Abstract
Spontaneous reversion of disease-causing mutations has been observed in some genetic disorders. In our clinical observations of severe generalized recessive dystrophic epidermolysis bullosa (RDEB), a currently incurable blistering genodermatosis caused by loss-of-function mutations in COL7A1 that results in a deficit of type VII collagen (C7), we have observed patches of healthy-appearing skin on some individuals. When biopsied, this skin revealed somatic mosaicism resulting from the self-correction of C7 deficiency. We believe this source of cells could represent an opportunity for translational “natural” gene therapy. We show that revertant RDEB keratinocytes expressing functional C7 can be reprogrammed into induced pluripotent stem cells (iPSCs) and that self-corrected RDEB iPSCs can be induced to differentiate into either epidermal or hematopoietic cell populations. Our results give proof in principle that an inexhaustible supply of functional patient-specific revertant cells can be obtained—potentially relevant to local wound therapy and systemic hematopoietic cell transplantation. This technology may also avoid some of the major limitations of other cell therapy strategies, e.g., immune rejection and insertional mutagenesis, which are associated with viral- and nonviral- mediated gene therapy. We believe this approach should be the starting point for autologous cellular therapies using “natural” gene therapy in RDEB and other diseases.
INTRODUCTION
Cells originating from bone marrow (BM) maintain functional integrity of multiple parenchymal tissues, typically as cells involved in local immunity (Kupper and Fuhlbrigge, 2004). In addition, BM cells have been shown to aid in tissue repair (Badiavas et al., 2003), and this concept has been applied therapeutically in BM transplantation for the inherited blistering skin disorder, recessive dystrophic epidermolysis bullosa (RDEB) (Fine et al., 2008; Tolar et al., 2009; Wagner et al., 2010). RDEB is caused by loss-of-function mutations in the collagen type VII gene, COL7A1 (Bruckner-Tuderman, 2010). First in animal models (Chino et al., 2008; Tolar et al., 2009) and later in a clinical trial (Wagner et al., 2010), BM and cord blood (CB) cells from healthy donors were shown to home to injured mucocutaneous membranes in RDEB patients and mediate wound healing, with histologic evidence for cross-correction of the type VII collagen (C7) deficiency in the skin. Hematopoietic cell transplantation (HCT) is not available for everyone, however, either because the patient does not have a suitably human leukocyte antigen-matched donor or is not sufficiently affected to justify the risks of the transplant procedure. Physical injury (e.g., renal and pulmonary toxicity) and immune injury (e.g., profound immunodeficiency) are common after allogeneic HCT. These complications can limit the clinical application of transplant therapy to RDEB and other extracellular matrix disorders.
Autologous HCT, however, would be expected to have significantly fewer risks. Gene correction of autologous cells would be a necessary prerequisite for such therapy. Because of the large size of the COL7A1 gene and the possibility that both viral- and non-viral-mediated gene therapies result in unwanted off-target side-effects, traditional gene therapy may be neither optimal nor desirable. Fortunately, nature provides an alternative. Some RDEB individuals develop skin patches that never blister because of a spontaneous COL7A1 gene correction in a sub-population of keratinocytes (not fibroblasts) (Almaani et al., 2010; Lai-Cheong et al., 2011; Pasmooij et al., 2010). Such “natural” gene therapy can result in normal expression of C7 protein in skin. We hypothesized that if these revertant keratinocytes could be expanded, they might represent an ideal cellular substrate for autologous cell therapy, with no need for gene correction and the additional risk of genotoxicity that can be associated with viral and non-viral gene delivery methods (Hacein-Bey-Abina et al., 2003; Howe et al., 2008). The mosaic skin cells can be used for local applications to skin wounds, but there are significant limitations to the benefits for the patient. 1) Highly proliferative skin cells appear to be depleted from mutant and mosaic skin of EB individuals (De Luca et al., 2009; Petrova et al., 2010). 2) Even if such cells were isolated, only a finite number of passages, i.e., a limited number of cells, can be derived from them. 3) Mucocutaneous wounds are often not readily accessible, so local wound treatment cannot provide the desired universal therapy. To solve these problems, we hypothesized that the revertant keratinocytes could be induced to pluripotency, and that both hematopoietic and non-hematopoietic cells of various lineages could be derived from them for a life-long supply of patient-specific cells for both systemic and local treatment of RDEB.
Here we show that induced pluripotent stem cells (iPSCs) can be derived from both mutant and revertant keratinocytes of the same RDEB individual. Both stratified skin epithelium and cells with hematopoietic characteristics were differentiated from these iPSC populations. Thus, in principle, the benefits of the “natural” gene therapy can be amplified by cellular reprogramming for future applications in autologous cell therapy in RDEB and other skin extracellular matrix disorders.
RESULTS
Spontaneous clinicopathologic reversion of a patch of RDEB skin
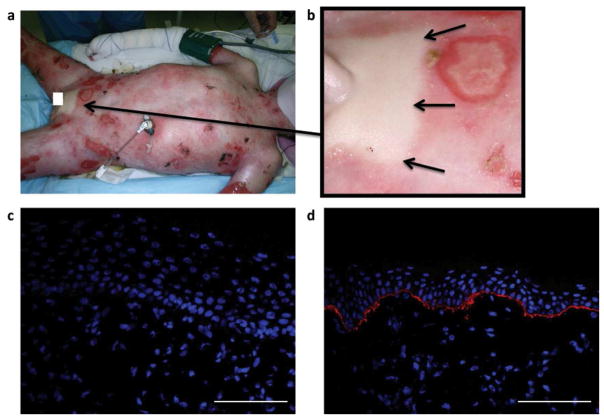
The subject, a 10-year-old male with generalized severe RDEB, had involvement of the oral and esophageal mucosa and a large cutaneous surface area that manifested as blisters and erosions (Figure 1a). Strikingly, a 7 × 11 cm patch of skin between the umbilicus and the pubis was entirely unaffected (Figure 1b) and had never blistered since birth. In contrast to immunofluorescence studies for expression of C7 in which no signal was detected at the dermal-epidermal junction (DEJ) (Figure 1c), strong continuous C7 staining was observed in a skin specimen obtained from the unaffected area (Figure 1d).
Figure 1. Mosaicism in RDEB.
(a) Blistered skin in a boy with generalized severe RDEB. Arrow points to area seen in 1b. (b) Small patch of unaffected skin in pubic area (arrows). (c) Blistered skin with no detectable expression of type VII collagen. (d) Non-blistered, pubic-area skin from the same individual with strong continuous linear labeling of type VII collagen (red) at the dermal-epidermal junction. Nuclei are stained with DAPI (blue). Scale bar = 50 μm.
In support of this data, keratinocytes isolated from this unaffected skin expressed C7 (Supplementary Figure 1) and, when examined by transmission electron microscopy, homotrimers of C7 (termed anchoring fibrils) and prominent C7 immunolabeling adjacent to the basement membrane were present in the healthy-appearing skin (Supplementary Figure 1), while absent in the blistered skin (data not shown). These clinical, biochemical, and ultrastructural observations were consistent with the possibility that a spontaneous correction of one of the mutations in the COL7A1 had occurred.
To investigate the etiology of this apparent somatic mosaicism, we performed allele-specific sequencing. Constitutionally, we found the boy to be a compound heterozygote for two loss-of-function mutations in COL7A1, a paternal frameshift mutation c. 3840delC (p.Thr1280Thrfs*44) and a maternal acceptor splice site mutation g.6751-2A>G (IVS85-2A>G). Therefore, one truncated (paternal) and one mis-spliced (maternal) COL7A1 mRNA products were generated. Direct cDNA sequencing revealed that the paternal c. 3840delC COL7A1 frameshift mutation was present in cells derived from blistered and from non-blistered (mosaic) skin (data not shown). However, cDNA PCR of the COL7A1 region surrounding the maternal g.6751-2A>G mutation located in intron 85 indicated skipping of exon 86 and generation of a presumably functional COL7A1 transcript (Supplementary Figure 2).
Revertant skin cells can be reprogrammed to induced pluripotent stem cells
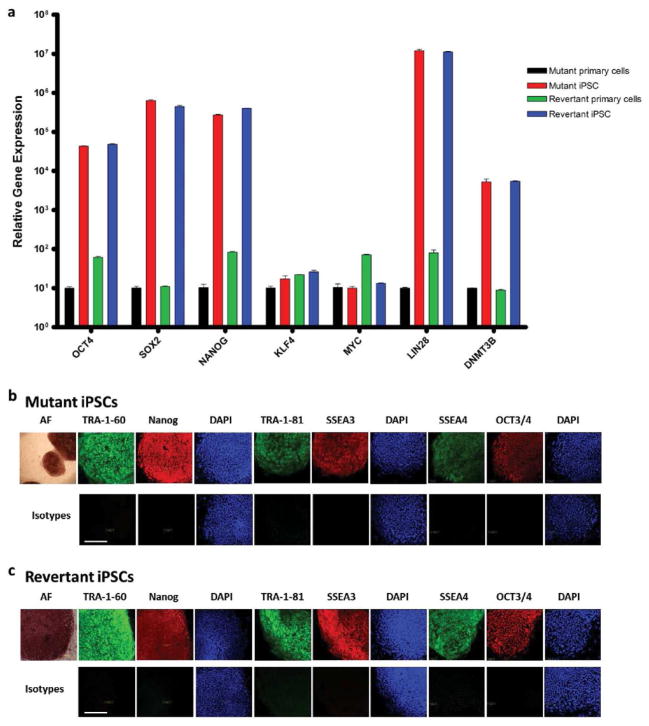
Restoration of normal biochemical (Figure 1 and Supplementary Figure 1) and ultrastructural phenotype (Supplementary Figure 1) immediately suggested the possibility of a treatment strategy using the patient-specific self-corrected cells. To provide preclinical evidence that such cells can be amplified into clinically meaningful numbers and phenotypes, we generated iPSCs from the COL7A1 revertant cells and, as a control, the COL7A1 mutant skin cells from the same individual. Following the well-established technology for development of iPSC-based disease models (Bilousova et al., 2011; Itoh et al., 2011; Park et al., 2008), we transduced COL7A1 revertant skin keratinocytes with OCT4-, SOX2-, KLF4-, and c-MYC-encoding retroviral vectors. After 3–6 weeks, 2–3 colonies of iPSCs per 100,000 primary skin cells emerged from the monolayer of supportive stromal cells. When compared with parental skin cells, iPSCs showed persistent mRNA expression of the OCT4 and SOX2 used for reprogramming, as well as other genes associated with pluripotency in iPSCs (such as NANOG, LIN28, and DNMT3b; Figure 2a).
Figure 2. Revertant iPSCs from RDEB skin.
(a) Persistent mRNA expression of OCT4, SOX2, NANOG, LIN28, and DNMT3b, and transient mRNA expression of c-MYC and KLF4, both consistent with fully reprogrammed mutant and revertant iPSCs. (b) Protein expression in mutant iPSCs of embryonic stem cell surface markers TRA-1-60, TRA-1-81, SSEA3, SSEA4, alkaline phosphatase (AF), and transcription factors Nanog and OCT3/4. (c) Same embryonic stem cell protein expression panel in revertant iPSCs. Nuclei are stained with DAPI (blue). Lower panels show corresponding isotype controls. Scale bar = 50 μm.
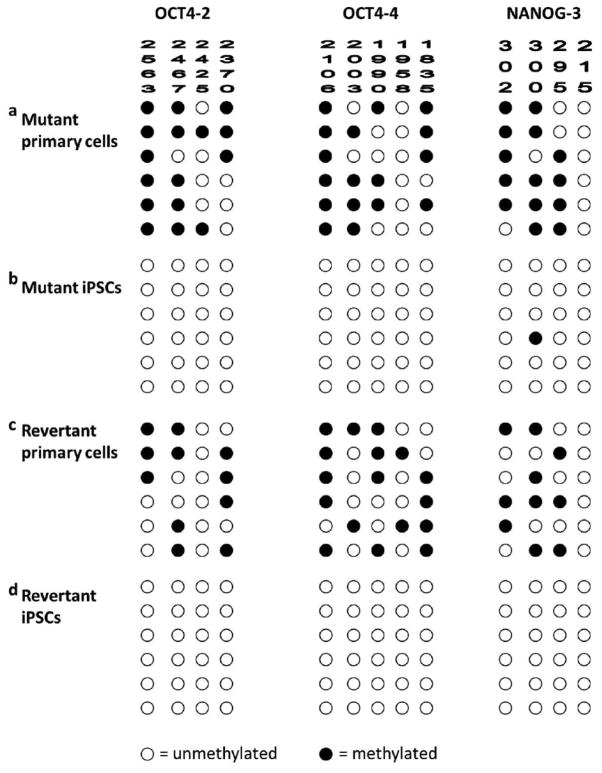
As would be expected to occur in wild-type (WT) iPSCs, there was transient mRNA expression of c-MYC and KLF4, also used to mediate reprogramming (Figure 2a). Also as expected, the exogenous OCT4 and SOX2 transgenes had been silenced in the iPSCs (Supplementary Figure 3a and 3b). Consistent with their acquired immaturity, the iPSCs expressed protein markers characteristic of fully reprogrammed iPSCs: TRA-1-81, stage-specific embryonic antigens-3 and - 4, OCT4, and NANOG (Figure 2b and 2c). As these reprogramming factors are believed to activate a network of transcriptional factors, which in turn induce epigenetic changes (Aasen et al., 2008; Freberg et al., 2007; Okita et al., 2008), we used bisulfite sequencing to confirm the methylation status of OCT4 and NANOG promoters in the iPSCs. A methylated pattern is indicative of gene silencing, whereas an unmethylated pattern indicates the potential for robust gene expression. Individual colonies of cells from mutant iPSCs and revertant iPSCs showed a methylation pattern characteristic of iPSCs in which OCT4 and NANOG promoter sequences are unmethylated. In contrast, parental skin cells had the expected pattern of partially methylated sequences (Figure 3a–d).
Figure 3. Epigenetic signature of RDEB iPSCs.
(a) A partially methylated pattern indicative of gene silencing of OCT4 and NANOG promoter sequences in mutant primary cells. (b) An unmethylated pattern in mutant iPSCs consistent with complete reprogramming into iPSCs. (c) A partially methylated pattern in revertant primary cells. (d) An unmethylated pattern in revertant iPSCs. Open circle indicates unmethylated site in the specified amplicon. Filled circle indicates methylated site in the same amplicon.
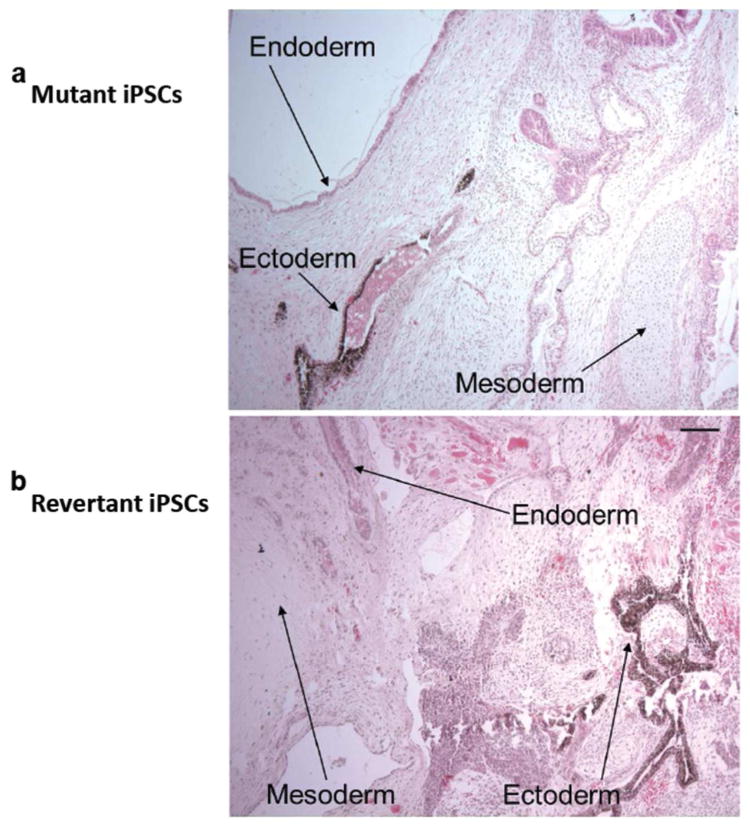
Both mutant iPSCs and revertant iPSCs had normal karyotypes as determined by high-resolution chromosomal G-banding (Supplementary Figure 4). To verify that the iPSCs originated from patient-derived primary cells, we performed polymerase chain reaction of variable number tandem repeats on genomic DNA isolated from both mutant and revertant iPSCs (data not shown). To provide further evidence of the identity of iPSCs on a functional level, iPSCs were injected intramuscularly into immune-deficient mice. Within 6–8 weeks, well-differentiated cystic teratomas were observed, confirming the phenotype-defining ability of iPSCs to differentiate in vivo into a wide array of cell lineages encompassing endodermal, mesodermal, and ectodermal origins (Figure 4). Collectively, the transcription profile, cellular phenotype, and in vivo behavior of the iPSCs were consistent with the morphological and phenotypical gain of cellular pluripotency.
Figure 4. In vivo pluripotentiality of RDEB iPSCs.
(a) Mutant iPSCs injected intramuscularly into immune-deficient mice yielded well-differentiated cystic teratomas with a wide array of cell lineages encompassing cells of endodermal, mesodermal, and ectodermal origins. (b) Same in vivo pluripotentiality was observed in teratomas generated from the revertant iPSCs. Hematoxylin-eosin staining. Scale bar = 50 μm.
Two cell types are most relevant for the preclinical modeling of revertant iPSC application for an skin extracellular matrix disorder such as RDEB: 1) hematopoietic cells with the potential to be used in the future for autologous HCT as efficacious as allogeneic HCT but with less toxicity, and 2) skin cells to be used in the local therapy of individual wounds, analogous to the skin injections of fibroblasts and mesenchymal stromal cells from unrelated donors that are already used in subjects with RDEB (Conget et al., 2010; Wong et al., 2008).
Blood cells and hematopoietic colonies can be derived from revertant iPSCs
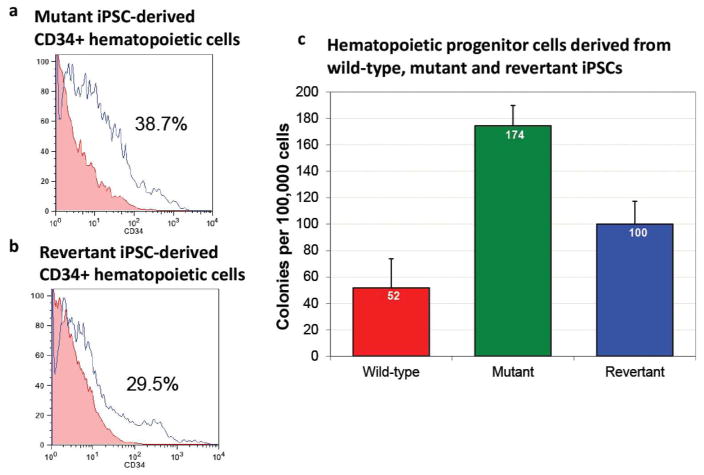
To test the blood-forming potential of the revertant iPSCs, we activated hematopoietic cell fate commitment in iPSCs using the formation of embryoid bodies and differentiation in medium containing human hematopoietic growth factors: stem cell factor, Flt3-ligand, interleukin-3, interleukin-6, granulocyte colony stimulating factor, and bone morphogenetic protein 4 (Tolar et al., 2011b; Tolar et al., 2011c). Consistent with conversion of iPSCs into hematopoietic progenitor cells, approximately one-third of the cells expressed an antigen found on hematopoietic stem cells, CD34 (Figure 5a and 5b). To further define the commitment of these cells expressing hematopoietic surface antigens to hematopoietic lineage, the capacity of hematopoietic progenitors from wild-type, mutant, and revertant iPSCs to function as colonyforming units was quantified by colony-formation assay (mean ± standard error of the mean, wild type 52 ± 22 colonies, mutant RDEB 174 ± 15 colonies, and revertant RDEB 100 ± 17 colonies). Consistent with the pattern we described previously for gene-corrected hematopoietic cells derived from iPSCs (Tolar et al., 2011b; Tolar et al., 2011c), the differences between wild-type and mutant cells, and revertant and mutant cells were statistically significant (p < 0.05), while the difference between wild-type and revertant cells were not (Figure 5c). We speculate that increased seeding capacity of the cells in semisolid medium is due to combination of higher proliferation rate (as evidenced by higher number of hematopoietic cells expressing stem cell marker CD34) and decreased connectivity among mutant cells due to lack of C7 in the extracellular matrix within the cellular colony of cells in semisolid medium.
Figure 5. Hematopoietic differentiation of RDEB iPSCs.
(a) Hematopoietic CD34-expressing stem cells derived from mutant iPSCs. (b) Hematopoietic CD34-expressing stem cells derived from revertant iPSCs. (c) Hematopoietic progenitors from wild-type, mutant, and revertant iPSCs quantified by colony-formation assay (mean ± standard error of the mean, wild type 52 ± 22 colonies, mutant RDEB 174 ± 15 colonies, and revertant RDEB 100 ± 17 colonies). The differences between wild-type and mutant cells, and revertant and mutant cells are statistically significant (p < 0.05), while the difference between wild-type and revertant cells is not.
Skin cells and three-dimensional skin can be derived from revertant iPSCs
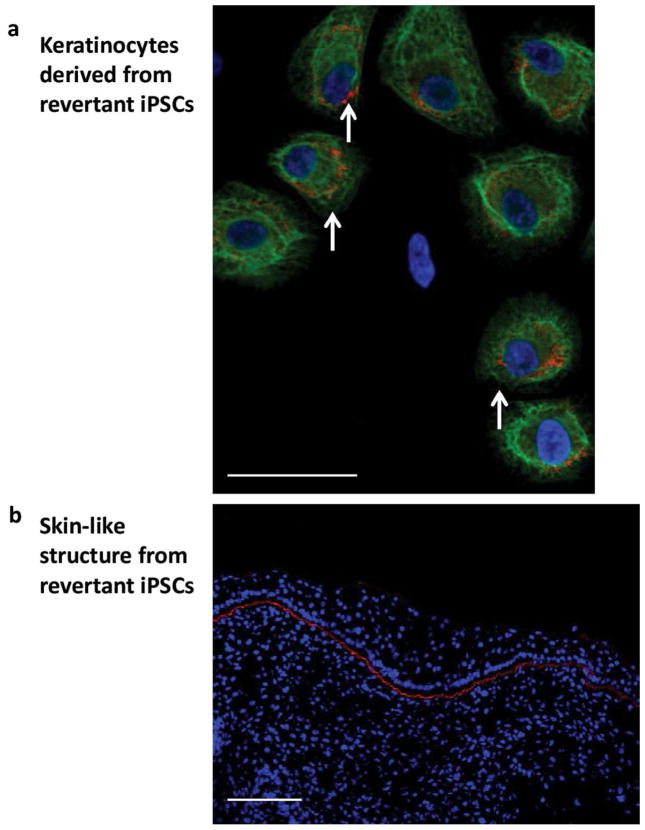
To demonstrate that, in principle, revertant iPSCs can be used as a resource for the unlimited production of disease-free, clinically-relevant cells, we derived keratinocytes from them as described (Tolar et al., 2011b; Tolar et al., 2012) and showed that they expressed C7 (Figure 6a). To further confirm functionality of the revertant iPSC-derived keratinocytes, we derived three-dimensional skin with epidermis-like layers of keratinocytes (expressing keratin 5) and a layer of C7 at the site analogous to the basement membrane in normal skin (Figure 6b) and comparable to the C7 expression in the revertant skin of this individual (Figure 1d).
Figure 6. Epidermal differentiation of RDEB iPSCs.
(a) Keratinocytes derived from revertant iPSCs express type VII collagen (red). Magnification 40x. (b) Three-dimensional skin from teratoma (Figure 4B) with epidermis-like layers of keratin 5 expressing keratinocytes (green) and linear type VII collagen immunoreactivity at the dermal-epidermal junction (red). Nuclei are stained with DAPI (blue). Scale bar = 50 μm.
In summary, these data show that iPSCs can be generated from revertant RDEB skin cells and that such iPSCs can differentiate into cells of hematopoietic and epidermal lineages.
DISCUSSION
We show that somatic mosaicism can form a platform for clinical intervention in RDEB and can be viewed as spontaneous “natural” gene therapy of this severe genodermatosis. Reversion in COL7A1 presumably occurs in a single skin cell, and selection of the new cellular phenotype then leads to growth expansion of progeny cells with the functional COL7A1 allele. Such cellular cloning in an EB individual, however, typically results in correction of only a small area of skin and may not substantively increase quality of life. Therefore, to make the available number of genetically distinct self-corrected skin cells clinically meaningful, we hypothesized that the mosaic cells could be amplified by reprogramming into pluripotency. Here we show that karyotypically stable and functional iPSCs can be derived from revertant RDEB skin cells. Although skin in the mosaic patch did not blister and appeared clinically normal (Figure 1), in principle, reversions of COL7A1 mutations could result in hypomorphic rather than fully functional C7. Nevertheless, we anticipate that iPSC-derived cells can be of major benefit to many people with RDEB in both local wound therapy (when differentiated to keratinocytes or fibroblasts), or systemic therapy (when differentiated to hematopoietic cells).
iPSCs are engineered stem cells that have the potential to develop into any other cell type in the body, much like embryonic stem cells (ESCs). As there are fewer ethical limitations associated with human iPSCs than human ESCs, iPSCs have become a popular tool for the investigation of tissue formation in health and disease, early stages of development, and drug interventions strategies (Taapken et al., 2011; Zhao et al., 2011; Zhu et al., 2011), all relevant to EB biology and treatment. Critical iPSC issues regarding transgene integration-free reprogramming, genomic fidelity, and transplantability (Anokye-Danso et al., 2011; Chen et al., 2011; Choi et al., 2011; Okita et al., 2011) are simultaneously being investigated in an effort to develop genomically stable, personalized iPSC-derived cells that are free of contamination with undifferentiated cells and less susceptible to immune rejection after transplantation. Critically, the data from iPSC biology can make possible the previously impossible advances in cell therapy tailored to specific clinical needs, such as expansion of disease-free cells from RDEB individuals with somatic mosaicism.
Any genetic disease can, in theory, be spontaneously corrected by a gene conversion, a compensatory mutation in cis, or by an intragenic crossover between maternal and paternal alleles with two different mutations in the same gene(Hirschhorn, 2003). Functional restoration of revertant cells has been reported in patients with a number of genetic diseases, such as epidermolysis bullosa, hereditary tyrosinemia type I, the BM failure syndromes Fanconi anemia and dyskeratosis congenita, and primary immunodeficiencies Wiskott-Aldrich syndrome, Bloom syndrome, X-linked severe combined immunodeficiency, and adenine deaminase-deficient severe combined immunodeficiency (Ariga et al., 1998; Darling et al., 1999; Ellis et al., 1995; Gregory et al., 2001; Gross et al., 2002; Hirschhorn et al., 1996; Jonkman et al., 1997; Lo Ten Foe et al., 1997; Schuilenga-Hut et al., 2002; Stephan et al., 1996). While the likelihood of reversion is presumably related to genetic instability in disorders of DNA repair, such as Fanconi anemia, ataxia telangiectasia, and Bloom syndrome, to our knowledge no such reason for increased opportunity for correction has been described for RDEB. Nevertheless, it is conceivable that the highly repetitive COL7A1 sequence and open chromatin structure during early stages of skin development combine to form a permissive environment for slipped DNA strand mispairing and other mechanisms underlying the somatic mosaicism. As these circumstances can equally well explain the introduction of pathogenic mutations in the first place, the favorable effect of the self-correcting mutation presumably becomes clinically obvious because of a selective growth advantage of revertant cells in vivo. Therefore it is likely that additional factors, such as early events associated with severe cellular stress, e.g., p53-dependent apoptosis (Liu et al., 2003), could contribute to the selection of these prenatal molecular events. At the present time, however, the cause and the time required for such selection remains unknown (Pasmooij et al., 2012).
The prognostic consequences of reversion may be difficult to predict, as they depend not only on the relative number of self-corrected cells, but also on how relevant the cell phenotype is to the primary disease. Both of these characteristics in turn depend on the proliferation potential (i.e., pluripotentiality, “stemness”) (Gregory et al., 2001; Mankad et al., 2006) and location of the revertant cells. Relevant to RDEB, even a less-than-complete level of mosaicism can be beneficial, as only 30–40% of the wild-type level of C7 expression may be necessary to prevent blistering (Fritsch et al., 2008; Kern et al., 2009). Furthermore, the mutation leading to restoration of function need not restore the original DNA sequence. Due to changes in post-translational modifications, the stability and ability to secrete the protein coded by the allele that has been repaired with a second site mutation may be only partially functional and still translate into a clinically meaningful change. Critically, a patient-specific combination of quantitative (the absolute level of C7 production) and qualitative (the ability of the partially functional C7 to polymerize into anchoring fibrils) factors will determine the prognostic consequences of somatic mosaicism in individuals with RDEB.
The somatic mosaicism in EB is visual, thus it is understandable to assume that only the healthy-appearing skin area is its full representation, with the rest of the mucocutaneous membranes carrying the biallelic germline pathogenic mutations and not being corrected by the reversion event. Since the frequency of somatic mosaicism is largely unknown, however, subclinical micromosaicism may exist in skin which could, in principle, contribute to the imperfect genotype-phenotype correlation in the EB individuals. Furthermore, as hematogenous cells from BM integrate into injured skin (Chino et al., 2008; Fu and Sun, 2009; Tolar et al., 2011b; Wagner et al., 2010), and as the reversions during early prenatal development have been described in hematologic genetic disorders (described above), it is possible that reversion in a multilineage BM cell can lead to repopulation of large areas of skin with C7-producing cells. This in turn can translate into long-term, albeit partial, restoration in skin integrity, and can provide another explanation for the phenotypical heterogeneity in various forms of EB (Fine, 2010; Kiritsi et al., 2011).
In summary, we show that iPSCs can be derived from mutant and revertant skin from an RDEB individual with somatic mosaicism in COL7A. From the perspective of skin pathology, this may provide a system to study mosaicism in a mechanistic manner. From the perspective of stem cell gene therapy in general, this approach may provide solutions to one major safety issue (the genotoxicity associated with viral-mediated gene therapy), and to one major technical roadblock in the development of technology that can rebuild injured skin (the paucity of adult stem cells in RDEB skin that has been exhausted by years of non-healing tissue repair). While this proof-of-concept supports the possibility of generating a limitless supply of patient-specific C7-producing cells for autologous cell therapy with self-corrected cells to ameliorate RDEB, the safety guidelines for such stem cell gene therapy will undoubtedly continue to be refined to deliver the benefits of this approach correctly, safely, and reliably. As further advances in RDEB research (Uitto et al., 2010) as well as cellular reprogramming, differentiation, and transplantation occur, each will be rapidly incorporated into a clinically meaningful design for the personalized cellular therapy of RDEB, one of the most painful and desperate of human conditions (Tolar et al., 2011a).
MATERIALS AND METHODS
Skin cells
After obtaining written, informed consent as approved by the Institutional Review Board of the Human Subjects Committee at the University of Minnesota, skin biopsies were collected from a healthy volunteer and from an individual with RDEB. All procedures adhered to the Helsinki Guidelines. Skin was cut into 2 × 2 mm pieces. Dermis was peeled from epidermis, placed in 6-well plates with each well containing 2 mL of medium, and immobilized under a sterile cover slip. Human fibroblast medium consisted of DMEM (Invitrogen, Grand Island, NY), 10% fetal bovine serum (Invitrogen) and penicillin/streptomycin (Invitrogen). Medium was changed every 2 days until the culture became 90% confluent, at which point the cells were lifted using 0.25% trypsin EDTA and serially replated. Human keratinocytes were grown in EpiLife medium (Cascade Biologics, Portland, OR) supplemented with 0.06 mM Ca2+, 1% EpiLife defined growth supplement, and 1% penicillin/streptomycin (Invitrogen). Cultures were maintained at 37°C in a humidified atmosphere of 5% CO2 and 95% air.
Histological evaluations
Skin biopsies were frozen in optimal cutting temperature (OCT, Sakura Finetek USA, Torrance, CA) and cut at 6 microns on a cryostat. Sections were fixed in room temperature acetone for 5 minutes followed by blocking with 10% normal donkey serum for 1 hour (Jackson Immunoresearch, West Grove, PA). Primary antibody specific for type VII collagen (1:250, lot 01591 30625, immunogen residues 952–1162, from BD Biosciences, San Jose, CA) was added for 1 hour, washed and then secondary antibody, donkey anti-mouse Cy3 (1:500, Jackson Immunoresearch, West Grove, PA) was applied for 1 hour. Slides were washed, coverslipped with hard set 4,6-diamidino-2-phenylindole (DAPI, Vector Labs, Burlingame, CA), and examined by confocal fluorescence microscopy (Olympus BX61, Olympus Optical, Tokyo, Japan). Ultrastructural skin examination was performed as described (Wagner et al., 2010).
Cellular reprogramming
About 50,000 keratinocytes were seeded per well in a 6-well plate and infected with a 1:1:1:1 mix of retroviral supernatants of pMIG containing OCT4, SOX2, KLF4, and c-MYC in the presence of 5 μg mL−1 protamine sulfate. Infection consisted of a 45-minute spin infection at 1800 revolutions per minute at room temperature, after which supernatants were left in contact with the cells for 24 hours at 37°C and 5% CO2. The next day cells were spininfected a second time using the same procedure. Five days after beginning the last round of infection, cells were trypsinized and seeded onto feeder layers of irradiated CF1 murine embryonic fibroblasts in the same culture medium. After 24 hours the medium was changed to human embryonic stem cell medium, consisting of DMEM/F12 (Invitrogen) supplemented with 10% KO-Serum Replacement (Invitrogen), 2 mM GlutaMAX (Invitrogen), 50 μM 2-mercaptoethanol (Invitrogen), 1× non-essential amino acids (Invitrogen), 50 U mL−1 penicillin and 50 mg mL−1 streptomycin and 10 ng mL−1 basic fibroblast growth factor (R&D Systems, Minneapolis, MN). Cultures were maintained at 37°C and 5% CO2, with daily medium changes. Colonies were picked based on morphology 20–30 days after the initial transduction.
Evaluations of induced pluripotent stem cells
Genome analyses (nucleic acid isolation, quantitative polymerase chain reactions, bisulfite sequencing, and karyotypes) were performed with standard techniques as described (Supplementary Methods and Tolar et al., 2012). For live staining, the TRA-1-60 antibody (1:400, Millipore, Billerica, MA) and secondary antibody Alexa 488-conjugated anti-mouse IgM (1:400, Invitrogen) were diluted in human embryonic stem cell medium and added into the culture plate. The plate was incubated at 37°C for 1 hour before medium was changed to fresh conditioned medium. TRA-1-60+ colonies were identified under a fluorescence microscope. For immunofluorescence evaluations, iPSCs grown on feeder cells in chamber slides were fixed with 4% paraformaldehyde for 15 minutes. If nuclear permeation was needed, cells were treated with 0.2% TritonX (Sigma-Aldrich, St. Louis, MO) in phosphate buffered saline for 30 minutes. Cell preparations were blocked in 3% bovine serum albumin in phosphate buffered saline for 2 hours, and incubated with primary antibody overnight at 4°C. The following antibodies were used: TRA1-60 (clone MAB4360, 1:400), TRA1-81 (clone MAB4381, 1:400), SSEA4 (clone MAB4304, 1:100) and SSEA3 (clone MAB-4303, 1:100) from Millipore; NANOG (clone EB06860, 1:100) from Everest Biotech, Upper Heyford, Oxfordshire, UK; OCT3/4 (clone AB27985, 1:200) from ABCAM, Cambridge, MA; and SOX2 (clone 630802, 1:500) from Biolegend, San Diego, CA. All secondary antibodies used were Alexa Fluor Series from Invitrogen (all 1:500) for 1 hour at room temperature. Images were taken using confocal microscope (Olympus BX61). Direct alkaline phosphatase activity was analyzed using Alkaline Phosphatase Staining Kit used according to the manufacturer’s recommendations (Millipore). For teratoma formation, young adult NOG mice were injected with 1 million cells re-suspended in a mixture of DMEM/F12 Matrigel (BD Biosciences), and type IV collagen (ratio 2:1:1, 40 μL per mouse) into the right quadriceps muscle. Tumors were harvested in 3–8 weeks and cryopreserved at −80°C in optimal cutting temperature medium (Sakura Finetek USA).
Hematopoietic and epidermal differentiation of induced pluripotent stem cells
For hematopoietic differentiation, embryoid bodies were produced by confluent cultures, harvested by enzymatic dissociation (Accutase, Invitrogen) to single cells, and added to AggreWell plates (StemCell Technologies, Vancouver, BC, Canada). According to the manufacturer’s recommendations, cells were differentiated for one day in basic embryoid body medium, containing Knockout (KO) Dulbecco modified Eagle medium (Invitrogen) supplemented with 20% fetal calf serum, 0.1 mM nonessential amino acids (Invitrogen), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), 1 mM L-glutamine (Invitrogen), 50 μg/mL ascorbic acid (Sigma-Aldrich), and 200 μg/mL human holo-transferrin (Sigma-Aldrich). After 24 hours, embryoid bodies were transferred to low-attachment dishes, and human stem cell factor (300 ng/mL), human Flt3-ligand (300 ng/mL), human interleukin-3 (10 ng/mL), human interleukin-6 (10 ng/mL), granulocyte colony-stimulating factor (50 ng/mL), and human bone morphogenetic protein 4 (50 ng/mL) were added to the cultures (all cytokines were from R&D Systems). At the time of analysis, embryoid bodies were collected and dissociated into single cells. Hematopoietic cells were characterized by flow cytometry analysis for CD34 expression and by colony-forming unit assay as described (Tolar et al., 2011b). For epidermal differentiation, keratinocytes were derived from iPSCs as described (Tolar et al., 2012).
Colony Forming Assay
The hematopoietic cells derived from iPSCs were counted using a hemocytometer and were re-suspended at a maximum concentration of 25,000 cells per mL in Iscove’s Modified Dulbecco’s Medium (IMDM) and 2% fetal bovine serum. 400 μL of the cell suspension was added to 4 mL aliquots of Human Methylcellulose Enriched Medium (catalog #HSC005, R&D Systems). The suspension was equally divided and placed in three 35 mm tissue culture plates and incubated at 37°C in 5% CO2 for 14 days. The plates were examined under a microscope, and CFU-GM and BFU-E colonies containing greater than 50 cells were scored as colonies.
Data analysis
Differences between measurements were evaluated with the Student t test, with P values < 0.05 considered significant.
Supplementary Material
Acknowledgments
The authors would like to thank the family of this child with RDEB, and all others who contributed to our efforts to develop safer therapy for genodermatoses. J.T. is supported by grants from the National Institutes of Health, Department of Defense, DebRA International, Jackson Gabriel Silver Fund, Epidermolysis Bullosa Medical Research Fund, and Children’s Cancer Research Fund, Minnesota. We would like to acknowledge the use of confocal microscopy made available through an NCRR Shared Instrumentation Grant (#1 S10 RR16851).
Abbreviations
- BM
bone marrow
- C7
type VII collagen
- CB
cord blood
- DEJ
dermal-epidermal junction
- EB
epidermolysis bullosa
- ESCs
embryonic stem cells
- HCT
hematopoietic cell transplantation
- iPSCs
induced pluripotent stem cells
- RDEB
recessive dystrophic epidermolysis bullosa
- WT
wild type
Footnotes
CONFLICT OF INTEREST
The authors state that they have no conflicts of interest.
References
- Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F, et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat Biotechnol. 2008;26:1276–84. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- Almaani N, Nagy N, Liu L, Dopping-Hepenstal PJ, Lai-Cheong JE, Clements SE, et al. Revertant mosaicism in recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2010;130:1937–40. doi: 10.1038/jid.2010.64. [DOI] [PubMed] [Google Scholar]
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell. 2011;8:376–88. doi: 10.1016/j.stem.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga T, Yamada M, Sakiyama Y, Tatsuzawa O. A case of Wiskott-Aldrich syndrome with dual mutations in exon 10 of the WASP gene: an additional de novo one-base insertion, which restores frame shift due to an inherent one-base deletion, detected in the major population of the patient’s peripheral blood lymphocytes. Blood. 1998;92:699–701. [PubMed] [Google Scholar]
- Badiavas EV, Abedi M, Butmarc J, Falanga V, Quesenberry P. Participation of bone marrow derived cells in cutaneous wound healing. J Cell Physiol. 2003;196:245–50. doi: 10.1002/jcp.10260. [DOI] [PubMed] [Google Scholar]
- Bilousova G, Chen J, Roop DR. Differentiation of mouse induced pluripotent stem cells into a multipotent keratinocyte lineage. J Invest Dermatol. 2011;131:857–64. doi: 10.1038/jid.2010.364. [DOI] [PubMed] [Google Scholar]
- Bruckner-Tuderman L. Dystrophic epidermolysis bullosa: pathogenesis and clinical features. Dermatol Clin. 2010;28:107–14. doi: 10.1016/j.det.2009.10.020. [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8:424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chino T, Tamai K, Yamazaki T, Otsuru S, Kikuchi Y, Nimura K, et al. Bone marrow cell transfer into fetal circulation can ameliorate genetic skin diseases by providing fibroblasts to the skin and inducing immune tolerance. Am J Pathol. 2008;173:803–14. doi: 10.2353/ajpath.2008.070977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KD, Vodyanik M, Slukvin Hematopoietic differentiation and production of mature myeloid cells from human pluripotent stem cells. Nat Protoc. 2011;6:296–313. doi: 10.1038/nprot.2010.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conget P, Rodriguez F, Kramer S, Allers C, Simon V, Palisson F, et al. Replenishment of type VII collagen and re-epithelialization of chronically ulcerated skin after intradermal administration of allogeneic mesenchymal stromal cells in two patients with recessive dystrophic epidermolysis bullosa. Cytotherapy. 2010;12:429–31. doi: 10.3109/14653241003587637. [DOI] [PubMed] [Google Scholar]
- Darling TN, Yee C, Bauer JW, Hintner H, Yancey KB. Revertant mosaicism: partial correction of a germ-line mutation in COL17A1 by a frame-restoring mutation. J Clin Invest. 1999;103:1371–7. doi: 10.1172/JCI4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca M, Pellegrini G, Mavilio F. Gene therapy of inherited skin adhesion disorders: a critical overview. Br J Dermatol. 2009;161:19–24. doi: 10.1111/j.1365-2133.2009.09243.x. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Lennon DJ, Proytcheva M, Alhadeff B, Henderson EE, German J. Somatic intragenic recombination within the mutated locus BLM can correct the high sister-chromatid exchange phenotype of Bloom syndrome cells. Am J Hum Genet. 1995;57:1019–27. [PMC free article] [PubMed] [Google Scholar]
- Fine JD. Inherited epidermolysis bullosa: past, present, and future. Ann N Y Acad Sci. 2010;1194:213–22. doi: 10.1111/j.1749-6632.2010.05463.x. [DOI] [PubMed] [Google Scholar]
- Fine JD, Eady RA, Bauer EA, Bauer JW, Bruckner-Tuderman L, Heagerty A, et al. The classification of inherited epidermolysis bullosa (EB): Report of the Third International Consensus Meeting on Diagnosis and Classification of EB. J Am Acad Dermatol. 2008;58:931–50. doi: 10.1016/j.jaad.2008.02.004. [DOI] [PubMed] [Google Scholar]
- Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell. 2007;18:1543–53. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch A, Loeckermann S, Kern JS, Braun A, Bosl MR, Bley TA, et al. A hypomorphic mouse model of dystrophic epidermolysis bullosa reveals mechanisms of disease and response to fibroblast therapy. J Clin Invest. 2008;118:1669–79. doi: 10.1172/JCI34292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Sun X. Can hematopoietic stem cells be an alternative source for skin regeneration? Ageing Res Rev. 2009;8:244–9. doi: 10.1016/j.arr.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Gregory JJ, Jr, Wagner JE, Verlander PC, Levran O, Batish SD, Eide CR, et al. Somatic mosaicism in Fanconi anemia: evidence of genotypic reversion in lymphohematopoietic stem cells. Proc Natl Acad Sci U S A. 2001;98:2532–7. doi: 10.1073/pnas.051609898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross M, Hanenberg H, Lobitz S, Friedl R, Herterich S, Dietrich R, et al. Reverse mosaicism in Fanconi anemia: natural gene therapy via molecular self-correction. Cytogenet Genome Res. 2002;98:126–35. doi: 10.1159/000069805. [DOI] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–9. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40:721–8. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn R, Yang DR, Puck JM, Huie ML, Jiang CK, Kurlandsky LE. Spontaneous in vivo reversion to normal of an inherited mutation in a patient with adenosine deaminase deficiency. Nat Genet. 1996;13:290–5. doi: 10.1038/ng0796-290. [DOI] [PubMed] [Google Scholar]
- Howe SJ, Mansour MR, Schwarzwaelder K, Bartholomae C, Hubank M, Kempski H, et al. Insertional mutagenesis combined with acquired somatic mutations causes leukemogenesis following gene therapy of SCID-X1 patients. J Clin Invest. 2008;118:3143–50. doi: 10.1172/JCI35798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh M, Kiuru M, Cairo MS, Christiano AM. Generation of keratinocytes from normal and recessive dystrophic epidermolysis bullosa-induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2011;108:8797–802. doi: 10.1073/pnas.1100332108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonkman MF, Scheffer H, Stulp R, Pas HH, Nijenhuis M, Heeres K, et al. Revertant mosaicism in epidermolysis bullosa caused by mitotic gene conversion. Cell. 1997;88:543–51. doi: 10.1016/s0092-8674(00)81894-2. [DOI] [PubMed] [Google Scholar]
- Kern JS, Loeckermann S, Fritsch A, Hausser I, Roth W, Magin TM, et al. Mechanisms of fibroblast cell therapy for dystrophic epidermolysis bullosa: high stability of collagen VII favors long-term skin integrity. Mol Ther. 2009;17:1605–15. doi: 10.1038/mt.2009.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiritsi D, Kern JS, Schumann H, Kohlhase J, Has C, Bruckner-Tuderman L. Molecular mechanisms of phenotypic variability in junctional epidermolysis bullosa. J Med Genet. 2011;48:450–7. doi: 10.1136/jmg.2010.086751. [DOI] [PubMed] [Google Scholar]
- Kupper TS, Fuhlbrigge RC. Immune surveillance in the skin: mechanisms and clinical consequences. Nat Rev Immunol. 2004;4:211–22. doi: 10.1038/nri1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai-Cheong JE, McGrath JA, Uitto J. Revertant mosaicism in skin: natural gene therapy. Trends Mol Med. 2011;17:140–8. doi: 10.1016/j.molmed.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu TX, Howlett NG, Deng M, Langenau DM, Hsu K, Rhodes J, et al. Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev Cell. 2003;5:903–14. doi: 10.1016/s1534-5807(03)00339-3. [DOI] [PubMed] [Google Scholar]
- Lo Ten Foe JR, Kwee ML, Rooimans MA, Oostra AB, Veerman AJ, van Weel M, et al. Somatic mosaicism in Fanconi anemia: molecular basis and clinical significance. Eur J Hum Genet. 1997;5:137–48. [PubMed] [Google Scholar]
- Mankad A, Taniguchi T, Cox B, Akkari Y, Rathbun RK, Lucas L, et al. Natural gene therapy in monozygotic twins with Fanconi anemia. Blood. 2006;107:3084–90. doi: 10.1182/blood-2005-07-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, et al. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–12. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–53. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- Park IH, Arora N, Huo H, Maherali N, Ahfeldt T, Shimamura A, et al. Disease-specific induced pluripotent stem cells. Cell. 2008;134:877–86. doi: 10.1016/j.cell.2008.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasmooij AM, Garcia M, Escamez MJ, Nijenhuis AM, Azon A, Cuadrado-Corrales N, et al. Revertant mosaicism due to a second-site mutation in COL7A1 in a patient with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2010;130:2407–11. doi: 10.1038/jid.2010.163. [DOI] [PubMed] [Google Scholar]
- Pasmooij AM, Jonkman MF, Uitto J. Revertant mosaicism in heritable skin diseases: mechanisms of natural gene therapy. Discovery medicine. 2012;14:167–79. [PubMed] [Google Scholar]
- Petrova A, Ilic D, McGrath JA. Stem cell therapies for recessive dystrophic epidermolysis bullosa. Br J Dermatol. 2010;163:1149–56. doi: 10.1111/j.1365-2133.2010.09981.x. [DOI] [PubMed] [Google Scholar]
- Schuilenga-Hut PH, Scheffer H, Pas HH, Nijenhuis M, Buys CH, Jonkman MF. Partial revertant mosaicism of keratin 14 in a patient with recessive epidermolysis bullosa simplex. J Invest Dermatol. 2002;118:626–30. doi: 10.1046/j.1523-1747.2002.01715.x. [DOI] [PubMed] [Google Scholar]
- Stephan V, Wahn V, Le Deist F, Dirksen U, Broker B, Muller-Fleckenstein I, et al. Atypical X-linked severe combined immunodeficiency due to possible spontaneous reversion of the genetic defect in T cells. N Engl J Med. 1996;335:1563–7. doi: 10.1056/NEJM199611213352104. [DOI] [PubMed] [Google Scholar]
- Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM, et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–4. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- Tolar J, Blazar BR, Wagner JE. Concise review: transplantation of human hematopoietic cells for extracellular matrix protein deficiency in epidermolysis bullosa. Stem Cells. 2011a;29:900–6. doi: 10.1002/stem.647. [DOI] [PubMed] [Google Scholar]
- Tolar J, Ishida-Yamamoto A, Riddle M, McElmurry RT, Osborn M, Xia L, et al. Amelioration of epidermolysis bullosa by transfer of wild-type bone marrow cells. Blood. 2009;113:1167–74. doi: 10.1182/blood-2008-06-161299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Park IH, Xia L, Lees CJ, Peacock B, Webber B, et al. Hematopoietic differentiation of induced pluripotent stem cells from patients with mucopolysaccharidosis type I (Hurler syndrome) Blood. 2011b;117:839–47. doi: 10.1182/blood-2010-05-287607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Lees CJ, Riddle M, McElroy A, Keene DR, et al. Keratinocytes from Induced Pluripotent Stem Cells in Junctional Epidermolysis Bullosa. J Invest Dermatol. 2012 doi: 10.1038/jid.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolar J, Xia L, Riddle MJ, Lees CJ, Eide CR, McElmurry RT, et al. Induced pluripotent stem cells from individuals with recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2011c;131:848–56. doi: 10.1038/jid.2010.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uitto J, McGrath JA, Rodeck U, Bruckner-Tuderman L, Robinson EC. Progress in epidermolysis bullosa research: toward treatment and cure. J Invest Dermatol. 2010;130:1778–84. doi: 10.1038/jid.2010.90. [DOI] [PubMed] [Google Scholar]
- Wagner JE, Ishida-Yamamoto A, McGrath JA, Hordinsky M, Keene DR, Woodley DT, et al. Bone marrow transplantation for recessive dystrophic epidermolysis bullosa. N Engl J Med. 2010;363:629–39. doi: 10.1056/NEJMoa0910501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong T, Gammon L, Liu L, Mellerio JE, Dopping-Hepenstal PJ, Pacy J, et al. Potential of fibroblast cell therapy for recessive dystrophic epidermolysis bullosa. J Invest Dermatol. 2008;128:2179–89. doi: 10.1038/jid.2008.78. [DOI] [PubMed] [Google Scholar]
- Zhao T, Zhang ZN, Rong Z, Xu Y. Immunogenicity of induced pluripotent stem cells. Nature. 2011;474:212–5. doi: 10.1038/nature10135. [DOI] [PubMed] [Google Scholar]
- Zhu H, Lensch MW, Cahan P, Daley GQ. Investigating monogenic and complex diseases with pluripotent stem cells. Nat Rev Genet. 2011;12:266–75. doi: 10.1038/nrg2951. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.