides Cx43p136–158 (C) or CaMKII peptide (D) in both patch pipettes at the indicated concentrations (1–2 Kd for CaM binding) also prevented the Ca2+-ionomycin induced reduction in Cx43 Gj. Adapted from Xu et al. [51] with Author’s permission from the American Physiological Society.
The inhibition of Cx44-mediated cell-to-cell dye transfer by Ca2+ was also shown to be regulated by CaM as this inhibition was prevented by prior incubation of Cx44-transfected cells with a CaM antagonist [50]. Cx50 was also shown to colocalize with CaM [51], and by measuring gap junctional conductance of Cx50-transfected N2a neuroblastoma cells, the reduction of Cx50-mediated junctional coupling was shown to be Ca2+-dependent. Junctional coupling mediated by either Cx50 or Cx43 was prevented by pre-incubation of transfected cells with a CaM inhibitor, indicating that the [Ca2+]i-dependent inhibition of Cx50 and Cx43 was CaM mediated. Furthermore, the Ca2+-dependent inhibition of gap junction permeability in Cx50-transfected cells was prevented by intracellular injection of a synthetic peptide encompassing the CaM binding domain of Cx50, while the scrambled peptide was without effect [52].
CaM regulation of the γ-connexin Cx45 was reported by Peracchia and coworkers by monitoring the sensitivity of Cx45 channels to CO2, and inhibiting CaM expression in oocytes [53]. Mouse Cx36, perch Cx35 and Cx34.7 that form electrical synapses have also been reported to bind CaM in a Ca2+-dependent manner using surface plasmon resonance assays and GST fusion proteins harboring the carboxyl-domain of these connexins [54, 55].
3. Molecular Basis of CaM Regulation of Connexins
There are several challenges in developing an understanding of the molecular mechanism by which CaM regulates the functional activity of connexins. Thus, the crystallization of these membrane proteins for determination of their high resolution atomic structure by X-ray crystallography is especially challenging. At this time, the structure of only one connexin, Cx26 in a presumed open state, has been resolved to 3.5 Å resolution [56, 57], although lower resolution electron microscopy crystallographic structures are available [58]. However, the intracellular loop and carboxyl terminus are largely “invisible” in these structures due to intrinsic disorder in these domains. In addition, in whole cell experiments, it is difficult to unambiguously distinguish between the effects of Ca2+ directly on the channel and the effects of Ca2+ mediated via CaM. CaM is a versatile and mobile trigger protein with many target proteins and is involved in both its direct and indirect regulations of its target proteins. To overcome these challenges, peptide models are often used to understand the molecular mechanism by which CaM interacts with its target proteins. Typically a number of questions are posed that include identification of the location(s) in the target proteins that bind CaM, whether the CaM binding to its target proteins is Ca2+-dependent or -independent, the consequences of the Ca2+ sensitivity of CaM on its interaction with its target protein, and the CaM binding modes by which it interacts with its target proteins.
3.1 CaM binds to Cx32 in β-connexin subfamily
Equilibrium binding studies using a fluorescent CaM derivative identified two CaM-binding domains in Cx32, a site in the N-terminal tail, and a site in the C-terminal tail region[59]. A peptide encompassing the N-terminal domain of Cx32 was shown to bind a fluorescent CaM derivative in a Ca2+-dependent manner with high affinity (Kd, 27 nM). A peptide encompassing a C-terminal region of Cx32 was shown to bind this fluorescent CaM derivative with a Kd of 1.2 uM [59]. The CaM binding domains of the N- and C-terminal regions of Cx32 were best defined as residues 1–21 (Cx32NT) and 216–227 (Cx32CT), respectively with the Cx32CT CaM binding region showing α-helical propensity indicating that CaM binding induced an α-helical structure in the Cx32CT and involved both the N- and C-lobes of this connexin. These data, showing separate functions of the N- and C-lobes of CaM in its interactions with Cx32, suggests trans-domain or trans-subunit bridging by CaM as a possible mechanism of gap junction gating [60]. Interestingly, both the N-terminal and the C-terminal CaM-binding domains of Cx32 are located close to the membrane and are contiguous with hydrophobic membrane-spanning sequences (Fig.1). Furthermore, although the C-terminal CaM binding domain in Cx32 represents only 15% of the C-terminal tail of Cx32, the removal of the other 85% had little effect on junctional permeability and chemical gating [61–63]. Recently, Stauch et al. further demonstrated that CaM binding could convert the intrinsically disordered C-terminal domain to an α-helical conformation that may enable Cx32 to interact with the protein partner synapse-associated protein 97 [64].
3.2 CaM binds to the γ-connexin subfamily
Using surface plasmon resonance with rapid kinetics, CaM was shown to bind in a concentration- and Ca2+-dependent manner to peptide fragments corresponding to a 10–30 amino acid segment at the beginning of the C-terminal intracellular domain of mouse Cx36, Cx35 from perch, and the related perch Cx34.7 [55]. Dissociation was also very rapid; Kds for CaM binding at a high-affinity site ranged from 11 to 72 nM (Table 1). No binding of CaM to the intracellular loops of these connexins was observed. The micromolar K½s, and the rapid on and off rates suggest that this interaction may change dynamically in neurons, and may occur transiently when [Ca2+]i is elevated to a level that would occur in the near vicinity of an activated synapse [54, 55].
Table 1.
CaM-binding affinities of peptides derived from different subfamilies of connexins.
| Peptide | Family | Position | Predicted Scorea |
Helicityb |
Kd (nM) |
Reference | |
|---|---|---|---|---|---|---|---|
| Ca2+ | EGTA | ||||||
| hCx43p138–157 | α | CL | 16 | 55.6 | 860 | nd | [48] |
| shCx44p129–150 | α | CL | 6 | 59.5 | 49 | >5000 | [50] |
| hCx50p141–166 | α | CL | 13 | 95.3 | 4.9 | >8000 | [52] |
| hCx32p1–21 | β | NT | 9 | nd | 27 | nd | [60] |
| rCx32p216–230 | β | CT | 0 | nd | 2100 | nd | [60] |
| mCx36p269–321 | γ | CT | 14 | nd | 11 | nd | [55] |
Predicted by CaM Target Database
Peptide helicity induced by 90% TFE
3.3 CaM binds to the α-connexin subfamily
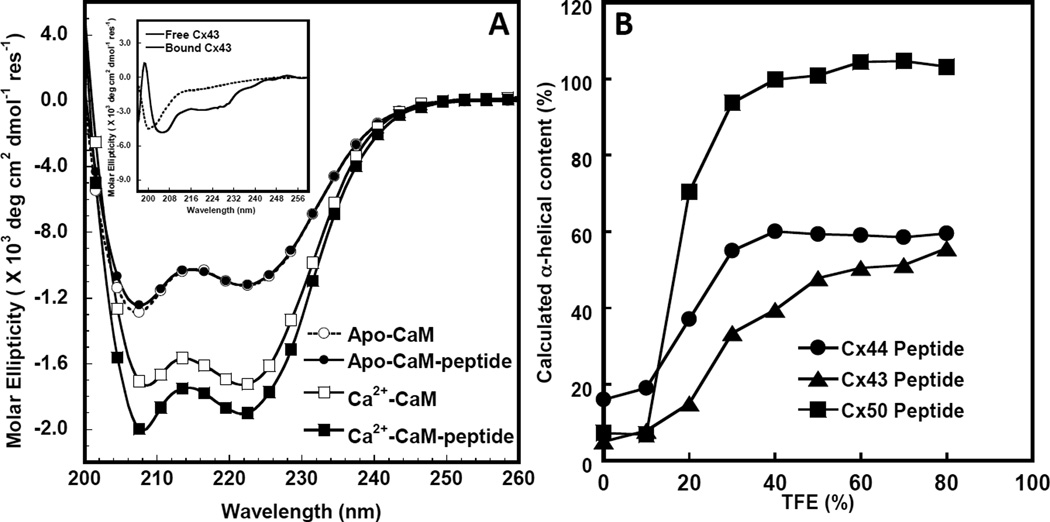
We have predicted that a CaM binding region resides in the central cytoplasmic loop of α-connexins that include Cx43 [48], Cx44 (the ovine analog of rodent Cx46) [50] and Cx50 [52, 65]. We used various biochemical approaches including circular dichroism (CD) nuclear magnetic resonance (NMR), isothermal titration calorimetry (ITC) and fluorescence spectroscopy to demonstrate that CaM binds to the cytoplasmic loop of Cx43 [48], Cx44 [50] and Cx50 [52] in a Ca2+-dependent manner. Figure 3A shows the far UV CD spectra of CaM in the absence and presence of a Cx43 cytoplasmic loop peptide. The addition of this Cx43 peptide increased the α-helical content likely due to the conversion of unstructured free peptide to helical conformation upon complex formation with CaM. On the other hand, based on the use of peptide models, no direct interaction was observed for other regions in this connexin such as the N-terminal region of Cx43 [59] and the C-terminal domain of Cx43 [66].
Figure 3. Circular dichroism studies of the interaction between Cx43p136–158 and Apo/Holo CaM.
A. Far UV circular dichroism spectrum of CaM in the presence of 1 mM EGTA (white circle) or 1 mM CaCl2 (white square) and a 1:1 CaM/synthetic peptide mixture with 1 mM EGTA (black circle) or 1 mM CaCl2 (black square) after subtracting the contribution from the buffer. Buffer conditions: 10 mM Tris, 100 mM KCl at pH=7.4 at room temperature. The insetdemonstratesthe far UVcircular dichroism spectra of Cx43p136–158 (dashed line) and the calculated difference spectrum (solid line) by subtracting the spectrum of Ca2+-CaM from that of the Ca2+-CaM-Cx43p136–158 mixture with 1 mM Ca2+ in a buffer consisting of 100 mM KCl, 10 mM Tris-HCl, at pH=7.4. B. TFE induced α-helical content increase of Cx43 (triangle), Cx44 (circle) and Cx50 (square) peptides corresponding to CaM-binding sites.
Studies in our laboratories identified CaM binding domains in Cx43 (residues 136–158) [48], Cx44 (residues 132–153) [50] and Cx50 (residues 141–166) [52]. NMR studies demonstrated all three Cx43-, Cx44- or Cx50-derived peptides (Cx43p136–158, residues from 136 to 158; Cx44p132–153, residues from 132 to 153; Cx50p141–166, residues from 141 to 166) bind CaM with a 1:1 stoichiometry. Such interactions of CaM with Cx43- or Cx50-derived peptides can also be observed using surface plasmon resonance and florescence anisotropy. Dansylated CaM florescence and high-resolution NMR studies demonstrated that the binding of a Cx43-derived peptide Cx43p136–158 to CaM in the presence but not the absence of Ca2+. The apparent dissociation constant binding of this peptide to CaM in physiologic K+ was 860 ± 20 nM [48]. On the other hand, the respective Kds for the peptides derived from Cx44 (Cx44p132–153) and Cx50 (Cx50p141–166) are 49 ± 3.0 and 4.9 ± 0.6 nM respectively in the presence of Ca2+. In the absence of Ca2+, both peptides bound to CaM with significantly reduced affinities (Kds greater than 5000 and 8000 nM, respectively). Thus, Cx50p141–166 exhibited a 10-fold higher binding affinity for CaM than Cx44p132–153, which was approximately 20-fold higher than Cx43p136–158.
Since a “ball-and-chain” or “particle-receptor” hypothesis involving the pH-dependent intramolecular interaction between the cytoplasmic domain and part of the intracellular loop of Cx43 has been proposed to explain the low pH-induced closure of gap junction [66, 67], we have also determined the CaM binding affinity of the Cx44 and Cx50 peptides to CaM as a function of pH. Both Cx44p132–153 and Cx50p141–166 peptides behaved similarly when pH was varied with binding affinities decreasing at pH values lower than 5.5 and greater than 8.5 [50]. However, over the pH range 5.5–8.5, CaM exhibits affinities for both peptides at nanomolar or submicromolar range. CaM binding to the peptide Cx50p141–166 was ~ 1 order of magnitude stronger than to the peptide Cx44p132–153 likely due to difference in electrostatic interactions.
It is interesting to note that Ca2+-CaM may directly or indirectly affect cytoplasmic loop/C tail interactions in a Cx43 hemichannel, either through its binding to the cytoplasmic loop of Cx43 which corresponds to amino acids 119–144 in the C-terminal portion of the cytoplasmic loop of Cx43, or through a Ca2+-dependent phosphorylation/dephosphorylation processes [29, 68, 69]. It is also possible that phosphorylation or other regulatory mechanisms such as pH could modulate this action of Ca2+-CaM. Our results provide direct evidence that CaM binds to a specific region of the intracellular loop region of the gap junction proteins Cx43, Cx44 and Cx50 in a Ca2+-dependent manner. Our data suggest a common mechanism by which the Ca2+-dependent inhibition of the α-class of gap junction proteins is mediated by the direct association of an intracellular loop region of these proteins with Ca2+-CaM.
3.4 Key factors contributing to the CaM binding affinity of the α-subfamily of connexins
Our results have also revealed that the α-helicity of the CaM binding peptide derived from connexin is an important factor in predicting their CaM binding affinities (Table 1) although the binding affinity of the full length connexin is yet available. The difference in CaM binding affinity of the Cx50 and Cx44 peptides may originate from intrinsic sequences encoded in the CaM binding regions of these different connexins especially at conserved residue positions 1 and 5 (Fig.1); Cx43 peptide contains two flexible Glys instead of the α-helical forming residues present in Cx44 and Cx50 (Fig.1). Such sequence variations, that affect the ability of these peptides to form α-helices, are likely important in determining their ability to interact with CaM [65]. In aqueous buffer, these peptides are largely unstructured in the absence of CaM. 2,2,2-Trifluoroethanol (TFE) is an organic solvent which when added to aqueous buffer can reveal the intrinsic helical propensity of a peptide [70]. We have shown that all these peptides form α-helical structures in the presence of TFE that follows the rank order Cx50p141–166> Cx44p132–153 ~ Cx43p136–158 (Fig.3B). The higher CaM binding affinity of Cx44 versus that of Cx43 may also be due in part to the relative helical content of these two peptides in solution indicating that the intrinsic helicity of the peptide may also contribute to this binding. The negatively charged E151 located close to R149 in Cx50 may stabilize the α-helical conformation of this peptide, while Cx43 with a positive charged R at the same location may destabilize the α-helix conformation of this connexin [48, 52].
There are several positively charged residues such as K147, R149 and R156 in Cx50 that are conserved in the α-connexin subfamily members Cx43, Cx44 and Cx50. Indeed, the observed pH dependence of CaM binding affinity to the cytosolic loop peptides of Cx44 and Cx50 suggests electrostatic interactions following the protonation of Asp and Glu, and the deprotonation of Lys, are important for the connexin-CaM complex formation [50, 52].
It is also noteworthy that the binding affinity of CaM to the connexin-derived CaM binding peptides derived from the cytosolic region of three α-subfamily connexins does not agree with their predicted CaM binding scores based on the CaM Target Database [65]. These predicted scores are based on parameters including average hydrophobicity, average hydrophobic moment, and average propensity for α-helix formation and are not intended to define the CaM binding affinity for the complex formation [65]. It is interesting to note that the reported CaM binding site in the N-terminal region of Cx32 (residue 1–21) is predicted to have a high score by the CaM target database. This region is likely α-helical since it is homologous to the NTH motif (N-terminal helix) in the X-ray-determined structure of Cx26 [57]; it is believed to form the channel “plug” in the β-subfamily of connexins [71]. However, the reported C-terminal CaM binding region of Cx32 is not predicted by the CaM target database and is largely undefined in the determined Cx26 X-ray-derived structure. Therefore it is important to perform detailed experimental studies to define the actual CaM binding sites in its target proteins especially for membrane channel proteins such as the connexins.
3.5 CaM binding modes to the α-connexin subfamily using a peptide model
One of the key aspects to understand the molecular mechanism by which CaM interacts with its target proteins is the CaM binding modes, by which it interacts with its target proteins. Currently, over 200 structures of CaM complexed with its receptors are reported in the protein data bank. CaM exhibits different target recognition modes that can be classified into two general binding styles, namely extended and collapsed [48]. In the extended mode, CaM has different recognition modes when it binds to different target proteins such as the IQ motif of myosin V Ca2+ channels [48], anthrax toxin [50], SK channels (Small conductance Ca2+-activated potassium channels) [52], Ca2+ pump [40], and glutamate decarboxylase [39]. In the collapsed binding style, CaM can be further divided into different binding modes depending on the spaces between conserved hydrophobic residues at the CaM binding regions. For example, CaM binds to N-methyl-D-aspartate receptors (NMDARs) in the C1 region of its NR1 subunit in a 1–7 binding mode [72]. A 1–10 binding mode has been reported for CaM binding to CaM Kinase II α [73] and the Ca2+ channel CaV1.2 [74]. A 1–5-10 CaM binding mode was observed for the CaM complex with myristoylated alanine-rich C-kinase substrate (MARCKS) and CaMKII [75]. A 1–14 binding mode was observed in the CaM complex with cyclic nucleotide-gated ion channels (CNG) [76], Endothelial nitric-oxide synthase (eNOS) [6], CaM kinase I [7], a myosin light chain kinase that is exclusively expressed in adult skeletal muscle (skMLCK) [8], and metabotropic glutamate receptors (mGluR7a) [9]. Ca2+/CaM-dependent protein kinase kinase (CaMKK) [7] has a 1–16 binding mode. The ryanodine receptor 1 (RyR1) [10] has 1–17 binding mode. The CaM binding regions of such CaM target proteins generally have a helical conformation on complex formation with CaM.
We have also used peptide models to understand the structural mechanisms driving CaM regulation of connexins with the advantage of obtaining site specific molecular recognition information. Several of our studies using various spectroscopic methods suggest that CaM binds to Cx50p141–166, Cx44p132–153, orCx43p136–158 with a 1–5-10 collapsed CaM binding mode as observed in the CaM-CaMKII complex. All three peptides are largely unstructured in aqueous buffer in the absence of CaM. Far UV circular dichroism studies have indicated that these CaM binding peptides likely become more helical upon binding CaM. Fluorescence spectroscopy revealed conformational changes of both the peptide and CaM following formation of the CaM:connexin-derived-peptide complex. Pulse-field gradient NMR studies demonstrated the hydrodynamic radius of CaM (22.6 ± 0.6 Å) was decreased by 23% upon formation of the CaM-Cx50p141–166 complexes. The determined size of the Ca2+/CaM–Cx50p141–166 complex of 17.3 ± 0.6 Å is comparable to the hydrodynamic radii of CaM in complex with CaM-CaMKIIα (PDB code 1CDM) [77]. Furthermore, chemical shift changes of 15N-labeled CaM have been used to show that these three connexin peptides are able to induce structural changes in both the N- and C-terminal domains of CaM as well as in the linker region. Using small angle X-ray scattering, Myllykoski et al. also observed that on binding a Cx43 cytoplasmic loop peptide (residues 144–158), CaM adopted a more globular conformation and collapsed the target [78]. In summary, these studies suggest that CaM likely embraces the peptides of α-connexins in a collapsed structure that involves the unwinding of the CaM central helix.
Because of its abundance, the mammalian water-channel aquaporin-0 (AQP0, or MIP26), was long considered to be the gap junction protein in the mammalian lens [79]. Like the connexins, aquaporin-0 channels were shown to close when bound with Ca/CaM [80–86]. More recent biochemical and NMR studies suggest that one CaM binds two AQP0 CBD peptides in a stepwise manner and two CaM molecules bind to a single AQP0 tetramer [83]. The 3D computational reconstruction of full-length AQP0-CaM complex based on the electron microscopy map and the determined X-ray structure of the plant glutamate decarboxylase (ptGAD)-CaM complex [87] suggest that two Ca2+-CaM are located directly under two of the AQP0 subunits and bind to two adjacent antiparallel C-terminal helices from two subunits of AQP0. This CaM binding mode to the water channel aquaporin-0 (AQP0) with a ratio of 1:2 is different from our observed CaM binding mode of the α-connexin subfamily which using peptide models have a 1:1 binding ratio. It would be interesting to observe how CaM is able to regulate the full length connexins using the same action mode. However, in the fully assembled gap junction, these connexin cytoplasmic loop CaM binding domains likely form a six-sided barrel near the M3 (third transmembrane) interface that may sterically limit the number of CaM molecules that may bind to this motif. The cytoplasmic loop and C terminal domains are thought to dimerize to close Cx43 gap junctions by the pH-dependent chemical gating mechanism [88]. Conversely, the six NT domains that fold into the cytoplasmic pore of the gap junction channel that contribute to the formation of the ion permeation pathway and transjunctional voltage (Vj) sensor apparently require the “gating” of only one NT domain to induce partial closure of the channel via the fast Vj-gating mechanism [57, 89, 90]. The connexin-CaM stoichiometry of binding required for gap junction channel closure remains to be determined, but is likely to require only 1–2 connexin subunits, based on current knowledge of the fast Vj- and slow chemical gating mechanisms.
3.6 Expanding the [Ca2+]i sensing range of CaM upon binding to connexins
We have shown that when connexin peptides bind to CaM, the apparent Kds of Ca2+ for CaM decreased and the Hill Coefficients increased. Isothermal titration studies of the Cx44p132–153 peptide suggest that its interaction with CaM is an exothermic event that is both enthalpically and entropically driven in which electrostatic interactions play an important role. The binding of Cx44129–150 peptide to CaM increases the CaM intradomain cooperativity, and further enhances the Ca2+-binding affinities of the C-domain of CaM by slowing the rate of Ca2+ release from the complex [50]. CaM is also able to decrease the Ca2+ affinity of the N-lobe of Cx50p141–166-bound CaM 2-fold, whereas the Ca2+ affinity of the C-lobe increased by ~20%. CaM responds to subtle changes in [Ca2+]i over a broader range of [Ca2+]i concentrations as a result of the binding of Cx50p141–166 [52].
4.0 Proposed Ca2+-CaM gating model
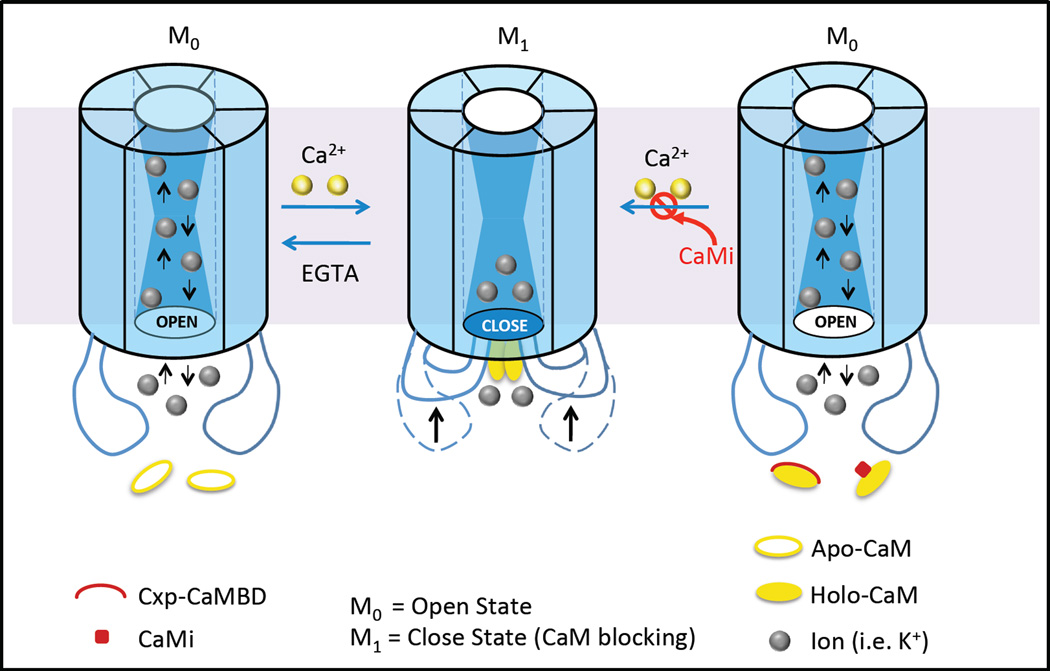
Results from our studies on the changes in Ca2+-binding kinetics, affinities, energies, dye-transfer and gap junction conductance between cells support the following working model of gap junction inhibition by Ca2+-CaM (Fig.4). Initially, an increase in [Ca2+]i is sensed by the C-domain of CaM which effects the interaction of CaM with the cytoplasmic loop of the α-connexin subfamily. This interaction enhances the efficiency and sensitivity of intracellular Ca2+ sensing by CaM because of an increase in both the Ca2+-binding affinity and intradomain cooperativity of the C-domain of CaM. The partially saturated, connexin-bound CaM might serve as an intermediate state to prevent the free diffusion of CaM into the cytoplasm. Further increase in [Ca2+]i to the near micromolar range allows the half-saturated CaM to rapidly respond to this further increase in [Ca2+]i such that it adopts a fully open conformation which is now able to inhibit gap junction mediated intercellular communication [48, 50]. Inhibition of the interaction of the cytosolic loop region with CaM by the addition of a CaM inhibitor (e.g. CDZ) or a Cx43 cytosolic loop peptide prevents the open (M0) to close (M1) state transition. The Ca2+-CaM induced gating response that closes the Cx43 gap junction is likely effected via a conformational change in the cytosolic loop of Cx43 [48]. It is also possible that, as proposed earlier by Peracchia and colleagues [91], Ca2+-CaM binds to the N and C-terminal domains of Cx32 to act as a “cork”, or it induces a conformational change in the cytosolic domain that occludes the cytosolic mouth of the gap junction channel, resulting in the restriction of the passage of current-carry ions [60, 92]. Unwin and Ennis proposed an alternative “iris” gating hypothesis that the extracellular Ca2+-induced closure of liver gap junctions is a result of a decrease in the tilt angle of six connexins within a connexon junction channel [93]. This model relied on the presence of the “bulky” phenylalanines in the third transmembrane domain (M3), the major channel lining domain. Unwin and Ennis’ idea was that the phenylalanine residues would gate the channel by obstructing its lumen, as they would be brought into the lumen by the M3 rotation. However, combined mutations of Cx32’s phenylalanine145 and phenylalanine149 residues (the most conserved phenylalanines among connexins) to valine (a much smaller hydrophobic residue) generated channels with gating sensitivities that were indistinguishable from Cx32 wild type channels [30].
Figure 4. A proposed gating model for the mechanism of the closure of Cx43 gap junction channels by direct Ca2+/CaM binding.
In this model, connexin has one open (Mo) state and open closed state (M1). The induction of a conformational change in the cytoplasmic loop (CL) of Cx43 through Ca2+/CaM binding, steric hindrance of CaM binding near the cytoplasmic opening of the transmembrane pore, or a combination of both steric hindrance and conformational gating mechanisms causes the closure of the channel. Addition of the Cx43-CL-derived peptides or a CaMi (CaM inhibitor e.g., CDZ), prevents the Ca2+/CaM-Cx43 cytoplasmic loop interaction which changes the status of the channel from closed (M1) to the open (M0) state.
In summary, due to the challenges associated with large oligomeric membrane proteins and the “invisibility” of the cytosolic loop and C-terminal domains within a X-ray-derived structures of these proteins [56, 57], the mechanism by which CaM regulates gap junction channels remains to be fully defined. However, several recent studies provide strong evidence for a direct role of CaM in the regulation of several classes of connexin gap junction channels. While CaM was reported to bind to the N and C–terminal regions of the β-subfamily Cx32, it interacts with the cytosolic loop region of three α-subfamily connexins Cx43, Cx44 and Cx50. Cx35 and Cx36, and Cx34.7 are regulated by CaM in a Ca2+-dependent manner through binding to only their C-terminal domains [55]. Furthermore, all these different connexins are regulated by CaM in a Ca2+-dependent manner. The gated closure of the α-subfamily of gap junction channels by Ca2+ is effected by Ca2+-CaM binding to connexins via an embracing CaM-binding mode with hydrophobic residues in the CaM-binding region at positions 1, 5, and 10. Electrostatic and hydrophobic interactions play an indispensable role in the intrinsic α-helicity of the CaM binding domains of α-subfamily connexins. Future studies are needed to address the stoichiometry of CaM binding to the full length connexin proteins, the intracellular Ca2+ affinity, and the connexin/CaM induced conformational changes required to induce the chemical gating mechanism. Furthermore, innovative approaches are required to reveal the novel mode of CaM action at the membrane interface via its association with flexible intracellular connexin regions resulting in the large conformational changes required for regulating the function of these membrane channels [94]. Since several spontaneous mutations have been identified in the CaM binding site of the cytosolic loop of Cx43 (such as in oculodentodigital dysplasia), understanding the mechanisms by which intracellular Ca2+-CaM regulates gap junctions is an essential step towards revealing the molecular bases of such connexin-linked diseases.
Acknowledgement
We thank Gina Sosinsky, and Yubin Zhou for their helpful discussions and Kathy Meenach for her help. Juan Zou is a fellow of Brain and Behavior Program at Georgia State University. This work was supported, in part, by NIH grants EY-05684 to CFL and JJY, HL-042220 to RDV and JJY, and GM-081749 to JJY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Klevit RE, Dalgarno DC, Levine BA, Williams RJ. 1H-NMR studies of calmodulin. The nature of the Ca2+-dependent conformational change. Eur J Biochem. 1984;139:109–114. doi: 10.1111/j.1432-1033.1984.tb07983.x. [DOI] [PubMed] [Google Scholar]
- 2.Wang CL. A note on Ca2+ binding to calmodulin. Biochem Biophys Res Commun. 1985;130:426–430. doi: 10.1016/0006-291x(85)90434-6. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharya S, Bunick CG, Chazin WJ. Target selectivity in EF-hand calcium binding proteins. Biochim Biophys Acta. 2004;1742:69–79. doi: 10.1016/j.bbamcr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Kawasaki H, Kretsinger RH. Calcium-binding proteins 1: EF-hands. Protein Profile. 1995;2:297–490. [PubMed] [Google Scholar]
- 5.Ikura M, Ames JB. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc Natl Acad Sci U S A. 2006;103:1159–1164. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu YT, Molday RS. Modulation of the cGMP-gated channel of rod photoreceptor cells by calmodulin. Nature. 1993;361:76–79. doi: 10.1038/361076a0. [DOI] [PubMed] [Google Scholar]
- 7.Ehlers MD, Zhang S, Bernhadt JP, Huganir RL. Inactivation of NMDA receptors by direct interaction of calmodulin with the NR1 subunit. Cell. 1996;84:745–755. doi: 10.1016/s0092-8674(00)81052-1. [DOI] [PubMed] [Google Scholar]
- 8.Rodney GG, Williams BY, Strasburg GM, Beckingham K, Hamilton SL. Regulation of RYR1 activity by Ca2+ and calmodulin. Biochemistry. 2000;39:7807–7812. doi: 10.1021/bi0005660. [DOI] [PubMed] [Google Scholar]
- 9.Xia XM, et al. Mechanism of calcium gating in small-conductance calcium-activated potassium channels. Nature. 1998;395:503–507. doi: 10.1038/26758. [DOI] [PubMed] [Google Scholar]
- 10.Fanger CM, et al. Calmodulin mediates calcium-dependent activation of the intermediate conductance KCa channel, IKCa1. J Biol Chem. 1999;274:5746–5754. doi: 10.1074/jbc.274.9.5746. [DOI] [PubMed] [Google Scholar]
- 11.Tang J, Lin Y, Zhang Z, Tikunova S, Birnbaumer L, Zhu MX. Identification of common binding sites for calmodulin and inositol 1,4,5-trisphosphate receptors on the carboxyl termini of trp channels. J Biol Chem. 2001;276:21303–21310. doi: 10.1074/jbc.M102316200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuhlke RD, Reuter H. Ca2+-sensitive inactivation of L-type Ca2+ channels depends on multiple cytoplasmic amino acid sequences of the alpha1C subunit. Proc Natl Acad Sci U S A. 1998;95:3287–3294. doi: 10.1073/pnas.95.6.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schonherr R, Lober K, Heinemann SH. Inhibition of human ether a go-go potassium channels by Ca2+/calmodulin. EMBO J. 2000;19:3263–3271. doi: 10.1093/emboj/19.13.3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bers DM, Grandi E. Calcium/calmodulin-dependent kinase II regulation of cardiac ion channels. J Cardiovasc Pharmacol. 2009;54:180–187. doi: 10.1097/FJC.0b013e3181a25078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai S, Hall DD, Hell JW. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol Rev. 2009;89:411–452. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tadross MR, Dick IE, Yue DT. Mechanism of local and global Ca2+ sensing by calmodulin in complex with a Ca2+ channel. Cell. 2008;133:1228–1240. doi: 10.1016/j.cell.2008.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sohl G, Willecke K. Gap junctions and the connexin protein family. Cardiovasc Res. 2004;62:228–232. doi: 10.1016/j.cardiores.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Eiberger J, Degen J, Romualdi A, Deutsch U, Willecke K, Sohl G. Connexin genes in the mouse and human genome. Cell Commun Adhes. 2001;8:163–165. doi: 10.3109/15419060109080717. [DOI] [PubMed] [Google Scholar]
- 19.Nielsen MS, Nygaard Axelsen L, Sorgen PL, Verma V, Delmar M, Holstein-Rathlou NH. Gap junctions. Compr Physiol. 2012;2:1981–2035. doi: 10.1002/cphy.c110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levin M. Gap junctional communication in morphogenesis. Prog Biophys Mol Biol. 2007;94:186–206. doi: 10.1016/j.pbiomolbio.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saez JC, Berthoud VM, Branes MC, Martinez AD, Beyer EC. Plasma membrane channels formed by connexins: their regulation and functions. Physiol Rev. 2003;83:1359–1400. doi: 10.1152/physrev.00007.2003. [DOI] [PubMed] [Google Scholar]
- 22.Sarieddine MZ, Scheckenbach KE, Foglia B, Maass K, Garcia I, Kwak BR, Chanson M. Connexin43 modulates neutrophil recruitment to the lung. J Cell Mol Med. 2009;13:4560–4570. doi: 10.1111/j.1582-4934.2008.00654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruger O, et al. Defective vascular development in connexin 45-deficient mice. Development. 2000;127:4179–4193. doi: 10.1242/dev.127.19.4179. [DOI] [PubMed] [Google Scholar]
- 24.Kumai M, Nishii K, Nakamura K, Takeda N, Suzuki M, Shibata Y. Loss of connexin45 causes a cushion defect in early cardiogenesis. Development. 2000;127:3501–3512. doi: 10.1242/dev.127.16.3501. [DOI] [PubMed] [Google Scholar]
- 25.Reaume AG, et al. Cardiac malformation in neonatal mice lacking connexin43. Science. 1995;267:1831–1834. doi: 10.1126/science.7892609. [DOI] [PubMed] [Google Scholar]
- 26.Lai-Cheong JE, Arita K, McGrath JA. Genetic diseases of junctions. J Invest Dermatol. 2007;127:2713–2725. doi: 10.1038/sj.jid.5700727. [DOI] [PubMed] [Google Scholar]
- 27.Laird DW. Closing the gap on autosomal dominant connexin-26 and connexin-43 mutants linked to human disease. J Biol Chem. 2008;283:2997–3001. doi: 10.1074/jbc.R700041200. [DOI] [PubMed] [Google Scholar]
- 28.Del Corsso C, Iglesias R, Zoidl G, Dermietzel R, Spray DC. Calmodulin dependent protein kinase increases conductance at gap junctions formed by the neuronal gap junction protein connexin36. Brain Res. 2012;1487:69–77. doi: 10.1016/j.brainres.2012.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bennett MV, Barrio LC, Bargiello TA, Spray DC, Hertzberg E, Saez JC. Gap junctions: new tools, new answers, new questions. Neuron. 1991;6:305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- 30.Peracchia C. Chemical gating of gap junction channels; roles of calcium, pH and calmodulin. Biochim Biophys Acta. 2004;1662:61–80. doi: 10.1016/j.bbamem.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 31.Deleze J. Electrophysiology of the Heart. In Calcium ions and the healing-over in heart fibers ed. Elmsford, New York: Pergamon Press; 1965. pp. 147–148. [Google Scholar]
- 32.Rose B, Simpson I, Lowenstein WR. Calcium ion produces graded changes in permeability of membrane channels in cell junctions. Nature (London) 1977;267:625–627. doi: 10.1038/267625a0. [DOI] [PubMed] [Google Scholar]
- 33.Cheung WG. Calmodulin plays a pivotal role in cellular regulation. Science. 1980;207:19–27. doi: 10.1126/science.6243188. [DOI] [PubMed] [Google Scholar]
- 34.Peracchia C, Bernardini G, Peracchia LL. Is calmodulin involved in the regulation of gap junction permeability? Pflugers Arch. 1983;399:152–154. doi: 10.1007/BF00663912. [DOI] [PubMed] [Google Scholar]
- 35.Peracchia C, Bernardini G. Gap junction structure and cell-to-cell coupling regulation: is there a calmodulin involvement? Fed Proc. 1984;43:2681–2691. [PubMed] [Google Scholar]
- 36.Peracchia C. Communicating junctions and calmodulin: inhibition of electrical uncoupling in Xenopus embryo by calmidazolium. J. Membr. Biol. 1984;81:49–58. doi: 10.1007/BF01868809. [DOI] [PubMed] [Google Scholar]
- 37.Peracchia C. Calmodulin-like proteins and communicating junctions. Electrical uncoupling of crayfish septate axons is inhibited by the calmodulin inhibitor W7 and is not affected by cyclic nucleotides. Pflugers Arch. EurJPhysiol. 1987;408:379–385. doi: 10.1007/BF00581132. [DOI] [PubMed] [Google Scholar]
- 38.Hertzberg EL, Gilula NB. Liver gap junctions and lens fiber junctions: comparative analysis and calmodulin interaction. Cold Spring Harb Symp Quant Biol 46 Pt. 1982;2:639–645. doi: 10.1101/sqb.1982.046.01.060. [DOI] [PubMed] [Google Scholar]
- 39.Van Eldik LJ, Hertzberg EL, Berdan RC, Gilula NB. Interaction of calmodulin and other calcium-modulated proteins with mammalian and arthropod junctional membrane proteins. Biochem Biophys Res Commun. 1985;126:825–832. doi: 10.1016/0006-291x(85)90259-1. [DOI] [PubMed] [Google Scholar]
- 40.Zimmer DB, Green CR, Evans WH, Gilula NB. Topological analysis of the major protein in isolated intact rat liver gap junctions and gap junction-derived single membrane structures. J Biol Chem. 1987;262:7751–7763. [PubMed] [Google Scholar]
- 41.Peracchia C, Wang X, Li L, Peracchia LL. Inhibition of calmodulin expression prevents low-pH-induced gap junction uncoupling in Xenopus oocytes. Pflugers Arch. 1996;431:379–387. doi: 10.1007/BF02207275. [DOI] [PubMed] [Google Scholar]
- 42.Sotkis A, Wang XG, Yasumura T, Peracchia LL, Persechini A, Rash JE, Peracchia C. Calmodulin colocalizes with connexins and plays a direct role in gap junction channel gating. Cell Commun Adhes. 2001;8:277–281. doi: 10.3109/15419060109080737. [DOI] [PubMed] [Google Scholar]
- 43.Blodow A, Ngezahayo A, Ernst A, Kolb HA. Calmodulin antagonists suppress gap junction coupling in isolated Hensen cells of the guinea pig cochlea. Pflugers Arch. 2003;446:36–41. doi: 10.1007/s00424-002-1004-9. [DOI] [PubMed] [Google Scholar]
- 44.Lurtz MM, Louis CF. Calmodulin and protein kinase C regulate gap junctional coupling in lens epithelial cells. Am J Physiol Cell Physiol. 2003;285:C1475–C1482. doi: 10.1152/ajpcell.00361.2002. [DOI] [PubMed] [Google Scholar]
- 45.Lurtz MM, Louis CF. Intracellular calcium regulation of connexin43. Am J Physiol Cell Physiol. 2007;293:C1806–C1813. doi: 10.1152/ajpcell.00630.2006. [DOI] [PubMed] [Google Scholar]
- 46.Churchill GC, Lurtz MM, Louis CF. Ca2+ regulation of gap junctional coupling in lens epithelial cells. AmJPhysiol. Cell Physiol. 2001;281:C972–C981. doi: 10.1152/ajpcell.2001.281.3.C972. [DOI] [PubMed] [Google Scholar]
- 47.Lurtz MM, Louis CF. Calmodulin and Protein Kinase C regulate gap junctional coupling in lens epithelial cells. AmJPhysiol. Cell. Physiol. 2003;285:C1475–C1482. doi: 10.1152/ajpcell.00361.2002. [DOI] [PubMed] [Google Scholar]
- 48.Zhou Y, et al. Identification of the calmodulin binding domain of connexin 43. J Biol Chem. 2007;282:35005–35017. doi: 10.1074/jbc.M707728200. [DOI] [PubMed] [Google Scholar]
- 49.Xu Q, Kopp RF, Chen Y, Yang JJ, Roe MW, Veenstra RD. Gating of connexin 43 gap junctions by a cytoplasmic loop calmodulin binding domain. Am J Physiol Cell Physiol. 2012;302:C1548–C1556. doi: 10.1152/ajpcell.00319.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou Y, Yang W, Lurtz MM, Chen Y, Jiang J, Huang Y, Louis CF, Yang JJ. Calmodulin mediates the Ca2+-dependent regulation of Cx44 gap junctions. Biophys J. 2009;96:2832–2848. doi: 10.1016/j.bpj.2008.12.3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Qi Y. Role of intramolecular interaction in connexin50: mediating the Ca2+-dependent binding of calmodulin to gap junction. Arch Biochem Biophys. 2005;440:111–117. doi: 10.1016/j.abb.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 52.Chen Y, et al. Molecular interaction and functional regulation of connexin50 gap junctions by calmodulin. Biochem J. 2011;435:711–722. doi: 10.1042/BJ20101726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peracchia C, Young KC, Wang XG, Peracchia LL. Is the voltage gate of connexins CO2-sensitive? Cx45 channels and inhibition of calmodulin expression. J Membr Biol. 2003;195:53–62. doi: 10.1007/s00232-003-2044-6. [DOI] [PubMed] [Google Scholar]
- 54.O'Brien J, Bruzzone R, White TW, Al-Ubaidi MR, Ripps H. Cloning and expression of two related connexins from the perch retina define a distinct subgroup of the connexin family. J Neurosci. 1998;18:7625–7637. doi: 10.1523/JNEUROSCI.18-19-07625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burr GS, Mitchell CK, Keflemariam YJ, Heidelberger R, O'Brien J. Calcium-dependent binding of calmodulin to neuronal gap junction proteins. Biochem Biophys Res Commun. 2005;335:1191–1198. doi: 10.1016/j.bbrc.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suga M, Maeda S, Nakagawa S, Yamashita E, Tsukihara T. A description of the structural determination procedures of a gap junction channel at 3.5 A resolution. Acta Crystallogr D Biol Crystallogr. 2009;65:758–766. doi: 10.1107/S0907444909014711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]
- 58.Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Three-dimensional structure of a human connexin26 gap junction channel reveals a plug in the vestibule. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:10034–10039. doi: 10.1073/pnas.0703704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Torok K, Stauffer K, Evans WH. Connexin 32 of gap junctions contains two cytoplasmic calmodulin-binding domains. Biochem J. 1997;326(Pt 2):479–483. doi: 10.1042/bj3260479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dodd R, Peracchia C, Stolady D, Torok K. Calmodulin association with connexin32-derived peptides suggests trans-domain interaction in chemical gating of gap junction channels. J Biol Chem. 2008;283:26911–26920. doi: 10.1074/jbc.M801434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Li L, Peracchia LL, Peracchia C. Chimeric evidence for a role of the connexin cytoplasmic loop in gap junction channel gating. Pflugers Arch. EurJPhysiol. 1996:844–852. doi: 10.1007/s004240050076. [DOI] [PubMed] [Google Scholar]
- 62.Werner R, Levine E, Rabadan-Diehl C, Dahl G. Gating properties of connexin32 cell-cell channels and their mutants expressed in Xenopus oocytes. Proc. Biol. Sci. 1991;243:5–11. doi: 10.1098/rspb.1991.0002. [DOI] [PubMed] [Google Scholar]
- 63.Wang X, Peracchia C. Positive charges of the initial C-terminus domain of Cx32 inhibit gap junction gating sensitivity to CO2. BiophysJ. 1997;73:C1743–C1749. doi: 10.1016/S0006-3495(97)78112-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stauch K, Kieken F, Sorgen P. Characterization of the structure and intermolecular interactions between the connexin 32 carboxyl-terminal domain and the protein partners synapse-associated protein 97 and calmodulin. J Biol Chem. 2012;287:27771–27788. doi: 10.1074/jbc.M112.382572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. Calmodulin target database. J Struct Funct Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 66.Duffy HS, Sorgen PL, Girvin ME, O'Donnell P, Coombs W, Taffet SM, Delmar M, Spray DC. pH-dependent intramolecular binding and structure involving Cx43 cytoplasmic domains. J Biol Chem. 2002;277:36706–36714. doi: 10.1074/jbc.M207016200. [DOI] [PubMed] [Google Scholar]
- 67.Morley GE, Taffet SM, Delmar M. Intramolecular interactions mediate pH regulation of connexin43 channels. Biophys J. 1996;70:1294–1302. doi: 10.1016/S0006-3495(96)79686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10:322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 69.De Vuyst E, et al. Ca2+ regulation of connexin 43 hemichannels in C6 glioma and glial cells. Cell Calcium. 2009;46:176–187. doi: 10.1016/j.ceca.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 70.Yang JJ, Buck M, Pitkeathly M, Kotik M, Haynie DT, Dobson CM, Radford SE. Conformational properties of four peptides spanning the sequence of hen lysozyme. J Mol Biol. 1995;252:483–491. doi: 10.1006/jmbi.1995.0513. [DOI] [PubMed] [Google Scholar]
- 71.Oshima A, Tani K, Hiroaki Y, Fujiyoshi Y, Sosinsky GE. Projection structure of a N-terminal deletion mutant of connexin 26 channel with decreased central pore density. Cell Commun Adhes. 2008;15:85–93. doi: 10.1080/15419060802013588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saimi Y, Kung C. Calmodulin as an ion channel subunit. Annu Rev Physiol. 2002;64:289–311. doi: 10.1146/annurev.physiol.64.100301.111649. [DOI] [PubMed] [Google Scholar]
- 73.Toutenhoofd SL, Strehler EE. The calmodulin multigene family as a unique case of genetic redundancy: multiple levels of regulation to provide spatial and temporal control of calmodulin pools? Cell Calcium. 2000;28:83–96. doi: 10.1054/ceca.2000.0136. [DOI] [PubMed] [Google Scholar]
- 74.Carafoli E. Intracellular calcium homeostasis. Annu Rev Biochem. 1987;56:395–433. doi: 10.1146/annurev.bi.56.070187.002143. [DOI] [PubMed] [Google Scholar]
- 75.Klee CB, Vanaman TC. Calmodulin. Adv Protein Chem. 1982;35:213–321. doi: 10.1016/s0065-3233(08)60470-2. [DOI] [PubMed] [Google Scholar]
- 76.Kakiuchi S, Yasuda S, Yamazaki R, Teshima Y, Kanda K, Kakiuchi R, Sobue K. Quantitative determinations of calmodulin in the supernatant and particulate fractions of mammalian tissues. J Biochem. 1982;92:1041–1048. doi: 10.1093/oxfordjournals.jbchem.a134019. [DOI] [PubMed] [Google Scholar]
- 77.Meador WE, Means AR, Quiocho FA. Modulation of calmodulin plasticity in molecular recognition on the basis of x-ray structures. Science. 1993;262:1718–1721. doi: 10.1126/science.8259515. [DOI] [PubMed] [Google Scholar]
- 78.Myllykoski M, Kuczera K, Kursula P. Complex formation between calmodulin and a peptide from the intracellular loop of the gap junction protein connexin43: Molecular conformation and energetics of binding. Biophys Chem. 2009;144:130–135. doi: 10.1016/j.bpc.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 79.Benedetti EL, Dunia I, Bentzel CJ, Vermorken AJ, Kibbelaar M, Bloemendal H. A portrait of plasma membrane specializations in eye lens epithelium and fibers. Biochim Biophys Acta. 1976;457:353–384. doi: 10.1016/0304-4157(76)90004-6. [DOI] [PubMed] [Google Scholar]
- 80.Girsch SJ, Peracchia C. Calmodulin interacts with a C-terminus peptide from the lens membrane protein MIP26. Curr Eye Res. 1991;10:839–849. doi: 10.3109/02713689109013880. [DOI] [PubMed] [Google Scholar]
- 81.Peracchia C, Girsch SJ. Calmodulin site at the C-terminus of the putative lens gap junction protein MIP26. Lens Eye Toxic Res. 1989;6:613–621. [PubMed] [Google Scholar]
- 82.Reichow SL, Gonen T. Noncanonical binding of calmodulin to aquaporin-0: implications for channel regulation. Structure. 2008;16:1389–1398. doi: 10.1016/j.str.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Reichow SL, Clemens DM, Freites JA, Nemeth-Cahalan KL, Heyden M, Tobias DJ, Hall JE, Gonen T. Allosteric mechanism of water-channel gating by Ca2+-calmodulin. Nat Struct Mol Biol. 2013;20:1085–1092. doi: 10.1038/nsmb.2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Peracchia C, Girsch SJ. Permeability and gating of lens gap junction channels incorporated into liposomes. Curr Eye Res. 1985;4:431–439. doi: 10.3109/02713688509025157. [DOI] [PubMed] [Google Scholar]
- 85.Girsch SJ, Peracchia C. Lens cell-to-cell channel protein: II. Conformational change in the presence of calmodulin. J Membr Biol. 1985;83:227–233. doi: 10.1007/BF01868697. [DOI] [PubMed] [Google Scholar]
- 86.Girsch SJ, Peracchia C. Lens cell-to-cell channel protein: I. Self-assembly into liposomes and permeability regulation by calmodulin. J Membr Biol. 1985;83:217–225. doi: 10.1007/BF01868696. [DOI] [PubMed] [Google Scholar]
- 87.Yap KL, Yuan T, Mal TK, Vogel HJ, Ikura M. Structural basis for simultaneous binding of two carboxy-terminal peptides of plant glutamate decarboxylase to calmodulin. J Mol Biol. 2003;328:193–204. doi: 10.1016/s0022-2836(03)00271-7. [DOI] [PubMed] [Google Scholar]
- 88.Hirst-Jensen BJ, Sahoo P, Kieken F, Delmar M, Sorgen PL. Characterization of the pH-dependent interaction between the gap junction protein connexin43 carboxyl terminus and cytoplasmic loop domains. J Biol Chem. 2007;282:5801–5813. doi: 10.1074/jbc.M605233200. [DOI] [PubMed] [Google Scholar]
- 89.Purnick PE, Benjamin DC, Verselis VK, Bargiello TA, Dowd TL. Structure of the amino terminus of a gap junction protein. Arch Biochem Biophys. 2000;381:181–190. doi: 10.1006/abbi.2000.1989. [DOI] [PubMed] [Google Scholar]
- 90.Oh S, Abrams CK, Verselis VK, Bargiello TA. Stoichiometry of transjunctional voltage-gating polarity reversal by a negative charge substitution in the amino terminus of a connexin32 chimera. J Gen Physiol. 2000;116:13–31. doi: 10.1085/jgp.116.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peracchia C, Wang XG, Peracchia LM. Gap junctions : molecular basis of cell communication in health and disease. San Diego: Academic Press; 2000. Behavior of chemical and slow voltage-gates of connexin channels. The cork gating hypothesis; pp. 271–295. [Google Scholar]
- 92.Peracchia C, Wang XG, Peracchia LL. Slow gating of gap junction channels and calmodulin. J Membr Biol. 2000;178:55–70. doi: 10.1007/s002320010015. [DOI] [PubMed] [Google Scholar]
- 93.Unwin PN, Ennis PD. Calcium-mediated changes in gap junction structure: evidence from the low angle X-ray pattern. J Cell Biol. 1983;97:1459–1466. doi: 10.1083/jcb.97.5.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gifford JL, Ishida H, Vogel HJ. Structural Insights into Calmodulin-regulated L-selectin Ectodomain Shedding. The Journal of biological chemistry. 2012;287:26513–26527. doi: 10.1074/jbc.M112.373373. [DOI] [PMC free article] [PubMed] [Google Scholar]