Abstract
Bacterial Lipopolysaccharide (LPS) is an essential cell envelope component for Gram-negative bacteria. As the most variable region of LPS, O-antigens serve as important virulence determinants for many bacteria and represent a promising carbohydrate source for glycoconjugate vaccines. In the Wzy-dependent O-antigen biosynthetic pathway, the integral membrane protein Wzy was shown to be the sole enzyme responsible for polymerization of O-repeat unit. Its catalytic mechanism, however, remains elusive. Herein, Wzy was successfully overexpressed in E. coli with an N-terminal His10-tag. Blue native polyacrylamide gel electrophoresis (BN-PAGE) revealed that the Wzy protein exists in its native confirmation as a dimer. Subsequently, we chemoenzymatically synthesized the substrates of Wzy, the lipid-PP linked repeat units. Together with an optimized O-antigen visualization method, we monitored the production of reaction intermediates at varying times. We present here our result as the first biochemical evidence that wzy functions in a distributive manner.
Keywords: Wzy, integral membrane protein, overexpression, distributive mechanism
Introduction
The lipopolysaccharide (LPS) on the cell surface of Gram-negative bacteria is critical for viability (Chng et al. 2012; Silhavy et al. 2010). On the outer leaflet of the outer membrane, LPS plays important roles in protecting bacteria from external environment and provides structural integrity to the bacterial cell (Silhavy et al. 2010). LPS typically consists of three structural regions: lipid A, a hydrophobic glycolipid which anchors LPS in the bacterial membrane; core oligosaccharide, a nonrepeating oligosaccharide which commonly contains sugars such as heptose and keto-deoxyoctulosonate (KDO); and the O-antigen, a polysaccharide with multiple oligosaccharide repeating units that are exposed on the surface of the bacterial cell envelope (Kim et al. 2010; Raetz and Whitfield 2002). Within any given species, variations of lipid A and core oligosaccharide are relatively limited. O-antigen, on the other hand, is very diverse and provides the major basis to serotype Gram-negative bacteria (Duda et al. 2011). The structural diversity of O-antigen comes from differing monosaccharides composition, the arrangement of the sugar moieties and the linkage between them (Guo et al. 2005; Zhao et al. 2007). So far, more than 180 E. coli O serotypes have been identified (Greenfield et al. 2012). O-antigen also provide a library of carbohydrate resources for several promising candidates of glycoconjugate vaccines (Ihssen et al. 2010; Wetter et al. 2013), which is proven to be attractive by several successful capsular polysaccharide based conjugate vaccines such as Prevnar 13 (Sucher et al. 2011).
Although O-antigens are remarkably diverse, their biosynthetic pathways are limited (Hug et al. 2010; Lerouge and Vanderleyden 2002; Wang et al. 2010). Thus far, only three major pathways have been defined including the Wzy-dependent, ATP-binding cassette (ABC) transporter dependent, and synthetase-dependent pathways. Of them, the Wzy-dependent pathway is responsible for the synthesis of most hetero-polysaccharides (Fig. 1). This pathway is initiated by transfer of a sugar phosphate residue to undecaprenyl-phosphate (Und-P) to form Und-PP-monosaccharide, from which the Und-PP-repeat unit (Und-PP-RU) is synthesized by the sequential addition of monosaccharides by the corresponding glycosyltransferases on the cytoplasmic face of the inner membrane (IM) (Yi et al. 2005). The Und-PP-RU is then flipped to the periplasmic side of IM by the flippase Wzx, and polymerized to Und-PP-linked O-antigen by Wzy. The strain-specific chain length distribution, or modality, is regulated by Wzz. Finally, the resultant O-antigen is transferred to lipid A core by the ligase WaaL (Han et al. 2012) to form the complete LPS molecule, and eventually transported to outer membrane (OM) through Lpt protein complex (Ruiz et al. 2009; Silhavy et al. 2010).
Figure 1. wzy-dependent pathway of lipopolysaccharide biosynthesis (E. coli O86:H2 O-polysaccharide as an example).
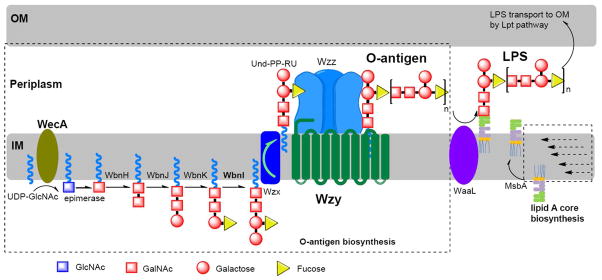
Biosynthesis is initiated with sequential assembly of repeat units on the cytoplasmic face of the inner membrane by glycosyltransferases (WbnH, WbnJ, WbnK and WbnI), after which translocation to the periplasmic face occurs. Polymerization of repeat units on the periplasmic face of the inner membrane then follows through the action of Wzy polymerase in a block transfer mechanism which is regulated by Wzz. The O-PS was transferred to lipid A core by WaaL to form the whole LPS which is then transported to outer membrane through Lpt pathway.
Due to the key role of the membrane proteins including Wzx, Wzy, Wzz and WaaL, understanding their mechanism would be vital in decoding LPS biosynthesis and their downstream applications in vaccines development. However, the membrane proteins are among the most challenging targets in structural and mechanistic studies (Carpenter et al. 2008) despite the fact that membrane proteins make up roughly 30% of the all proteins in currently sequenced genomes; and they present more than 60% of drug targets underscoring their importance in nature (Arinaminpathy et al. 2009). The hydrophobic nature of membrane proteins usually brings technical difficulties into experimental operations because it can cause unfolding, aggregation and proteolysis. In addition, improper insertion of membrane proteins into the cellular membrane is sometimes problematic and often is the bottlenecks in overexpression. The toxic effect of membrane proteins could also compromise the expression of membrane proteins in a large amount (Schwarz et al. 2008). Wzy of E. coli O86 is such a membrane protein with 10 predicted transmembrane helixes (Fig. 1). It belongs to the O-antigen polymerase superfamily (Pfam 14296) in which the enzymes vary in sequence and specificity. Although the attempts to express the enzyme have been started a decade ago, few successful cases have been reported (Daniels et al. 1998; Woodward et al. 2010). In addition to the common problems facing all of the membrane proteins, rare codons at the N-terminus and the poor translation initiation from an undetectable ribosome-binding site (RBS) may also lead to the extremely low expression of Wzy (Daniels et al. 1998). Therefore, the codon optimization likely contributed partially to its heterogeneous expression (Woodward et al. 2010). Due to the difficulty in obtaining pure protein, the mechanistic study on O-antigen polymerization has been greatly hindered, and most of our knowledge on Wzy is obtained through genetic studies (Islam et al. 2010; Kim et al. 2010). However, the catalytic mechanism of Wzy remains unknown to a large extent.
We recently obtained small amount of pure Wzy and developed an in vitro assay to investigate the function of Wzy (Woodward et al. 2010). Following these achievements, we, in this study, applied a recent finding (Kudla et al. 2009) about the effect of N-terminal sequence on protein expression to Wzy to further increase its expression. The purified Wzy can polymerize the chemically synthetic substrate in vitro. By monitoring the polymerization in varying reaction times, we here present the first biochemical evidence that Wzy functions in a distributive manner.
Materials and methods
Bacterial strains, plasmids and materials
E. coli O86:K61:B7 (ATCC 12701) was obtained from American Type Culture Collection (Rockville, MD, USA). E. coli DH5 [lacZΔM15 hsdR recA] competent cells were obtained from Gibco-BRL Life Technology. E. coli C43 (DE3) competent cells were obtained from Lucigen Co. (Middleton, WI). Nitrocellulose membrane, PCR reagents, and pBAD/Myc-His A plasmid were obtained from Invitrogen (Carlsbad, CA). Detergents were purchased from Anatrace (Maumee, OH). Ni-NTA agarose was from Qiagen (Valencia, CA). Anti-E. coli O86 antiserum (from Rabbit) was from Tianjin Biochip Corporation (Tianjin, China). HRP-linked anti-mouse IgG, anti-rabbit IgG, and ECL plus Western Blot detection reagents were purchased from GE healthcare (Piscataway, NJ). FastDigest restriction enzymes were purchased from Fermentas (Glen Burnie, MD). All other chemicals and solvents were purchased from Sigma-Aldrich.
Clone and expression of Wzy
The gene was amplified from the optimized wzy sequence (GeneBank access no. AB863203) (Woodward et al. 2010). An N-terminal His10 tag was inserted into pBAD-A vector using primers: forward primer: TGACCATGGCTCACCATCATCACCACCATCATCATCACCACATGGTGATTAGT CGTAGCAACTATC (NcoI) and reverse primer: GAGGGAATTCTTAATGACATTTTTTGATATTCAGGGCA (EcoRI). The recombinant plasmid was co-transformed with pGro7 vector into E. coli O86 (ΔwecA, ΔwaaL) for protein expression. For screening detergents, cells were cultured and the membrane fractions were isolated by ultracentrifugation (150,000 g, 60 min, 4 °C) followed by suspending in 5 mL Lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl). Each 100 μL of suspension was mixed with equal amount of different detergents overnight at 4 °C. The insoluble membrane materials were removed by ultracentrifugation (150,000 g, 60 min, 4 °C). The supernatant was collected and analyzed by SDS-PAGE and western blot.
The purification of His10-Wzy was similar to described previously (Woodward et al. 2010). Briefly, six liters of cell broth containing the recombinant wzy plasmid were cultured and harvested by centrifugation (4000 g, 30 min, 4 °C). The cells were suspended in the lysis buffer (20 mM Tris-HCl, pH 7.5, 1 mM EDTA), and disrupted by cell disruptor. The membrane fractions were isolated by ultracentrifugation (150,000 g, 60 min, 4 °C) followed by re-dissolved in 5 mL membrane dissolving buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA) with 4.2% (10 × CMC) SDDS. Insoluble material was removed by ultracentrifugation (150,000 g, 60 min, 4 °C), after which soluble material containing Wzy was loaded on the pre-equilibrated Ni-NTA resin (Qiagen). After washing with 100 mL of binding/washing buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 20 mM Imidazole, 0.1% DDM, 10% glycerol), bound proteins were eluted with 15 mL of elution buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 250 mM imidazole, 0.1% DDM, 10% glycerol). The protein was concentrated and dialyzed against the storage buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM DTT, 0.1% DDM, 10% glycerol).
Activity assay
The activity of wzy was analyzed as previously reported (Woodward et al. 2010). Briefly, the 50 μL reaction was set up containing 50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 1 mM RU-PP-Und, and Wzy. The reaction was performed at room temperature. The reaction mixture was quenched by adding equal volumes of iced-methanol at different reaction time. The pellet was removed by centrifugation at 16,000g for 30 min. The supernatant was dried with a speedvac and re-dissolved in 15 μL water. The reaction product was detected by autoradiography (Woodward et al. 2010) or western blot.
Isolation of LPS
Hot water-phenol method was used as described (Yi et al. 2009). Briefly, E.coli O86:H2 wide-type strains were collected from 1.5 mL overnight culture. After being washed with water, cells were resuspended in 0.5 mL ddH2O followed by addition of 0.5 mL liquefied phenol. The tube was incubated in 65 °C water bath for 40 min with vigorous votexing every 10 min. The mixture was then centrifuged at 16,100×g for 30 min. The top aqueous phase was transferred into a new tube and the organic phase was reextracted with 0.5 mL ddH2O. The combined fractions were concentrated to 100 μL. 150 μL of 100% ethanol was added into the fraction and LPS was precipitated by centrifugation for 45 min. The pellet was dried in 1 mL ddH2O and freeze-dried.
Western-blot of LPS
The wet-transfer method was performed as repoted previously (Renart et al. 1979; Towbin et al. 1979). The semi-dry transfer method was performed using iBlot system as manufacture’s instruction (Invitrogen).
Results
Overexpression and purification of Wzy
According to a recent finding by Kuldla et al, the 5′ end of a gene sequence (i.e. nucleotide (nt) −4 to nt +37 bp) can significantly affect the expression level of the target protein since this area determines the translation initiation and stability of mRNA near the ribosomal binding site (Kudla et al. 2009). They found that a stronger mRNA folding energy near the ribosomal binding site usually causes lower GFP fluorescence, i.e. lower protein expression. This finding also addressed why synonymous mutations that do not alter the encoded protein can influence protein expression. In this study, we analyzed seven common fusion tags using mfold web server (Mathews et al. 1999; Walter et al. 1994; Zuker 2003). The mRNA free energy of the nucleotide (nt) −4 (from expression vector pBAD/myc-his A) to +37 (from each fusion tag) was calculated and the mRNA structures were predicted. Of all candidates, a decahistidine tag (ten tandem histidines, His10) exhibits the weakest free folding energy (−1.9 kcal/mol) (Supplementary Fig. S1). Therefore, we subsequently inserted this his-tag into the 5′ end of the optimized wzy sequence. The expression strain and chaperon plasmid used here are the same as those in our previous study (Woodward et al. 2010). His10-tagged Wzy was expressed by induction with 0.1% L-arabinose at room temperature. The membrane fraction containing recombinant Wzy was collected after ultracentrifugation. To optimize membrane protein solubility, we screened twenty two detergents, including 5 ionic detergents, 9 non-inoic detergents, 6 zwitterionic detergents and 2 synthetic lipids (Supplementary Table S1). Six potential detergent candidates were selected through the initial screening (Supplementary Fig. S2). A second batch of His-tagged Wzy was solubilized by each of these six detergents and purified by Ni-NTA column. SDS-PAGE and immunoblotting results showed that sodium dodecanoyl sarcosine (SDDS) had the strongest capacity in solubilizing Wzy from the membrane (Supplementary Fig. S3).
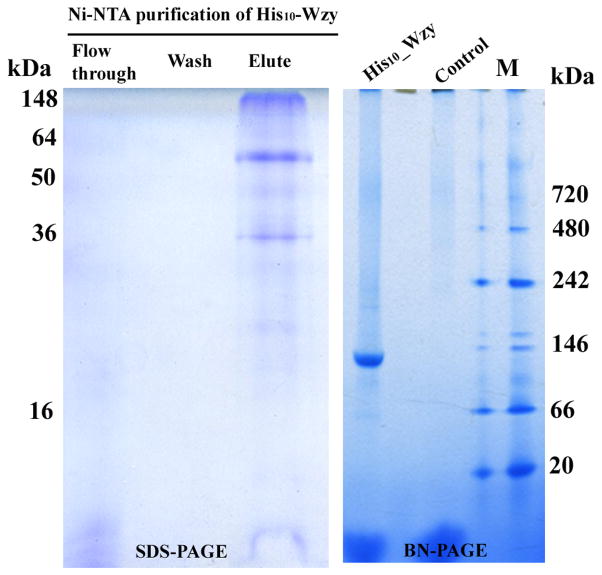
After extracted from the membrane by SDDS, Wzy was exchanged into the buffer containing n-Dodecyl β-D-maltoside (DDM) during the purification process when large-scale purification of Wzy was performed due to the high demand on SDDS resulting from its high CMC value. As previously observed with purified Wzy with C-terminal his-tag (Woodward et al. 2010) (Supplementary Fig. S4), His10-Wzy also exhibits three bands on the SDS-PAGE gel, corresponding to monomer, dimer and aggregate, respectively (Fig. 2). Compared to the previously purified Wzy, His10-Wzy has more dimeric protein. Pierce BCA (bicinchoninic acid) protein assay showed that the expression of His10-Wzy (12.5 μg/L broth) is improved by over three times than that of its C-terminal equivalent (3.75 μg/L broth). To evaluate the oligomeric state of Wzy under native condition, blue native polyacrylamide gel electrophoresis (BN-PAGE) is employed. The purified Wzy exhibited only one strong band with the apparent molecular weight about 130 kDa, suggesting the homogeneity of the purified Wzy (Fig. 2). Given the micelle size of DDM (~ 60 kDa), the native Wzy protein (~ 70 kDa) is likely a dimer.
Figure 2. Expression and detection of Wzy on SDS-PAGE and Blue native (BN)-PAGE.
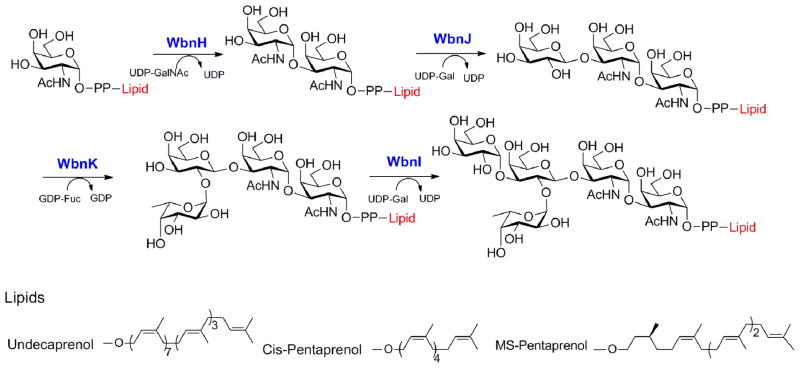
Chemo-enzymatic synthesis of lipid-PP-RU and activity determination of Wzy
According to our previous study on Wzy substrate specificity (Woodward et al. 2010), two Wzy substrates, cis-pentaprenol-PP-RU and Und-PP-RU, were synthesized (Fig. 3). The final product was determined by ESI-MS or MALDI-TOF-MS. The detected m/z values of cis-pentaprenol-PP linked RU ([M-H]−; 1393.5) and Und-PP linked RU ([M-2H]2−; 900.4) are both consistent with the calculated value. Monosaturated pentaprenol (MS-Pent, [M-2H]2− 697.3.), a saturated form of α-isoprene unit, was also synthesized as the negative control (Supplementary Fig. S5).
Figure 3. Chemo-enzymatic synthesis of lipid-PP-linked repeat units.
To check if purified Wzy was active, we carried out an in vitro reaction which was monitored using a radioactivity-based assay (Woodward et al. 2010). Cis-pentaprenol-PP-RU was labeled with 3H in the final enzymatic reaction catalyzed by WbnI in the presence of UDP-[3H] Gal. The radio-labeled substrate was then incubated overnight at room temperature with purified Wzy. The reaction mixture was subjected to SDS-PAGE and detection by autoradiography. In the absence of Wzy, only one band was detected with high mobility, corresponding to the cis-pentaprenol-PP-RU. In the reaction containing Wzy, a significant amount of radioactivity appeared at positions with low mobility, consistent with the formation of compounds containing multiple repeating units. Thus, His10-Wzy is active and capable of catalyzing the polymerization of repeating units (Supplementary Fig. S6).
Wzy polymerizes the repeating unit by a distributive mechanism
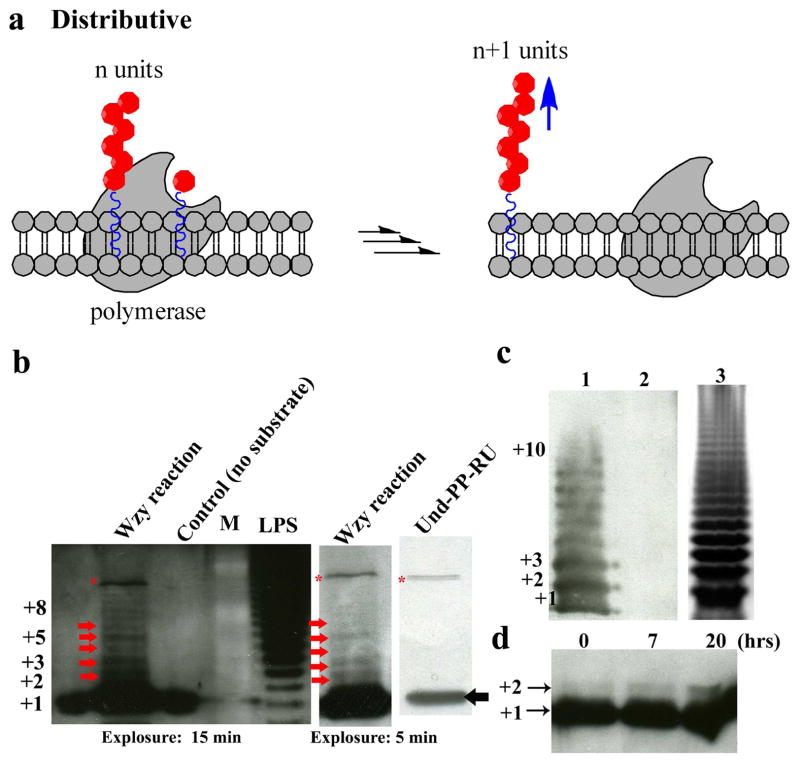
Processive and distributive mechanisms are usually used to describe the catalytic behavior of polymerase. A processive enzyme binds to the polymer end at which it works, catalyzes one round of reaction and remains bound to catalyze subsequent rounds. In contrast, a distributive enzyme will dissociate after each catalytic round and randomly bind to other substrates in the reaction thereafter (Fig. 4a). As such, processive and distributive eznymes will produce polymers with different patterns, by which a polymerase can be defined. For instance, class I Hyaluronan (HA) synthase (streptococcus, virus or avian) (Kumari and Weigel 1997), Aquifex aeolicus peptidoglycan glycosyltransferase (Yuan et al. 2007), and Mycobacterium tuberculosis galactofuranosyltransferase GlfT2 (May et al. 2009; Wheatley et al. 2012) are processive; while class II (such as P. multocida) HA synthase (DeAngelis et al. 2003) is distributive. The study of the mechanism of Wzy necessitates the enzyme and substrates for the in vitro reaction; but the expression and purification of membrane protein and the synthesis of oligosaccharide remain challenging.
Figure 4. Wzy adopts distributive mechanism for polymerization.
(a) The process of distributive mechanims; (b) The Wzy reaction with different exposure time (15 min vs 5 min); (c) The LPS from E. coli (Δwzy) (lane 2), from Δwzy with complementary wzy gene (Lane 1) and wzz knockout strain (Lane 3); (d) the formation of Und-PP-linked di-unit. *: contaminant band coming from synthetic Und-PP-RU; The read arrow shows the intermediates of wzy reaction; the black arrow indicated the synthetic Und-PP-RU.
In this work, after we made the enzyme and its substrates available, we performed the polymerase reaction in vitro to characterized Wzy. We used western blotting as the primary analytical method. In order to see clear ladder-like pattern of O-polysaccharide, we optimized the blotting condition by comparing two commonly used blotting methods. Our results suggested that wet transfer is ten times more efficient than semi-dry transfer in polysaccharide detection (Supplementary Fig. S7). The size of the reaction intermediates, i.e. the number of RU in each O-polysaccharide chain, was evaluated by comparing the interval between the product bands and Und-PP-RU (+1 RU) with that of LPS bands. By monitoring the reaction process at different times, we observed multiple product bands at 50 (Supplementary Fig. S8 A) and 120 hours (Fig. 4b). The intermediates bands at 50 hours were weak, but became stronger at 120 hours, especially for those at low molecular weight. It was impressive that in both reactions more short intermediate products such as +2 units and +3 units and less long products such as +4 units, +5 units, +6 units or more were observed. Comparing to the LPS we isolated from E. coli (Δwzy) plus complementary His10-Wzy plasmid (Fig. 4c lane 1, 2), and from E. coli (Δwzz) (Fig. 4c, lane 3), which represent the bands profile from a completed reaction, we reasoned that Wzy adopted a distributive mechanism. On the basis of the principle of distributive mechanism, the reaction product should leave the active site of the polymerase after each round of reaction. Thus, low molecular weight intermediates (such as +2 units), but not high molecular weight products, would be accumulative before next round of reactions. Therefore, we performed a shorter time reaction and observed that the +2 units can be detected within 7–20 hours without any detectable higher MW products (Fig. 4d). Taken together, our results definitely support a distributive mechanism of Wzy in O-antigen synthesis.
Discussion
Wzy-dependent O-antigen biosynthetic pathway is widely distributed in Gram-negative bacteria. A putative inner membrane (IM) protein complex including Wzx, Wzy and Wzz is involved in this pathway (Marolda et al. 2006). Their functions are known through genetic and biochemical analyses. However, the difficulty in heterogeneous expression of these membrane proteins represents a challenge to further characterize them. In this work, we attempted to improve overexpression and purification of Wzy under the direction of a recent finding. Successful overexpression of Wzy allowed us to study its catalytic mechanism in vitro. A recent study provided a possible structural basis that is compatible with the distributive mechanism we suggested in this study. Through mutagenesis studies (Islam et al. 2010), Islam et al uncovered two larger periplasmic loops (PL3 and PL5) in PAO1 Wzy protein that were essential to Wzy catalyzed reaction. More interestingly, they noticed that those two loops have conserved amino acids sequence but dramatically different overall isoelectronic value (pI 8.59 for PL3 and pI 5.49 for PL5). In a model called “catch and release”, PL3, due to its net-negative charge, was proposed to “capture” the donor molecule; while PL5, the largest loops with RX10G motif and net-positive charge, was suggested to function as the active site as well as the product release site. E. coli O86 Wzy is different from those from PAO1 in sequence length and number of transmembrane helix. According to the TMHMM v2.0 prediction (Krogh et al. 2001; Sonnhammer et al. 1998), the O86 Wzy have only 10 transmembrane domain which is much less than PAO1 Wzy (14 TM domains); and there is not significant amino acid similarity between PO1 and O86 Wzy. However, we observed the similar topological components in E. coli O86 Wzy. The third and fourth periplasmic loops in O86 Wzy are the two largest ones. The isoelectronic point analysis showed that the pI value of the third loops is 9.08 (equivalent of PL3 in PAO1 Wzy) and that of the fourth loops is 4.29 (equivalent of PL5 in PAO1 Wzy) (Supplementary Fig. S9). This similarity on membrane topology implies that they adopt the same catalytic mechanism.
A previous study demonstrated that the chain length of O-antigen is regulated in the polymerization step (Daniels et al. 2002). The in vitro assay further shows that Wzz and Wzy are sufficient to produce O-antigen with specific chain length distribution (Woodward et al. 2010). We further speculated on the basis of our recent results that the polymerization occurs inside Wzz complex until the mature O-antigen formed (Guohui Zhao et al, unpublished results). Thus, the distributive mechanism of Wzy would have helped us to understand the role of Wzz. Given that Wzy is distributive, the reaction intermediates leaving Wzy after each round of reaction will either re-enter Wzy for polymerization or be captured by WaaL for ligation forming LPS in the absence of Wzz oligomeric protein complex, leading to the stochastic distribution of LPS. However, the presence of Wzz complex that can form a spacious intramolecular cavity for Wzy reaction may favor the polymerization and regulate to form a long but unique length of O-antigen. In the regard, the presence of Wzz protein complex may be required by the catalytic mechanism of Wzy in order to produce evolutionarily favorable specific length of O-antigen.
In summary, we have improved the overexpression and purification of Wzy by fusing a N-terminal tag with weak mRNA folding free energy. BN-PAGE suggests that it is a dimer under native conditions. Our enzymatic assay with varying reaction times provided the first direct biochemical evidence that indicates that Wzy adopts a distributive mechanism to synthesize O-antigens.
Supplementary Material
Acknowledgments
P.G.W. acknowledges the NIH R01 GM085267 for financial support.
Footnotes
CONFLICT OF INTEREST
The authors declare that they have no conflict of interest.
References
- Arinaminpathy Y, Khurana E, Engelman DM, Gerstein MB. Computational analysis of membrane proteins: the largest class of drug targets. Drug Discov Today. 2009;14(23–24):1130–5. doi: 10.1016/j.drudis.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter EP, Beis K, Cameron AD, Iwata S. Overcoming the challenges of membrane protein crystallography. Curr Opin Struct Biol. 2008;18(5):581–6. doi: 10.1016/j.sbi.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng SS, Xue M, Garner RA, Kadokura H, Boyd D, Beckwith J, Kahne D. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science. 2012;337(6102):1665–8. doi: 10.1126/science.1227215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels C, Griffiths C, Cowles B, Lam JS. Pseudomonas aeruginosa O-antigen chain length is determined before ligation to lipid A core. Environ Microbiol. 2002;4(12):883–97. doi: 10.1046/j.1462-2920.2002.00288.x. [DOI] [PubMed] [Google Scholar]
- Daniels C, Vindurampulle C, Morona R. Overexpression and topology of the Shigella flexneri O-antigen polymerase (Rfc/Wzy) Mol Microbiol. 1998;28(6):1211–22. doi: 10.1046/j.1365-2958.1998.00884.x. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL, Oatman LC, Gay DF. Rapid chemoenzymatic synthesis of monodisperse hyaluronan oligosaccharides with immobilized enzyme reactors. J Biol Chem. 2003;278(37):35199–203. doi: 10.1074/jbc.M306431200. [DOI] [PubMed] [Google Scholar]
- Duda KA, Lindner B, Brade H, Leimbach A, Brzuszkiewicz E, Dobrindt U, Holst O. The lipopolysaccharide of the mastitis isolate Escherichia coli strain 1303 comprises a novel O-antigen and the rare K-12 core type. Microbiology. 2011;157(Pt 6):1750–60. doi: 10.1099/mic.0.046912-0. [DOI] [PubMed] [Google Scholar]
- Greenfield LK, Richards MR, Li J, Wakarchuk WW, Lowary TL, Whitfield C. Biosynthesis of the polymannose lipopolysaccharide O antigens from Escherichia coli serotypes O8 and O9a requires a unique combination of single- and multi-active site mannosyltransferases. J Biol Chem. 2012 doi: 10.1074/jbc.M112.401000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yi W, Shao J, Lu Y, Zhang W, Song J, Wang PG. Molecular analysis of the O-antigen gene cluster of Escherichia coli O86:B7 and characterization of the chain length determinant gene (wzz) Appl Environ Microbiol. 2005;71(12):7995–8001. doi: 10.1128/AEM.71.12.7995-8001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Wu B, Li L, Zhao G, Woodward R, Pettit N, Cai L, Thon V, Wang PG. Defining function of lipopolysaccharide O-antigen ligase WaaL using chemoenzymatically synthesized substrates. J Biol Chem. 2012;287(8):5357–65. doi: 10.1074/jbc.M111.308486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug I, Couturier MR, Rooker MM, Taylor DE, Stein M, Feldman MF. Helicobacter pylori lipopolysaccharide is synthesized via a novel pathway with an evolutionary connection to protein N-glycosylation. PLoS Pathog. 2010;6(3):e1000819. doi: 10.1371/journal.ppat.1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihssen J, Kowarik M, Dilettoso S, Tanner C, Wacker M, Thony-Meyer L. Production of glycoprotein vaccines in Escherichia coli. Microb Cell Fact. 2010;9:61. doi: 10.1186/1475-2859-9-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam ST, Taylor VL, Qi M, Lam JS. Membrane topology mapping of the O-antigen flippase (Wzx), polymerase (Wzy), and ligase (WaaL) from Pseudomonas aeruginosa PAO1 reveals novel domain architectures. MBio. 2010;1(3) doi: 10.1128/mBio.00189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Sebastian S, Pinkham JT, Ross RA, Blalock LT, Kasper DL. Characterization of the O-antigen polymerase (Wzy) of Francisella tularensis. J Biol Chem. 2010;285(36):27839–49. doi: 10.1074/jbc.M110.143859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305(3):567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324(5924):255–8. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari K, Weigel PH. Molecular cloning, expression, and characterization of the authentic hyaluronan synthase from group C Streptococcus equisimilis. J Biol Chem. 1997;272(51):32539–46. doi: 10.1074/jbc.272.51.32539. [DOI] [PubMed] [Google Scholar]
- Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol Rev. 2002;26(1):17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- Marolda CL, Tatar LD, Alaimo C, Aebi M, Valvano MA. Interplay of the Wzx translocase and the corresponding polymerase and chain length regulator proteins in the translocation and periplasmic assembly of lipopolysaccharide o antigen. J Bacteriol. 2006;188(14):5124–35. doi: 10.1128/JB.00461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J Mol Biol. 1999;288(5):911–40. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- May JF, Splain RA, Brotschi C, Kiessling LL. A tethering mechanism for length control in a processive carbohydrate polymerization. Proc Natl Acad Sci U S A. 2009;106(29):11851–6. doi: 10.1073/pnas.0901407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renart J, Reiser J, Stark GR. Transfer of proteins from gels to diazobenzyloxymethyl-paper and detection with antisera: a method for studying antibody specificity and antigen structure. Proc Natl Acad Sci U S A. 1979;76(7):3116–20. doi: 10.1073/pnas.76.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz N, Kahne D, Silhavy TJ. Transport of lipopolysaccharide across the cell envelope: the long road of discovery. Nat Rev Microbiol. 2009;7(9):677–83. doi: 10.1038/nrmicro2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz D, Dotsch V, Bernhard F. Production of membrane proteins using cell-free expression systems. Proteomics. 2008;8(19):3933–46. doi: 10.1002/pmic.200800171. [DOI] [PubMed] [Google Scholar]
- Silhavy TJ, Kahne D, Walker S. The bacterial cell envelope. Cold Spring Harb Perspect Biol. 2010;2(5):a000414. doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- Sucher AJ, Chahine EB, Nelson M, Sucher BJ. Prevnar 13, the new 13-valent pneumococcal conjugate vaccine. Ann Pharmacother. 2011;45(12):1516–24. doi: 10.1345/aph.1Q347. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76(9):4350–4. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter AE, Turner DH, Kim J, Lyttle MH, Muller P, Mathews DH, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc Natl Acad Sci U S A. 1994;91(20):9218–22. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang Q, Reeves PR. The variation of O antigens in gram-negative bacteria. Subcell Biochem. 2010;53:123–52. doi: 10.1007/978-90-481-9078-2_6. [DOI] [PubMed] [Google Scholar]
- Wetter M, Kowarik M, Steffen M, Carranza P, Corradin G, Wacker M. Engineering, conjugation, and immunogenicity assessment of Escherichia coli O121 O antigen for its potential use as a typhoid vaccine component. Glycoconj J. 2013;30(5):511–22. doi: 10.1007/s10719-012-9451-9. [DOI] [PubMed] [Google Scholar]
- Wheatley RW, Zheng RB, Richards MR, Lowary TL, Ng KK. Tetrameric Structure of the GlfT2 Galactofuranosyltransferase Reveals a Scaffold for the Assembly of Mycobacterial Arabinogalactan. J Biol Chem. 2012;287(33):28132–43. doi: 10.1074/jbc.M112.347484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward R, Yi W, Li L, Zhao G, Eguchi H, Sridhar PR, Guo H, Song JK, Motari E, Cai L, Kelleher P, Liu X, Han W, Zhang W, Ding Y, Li M, Wang PG. In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat Chem Biol. 2010;6(6):418–23. doi: 10.1038/nchembio.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Liu X, Li Y, Li J, Xia C, Zhou G, Zhang W, Zhao W, Chen X, Wang PG. Remodeling bacterial polysaccharides by metabolic pathway engineering. Proc Natl Acad Sci U S A. 2009;106(11):4207–12. doi: 10.1073/pnas.0812432106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W, Shao J, Zhu L, Li M, Singh M, Lu Y, Lin S, Li H, Ryu K, Shen J, Guo H, Yao Q, Bush CA, Wang PG. Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J Am Chem Soc. 2005;127(7):2040–1. doi: 10.1021/ja045021y. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Barrett D, Zhang Y, Kahne D, Sliz P, Walker S. Crystal structure of a peptidoglycan glycosyltransferase suggests a model for processive glycan chain synthesis. Proc Natl Acad Sci U S A. 2007;104(13):5348–53. doi: 10.1073/pnas.0701160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Liu J, Liu X, Chen M, Zhang H, Wang PG. Cloning and characterization of GDP-perosamine synthetase (Per) from Escherichia coli O157:H7 and synthesis of GDP-perosamine in vitro. Biochem Biophys Res Commun. 2007;363(3):525–30. doi: 10.1016/j.bbrc.2007.08.184. [DOI] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–15. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.