Abstract
Purpose
The SET oncoprotein, a potent inhibitor of the protein phosphatase 2A (PP2A), is overexpressed in leukemia. We evaluated the efficacy of SET antagonism in chronic myeloid leukemia (CML) and acute myeloid leukemia (AML) cell lines, a murine leukemia model, and primary patient samples using OP449, a specific, cell-penetrating peptide that antagonizes SET's inhibition of PP2A.
Experimental Design
In vitro cytotoxicity and specificity of OP449 in CML and AML cell lines and primary samples were measured using proliferation, apoptosis and colonogenic assays. Efficacy of target inhibition by OP449 is evaluated by immunoblotting and PP2A assay. In vivo antitumor efficacy of OP449 was measured in human HL-60 xenografted murine model.
Results
We observed that OP449 inhibited growth of CML cells including those from patients with blastic phase disease and patients harboring highly drug-resistant BCR-ABL1 mutations. Combined treatment with OP449 and ABL1 tyrosine kinase inhibitors was significantly more cytotoxic to K562 cells and primary CD34+ CML cells. SET protein levels remained unchanged with OP449 treatment, but BCR-ABL1-mediated downstream signaling was significantly inhibited with the degradation of key signaling molecules such as BCR-ABL1, STAT5, and AKT. Similarly, AML cell lines and primary patient samples with various genetic lesions showed inhibition of cell growth after treatment with OP449 alone or in combination with respective kinase inhibitors. Finally, OP449 reduced the tumor burden of mice xenografted with human leukemia cells.
Conclusions
We demonstrate a novel therapeutic paradigm of SET antagonism using OP449 in combination with tyrosine kinase inhibitors for the treatment of CML and AML.
Keywords: CML, AML, SET, PP2A, Tyrosine Kinase inhibitors
Introduction
Tyrosine kinases play critical biological roles in the pathogenesis of chronic and acute leukemia. A ground-breaking advance came with the identification of the constitutively active fusion tyrosine kinase, BCR-ABL1, which causes chronic myeloid leukemia (CML) (reviewed in (1)). Similarly, most acute myeloid leukemia (AML) cells exhibit constitutive phosphorylation of signal transducer and activator of transcription 5 (STAT5), a marker for tyrosine kinase activity (2). The mechanism of STAT5 activation is explained by genetic abnormalities in FLT3, KIT, PDGFR, JAK1, and JAK2 kinases in only 35% of AML cases, which suggests that unidentified mechanisms of kinase dysregulation are active in the remainder of these patients.
Clinically, the most successful example of targeted therapy for any cancer has been imatinib (Gleevec; STI571), a small molecule ABL1 tyrosine kinase inhibitor that has been frontline treatment for CML for over a decade. More than 80% of newly diagnosed chronic phase CML patients achieve durable complete cytogenetic response (CCyR) on imatinib therapy (3). However, 20-25% of chronic phase patients exhibit primary resistance to imatinib or relapse after an initial response. Furthermore, among patients who progress to accelerated or blastic phase disease, responses to imatinib are significantly less frequent and almost always transient. Various mechanisms have been found to account for the resistance to imatinib including BCR-ABL1 kinase-dependent mechanisms (4-6) or BCR-ABL1 kinase-independent mechanisms (7-9). The additional ABL1 kinase inhibitors dasatinib (10, 11) and nilotinib (12-14) have been shown to inhibit many kinase domain-mutant forms of BCR-ABL1 that are resistant to imatinib (15), and recently ponatinib has proven effective in patients carrying the highly recalcitrant T315I mutation (16, 17). However, selected BCR-ABL1 compound mutations (two or more kinase domain point mutations in the same BCR-ABL1 molecule) have been implicated in resistance to all current clinical ABL1 kinase inhibitors (16, 18, 19).
The treatment of patients with AML has proven to be more challenging, primarily due to the significant heterogeneity of molecular abnormalities driving the disease (20). Indeed, the majority of disease-causing aberrant molecular pathways that could serve as therapeutic targets in AML remain unknown. Despite significant progress in the treatment of AML, most patients still do not achieve complete remission (CR) and about 40-50% of patients who have reached CR eventually relapse (20).
Emerging evidence suggests that there is a tight regulation of phosphatase and kinase activity in cancer cells (21). Accordingly, protein phosphatase 2A (PP2A) represents a novel potential therapeutic target in various leukemias (22-29). The PP2A enzyme is a serine/threonine phosphatase that acts as a tumor suppressor and plays a critical role in the regulation of cell cycle progression, survival, and differentiation (30). It has been shown that PP2A activity is significantly reduced in patients with blastic phase CML, Philadelphia chromosome-positive acute lymphoblastic leukemia (Ph+ ALL), and AML (22-25). Inactivation of PP2A in these cells is due in a subset of cases to increased accumulation of the SET oncoprotein, an endogenous inhibitor of PP2A, and accounts for increased and sustained kinase activity in leukemic cells. Notably, restoration of PP2A activity in both CML and AML cells to normal levels through overexpression of the PP2A catalytic subunit (PP2Ac), pharmacological activation by FTY720 (fingolimod), or silencing of SET resulted in reduced tumorigenesis in vitro and in vivo (22-24). Therefore, given the central role of PP2A and SET in regulating various downstream signaling pathways, pharmacological activation of PP2A via SET antagonism may provide a potential therapeutic approach for patients with high expression of SET including patients with drug-resistant leukemias.
Recently, we reported the discovery of a novel compound known as OP449 (formerly COG449) which is a specific, physiologically stable, cell-penetrating peptide that binds to SET and antagonizes SET's inhibition of PP2A (29, 31, 32). Additionally, SET levels is significantly increased in AML samples and its expression is correlated with a poor disease outcome (33). These evidence lead to our combined efforts to analyze the effect to SET antagonism using OP449 for the treatment of CML and AML. Furthermore, we evaluated the effects of SET antagonism utilizing OP449 in combination with specific tyrosine kinase inhibitors in CML and AML cells. We found that OP449 is selectively cytotoxic to leukemic cell lines as well as primary patient cells including cells with tyrosine kinase inhibitor-resistant BCR-ABL1 kinase mutations. Our results demonstrate that SET antagonism with OP449 in combination with tyrosine kinases inhibitors provides more efficient and selective inhibition of leukemia cell growth for a broad range of oncogenic lesions as compared to normal cells. To our knowledge, this is the first report showing that SET antagonism in combination with standard targeted therapies may provide an improved treatment option for leukemia patients.
Materials and Methods
Cell culture
Certified K562, and LAMA cells were obtained from the American Type Culture Collection. CMK, HL-60, GDM-1, and UT-7 were obtained from the German National Resource Center for Biological Material. MOLM-14 cells were generously provided by Dr. Yoshinobu Matsuo (Fujisaki Cell Center, Hayashibara Biochemical Labs, Okayama, Japan) (34). Parental Ba/F3 cells were maintained in RPMI 1640 medium supplemented with 2 mM L-glutamine, 10% FCS, 100 U/mL penicillin, 100 μg/mL streptomycin and 10% WEHI conditioned media for source of IL-3. Ba/F3 transfectants expressing wild-type BCR-ABL1, BCR-ABL1T315I, or BCR-ABL1E255V/T315I were generated and maintained as described previously (16, 19, 35). None of the cell lines used in this study was cultured for longer than 6 months from initial purchase or characterization. All cell lines were cultured in the recommended culture medium at 37°C in 5% CO2. OP449 (Molecular weight is 9223 g/mol) is reconstituted in PBS as a 10 mM stock. OP449 was prepared as previously described (29). For convenience we have also included peptide sequence and synthesis details in the supplemental data. For proliferation assays, cells were incubated for 72 hrs in the presence or absence of the tyrosine kinase inhibitors including imatinib, nilotinib, dasatinib or ponatinib (LC Lab, Woburn, MA), OP449 with or without okadaic acid (LC Labs), FTY720 (Cayman Chemical, Ann Arbor, MI), JAK Inhibitor I (Millipore, Billerica, MA), or AC220 (Selleck, Houston, TX). The number of viable cells was determined by Cell Titer 96 Aqueous One solution cell proliferation assay (Promega, Madison, WI). For apoptosis assays, cells were incubated for 48 hrs in the presence of the indicated inhibitor(s), and the number of Annexin V+ cells was determined using the Guava Nexin kit (Millipore).
PP2A Assay: PP2A activity was determined as previously described (22, 23, 29, 36) using commercially available assay (Upstate Biotechnology, 17-313). Briefly, protein lysates were prepared in 20 mM imidazole-HCl, 2 mM EDTA, 2 mM EGTA, pH 7.0 with 10 μg/ml each of aprotinin leupeptin, pepstatin, 1 mM benzamidine, 1mM PMSF and phosphatase inhibitors tablets (Roche). 50 μg of protein was immunoprecipitated with 4 μg of anti-PP2A antibody (1D6; Upstate Biotechnology) and 50 μL of protein-A-agarose beads for 2 hrs at 4°C. Beads were washed extensively with lysis buffer first and with Ser/Thr assay buffer last and than used in the phosphatase reaction for measuring dephosphorylation of the phosphopeptide (K-R-pT-I-R-R) according to the manufacturer's protocol using malachite green phosphate detection solution. The level of free phosphate is normalized to total amount of PP2Ac immunoprecipitated as measured by densitometry analysis of immunoblots.
Patient samples
All CML and AML samples were obtained after patients have provided written and oral informed consents prior to their participation in the study (Table S1 and S2). The study was reviewed and approved by the institutional review boards at the Oregon Health & Science University; University of Texas Southwestern Medical Center, Dallas; and M. D. Anderson Cancer Center, Houston. Bone marrow from normal donors was purchased commercially (Lonza, Walkersville, MD). Mononuclear cells (MNCs) were isolated by centrifugation through a Ficoll gradient, and red blood cells were lysed using an aqueous solution of 0.15 M NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA. CD34+ cells were enriched using an immunomagnetic column according to the manufacturer's protocol (Miltenyi Biotech) and purity was determined by fluorescence activated cell sorting (FACS) using a FACSAria instrument (BD Biosciences, San Jose, CA). For proliferation assays, patient sample MNCs were maintained in RPMI-1640 medium supplemented with 2 mM L-glutamine, 10% FBS, penicillin (100 U/mL), streptomycin (100 μg/mL), and 100 μM β-mercaptoethanol.
Colony assays
CML and normal CD34+ cells were plated in triplicate in Methocult H4534 (Stem Cell Technologies, Vancouver, BC, Canada) with varying concentrations of tyrosine kinase inhibitors and OP449 alone or in combination. Colony forming units–granulocyte macrophage (CFU-GM) colonies were counted after 2 weeks of incubation at 37°C in 5% CO2.
Immunoblotting
CML and AML cell lines were cultured in the presence or absence of OP449. Following the indicated drug exposure time, cells were washed in PBS and lysed in 50 μL lysis buffer (Cell Signaling, Boston, MA) supplemented with complete protease inhibitor and phosphatase inhibitor cocktail-2 (Sigma-Aldrich, St. Louis, MO). Equal amounts of protein were fractionated on 4-15% Tris-glycine polyacrylamide gels (Bio-Rad, Hercules, CA), transferred to PVDF membranes, and probed with the indicated antibodies: BCR, pABL, STAT5, pSTAT5, AKT, pAKT (S473), pAKT (T308), ERK1/2, pERK1/2, P38/MAPK, pP38/MAPK, S6, pS6 ribosomal (S235/236) (Cell Signaling), SET, PP2Ac (Millipore), and α-tubulin (Sigma-Aldrich).
Animal experiments
RAG2-/-γc-/- mice (20 g) were bred in-house. Animals received a standard diet and water in the Animal Core Facilities of the Center for Applied Medical Research (University of Navarra). The protocol for animal experiments was approved by the University of Navarra Animal Experimentation Ethics Committee. Xenografts were established after injecting HL-60 cells (4×105 cells/animal) in non-irradiated mice. Treatment was initiated 7 days post-implantation of HL-60 cells with 5mg/kg OP449 or lactated ringer solution control by intraperitoneal injection every 3 days.
Statistical methods
Continuous variables were compared by pairwise Student's t-test for two independent samples using Excel software. IC50 values were generated using GraphPad Prism software. Combination indices (CI) were calculated using Calcusyn software. A CI value less than 1.0 is represents a synergistic drug combination. A p value less than 0.05 was considered statistically significant. * denotes p<0.05, ** denotes p<0.01, and *** denotes p<0.001.
Results
OP449, a peptide antagonist of SET, inhibits growth of CML cell lines including cells harboring drug-resistant BCR-ABL1 kinase domain mutations
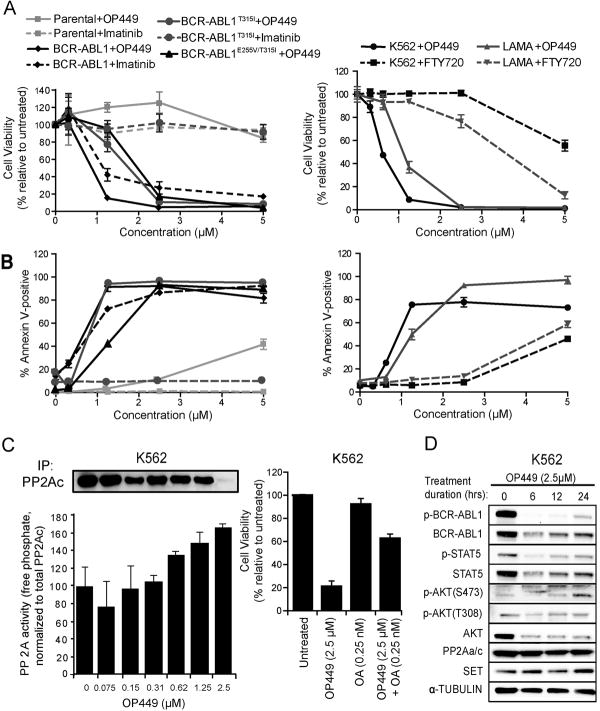
The SET oncoprotein contributes to various cancers by inhibiting the tumor suppressor PP2A (reviewed in (26)). Recently we reported that OP449 and its precursors selectively binds to SET and increased PP2A activity in leukemia cells (29, 31, 32). Therefore, we first evaluated the efficacy of OP449 against BCR-ABL1-positive CML cells. OP449 treatment resulted in selective, dose-dependent inhibition of murine Ba/F3 cells expressing wild-type BCR-ABL1 (IC50: 0.89 μM), while parental Ba/F3 cells exhibited no measurable cytotoxicity (Fig. 1A). Importantly, OP449 also demonstrated activity against both the imatinib-insensitive BCR-ABL1T315I point mutant and the BCR-ABL1E255V/T315I compound mutant (IC50: 1.62 and 1.97 μM, respectively), the latter of which confers high-level resistance to all currently available ABL1 kinase inhibitors including ponatinib (16, 19). The efficacy of OP449 for BCR-ABL1-expressing cells extended to the human CML lines K562 and LAMA (IC50: 0.60 and 1.11 μM, respectively). Further, we found that OP449 was approximately three to eight-fold more potent than FTY720 (a previously tested pharmacological activator of PP2A)(22, 24, 28) at inhibiting the growth of K562 cells (IC50: 0.60 vs. 4.68 μM, respectively) and LAMA cells (IC50: 1.11 vs. 3.25 μM, respectively; Fig. 1A,B, right panels). Reduction in viability of BCR-ABL1-positive cells after treatment with OP449 also correlated with induction of apoptosis in both murine and human CML cell lines (Fig. 1B). Importantly, we found that the treatment of K562 cells with OP449 increased PP2A activity in a dose-dependent manner. For example, a 67% increase in PP2A activity was observed upon treatment with 2.5 μM OP449 (p= 0.01) (Fig. 1C, left panel). In addition, we confirmed that OP449 activity was mediated through activation of PP2A as the effect of OP449 on the viability of K562 cells was significantly rescued by co-treatment with okadaic acid, a known PP2A inhibitor (37) (Fig. 1C, right panel).
Figure 1. OP449 inhibits growth, increases apoptosis, and decreases BCR-ABL1 signaling activity in CML cells by specifically activating PP2A.
(A) Effect of OP449 on growth of BCR-ABL1-positive cells. Murine Ba/F3 cells expressing BCR-ABL1, BCR-ABL1T315I or BCR-ABL1E255V/T315I and human CML cells (K562 and LAMA) were cultured in graded concentrations of OP449, imatinib or FTY720 and cell viability was measured at 72 hrs by standard MTS assay. Parental Ba/F3 cells were included as a control. Results are graphed as the mean percent viability relative to untreated cells ± standard deviation. (B) Induction of apoptosis by OP449 in BCR-ABL1-positive cells. Murine Ba/F3, Ba/F3-BCR-ABL1, Ba/F3-BCR-ABL1T315I and Ba/F3-BCR-ABL1E255V/T315I cells and human CML cells (K562 and LAMA) were cultured in graded concentrations of OP449, imatinib or FTY720 and apoptosis was measured at 72 hrs. Results are graphed as the mean percent annexin-positive cells ± standard deviation. (C) Left Panel: Reactivation of PP2A in K562 cells following OP449 treatment. K562 cells were exposed to graded concentrations of OP449 for 24 h and free phosphate levels were measured. Results are presented as free phosphate levels normalized to the total amount of immunoprecipitated PP2Ac protein as detected by western blot analysis± standard deviation. Right Panel: Rescue of OP449-mediated inhibition by okadaic acid. K562 cells were treated with OP449 and okadaic acid (OA) either alone or in combination for 72 hrs and cell viability was measured by standard MTS assay. Bars represent the mean percent viability relative to untreated cells ± standard deviation. (D) Inhibition of BCR-ABL1 signaling by OP449. K562 cells were cultured for the indicated times in the presence of OP449, and whole cell lysates were subjected to SDS-PAGE and immunoblotted using the indicated antibodies. All experiments shown are representative of three independent experiments.
To determine if OP449-mediated activation of PP2A modulates the BCR-ABL1 kinase signaling pathway, we treated K562 cells with OP449 in a time-dependent manner and found significantly reduced levels of both phosphorylated and total BCR-ABL1, AKT, and STAT5 (Fig. 1D). Notably, signaling appeared to be partially restored after 24 hrs treatment, suggesting potential depletion or degradation of functional OP449 by this time point. In addition, after 24 hrs of OP449 treatment, phosphorylation of AKT at S473 but not at T308 was slightly increased as compared to the basal levels despite of the reduced total protein level. The reason for this differential regulation is not clear. The ability of OP449 to decrease tyrosine phosphorylation and degradation of BCR-ABL1 is consistent with previous studies where activation of PP2A results in dephosphorylation of BCR-ABL1 through a putative SET/PP2A/SHP-1 pathway (22, 23). Together, our results demonstrate that the SET antagonism-mediated increase in PP2A activity by OP449 efficiently and specifically inhibits the growth of CML cells by attenuating BCR-ABL1 kinase signaling and leading to BCR-ABL1 degradation.
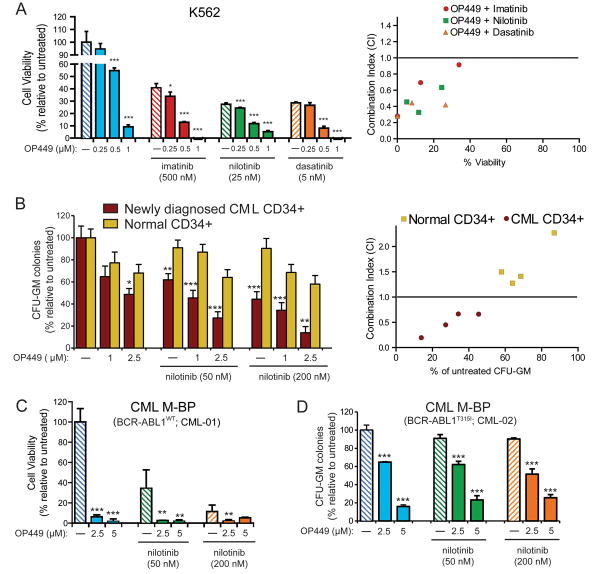
OP449 increases efficacy of ABL1 tyrosine kinase inhibitors in CML cell lines and primary patient samples
A fine balance between kinase and phosphatase activity is required for normal cell growth, and deregulation of this balance may lead to oncogenesis (21, 26). Therefore, we hypothesized that the combined targeting of both pathways might provide therapeutic advantage for targeting leukemic cells. We found that the treatment of K562 cells with OP449 in combination with the ABL1 tyrosine kinase inhibitors imatinib, nilotinib, dasatinib, or ponatinib showed significantly increased cytotoxicity as compared to each compound alone (Fig. 2A, Supplemental figure 1B). For example, while treatment of K562 cells with 500 nM imatinib or 500 nM OP449 alone, reduced viability of K562 cells by approximately 60% and 45%, respectively, the combination treatment at these concentrations resulted in significant and synergistic (CI value: 0.695) reduction in cell viability by 87% (Fig 2A, Table S3). Combination of OP449 and ABL1 kinase inhibitors in K562 cells also demonstrated enhanced inhibition of BCR-ABL1 signaling activity, though combination treatment compared to single agent ABL1 inhibitor treatment afforded only a slight increase over the already thorough signaling inhibition achievable with ABL tyrosine kinase inhibitors. In addition, the treatment of K562 cell with OP449 reduced BCR-ABL1 protein levels (Fig. S1A). Therefore, from a mechanistic point of view, the primary advantage of combining OP449 treatment with ABL1 tyrosine kinase inhibitors in CML cells is simultaneous reduction in BCR-ABL1 protein levels that contributes to increased efficacy of combination therapy.
Figure 2. The combination of OP449 with ABL1 tyrosine kinase inhibitors is more effective than single treatments and selective for CML cells over normal cells.
(A) Combined effects of OP449 and ABL1 kinase inhibitors on proliferation of CML cells. K562 cells were cultured in the presence of OP449 alone or in combination with imatinib, nilotinib, or dasatinib, and viability was measured at 72 hrs by standard MTS assay. Bars represent the mean percent viability relative to untreated cells ± standard deviation. A combination index (CI) value less than 1.0 indicates drug combinations which have a synergistic effect on inhibition of cell growth. (B) Effect of OP449 in combination with nilotinib on primary CML cells. CD34+ cells isolated from newly diagnosed CML patients (N=4) or healthy donors (N=4) were plated in methocult containing cytokines in the presence of OP449 and nilotinib alone or in combination, and colonies were counted after 2 weeks. Bars represent the mean percent of untreated ± standard deviation. * denotes p<0.05, ** denotes p<0.01 and *** denotes p<0.001, where the effect of treatments on CML CD34+ cells are compared with the respective treatments in normal CD34+ cells. A combination index (CI) value less than 1.0 indicates drug combinations which have a synergistic effect on inhibition of cell growth. (C) Efficacy of OP449 in primary myeloid blastic phase (M-BP) CML samples harboring wild-type BCR-ABL1. Samples were cultured in the presence of OP449 alone or in combination with nilotinib and proliferation was measured at 72 hrs by standard MTS assay. Bars represent the mean percent of untreated ± standard deviation. (D) Efficacy of OP449 in primary ABL1 kinase inhibitor-resistant CML cells. Similar CFU-GM colony assays were performed using CD34+ cells isolated from a CML M-BP patient harboring a BCR-ABL1T315I mutation and treated with OP449 and nilotinib alone and in combination. Bars represent the mean percent of untreated ± standard deviation. For panels A, C, and D, * denotes p<0.05, ** denotes p<0.01 and *** denotes p<0.001 when treatment with OP449 alone was compared to untreated or combination treatment was compared to the respective tyrosine kinase inhibitors alone.
Although most of the ABL1 inhibitor-OP449 combinations tested achieved comparable levels of cytotoxicity, given nilotinib's highly potent BCR-ABL1 inhibition and its narrow kinase target selectivity profile (12, 13), we performed further combination experiments with this compound in primary CML cells. While treatment of primary CD34+ CML cells with either 2.5 μM OP449 or 200 nM nilotinib individually resulted in a roughly 50% reduction in colony formation, combination of OP449 and nilotinib at these concentrations reduced colony formation by approximately 87%, suggesting synergistic reduction of clonogenicity by the combination (CI value: 0.195; Fig. 2B, Table S3). Importantly, we compared the effect of OP449 on cell growth of normal and CML human CD34+ cells and observed significantly enhanced reduction in colony formation of primary CML CD34+ cells as compared to normal controls at all tested combination treatment doses (Fig. 2B). These findings suggest a significant potential therapeutic window of selective efficacy of OP449-ABL1 inhibitor combination treatment for CML cells. OP449 was also highly effective at inhibiting growth of primary cells from CML blastic phase patients harboring either wild-type BCR-ABL1 (Fig. 2C) or BCR-ABL1T315I (Fig. 2D), with 2.5 μM OP449 affording ∼90% and ∼35% inhibition, respectively, and combination of OP449 with nilotinib provided additional inhibition only in those samples harboring wild-type BCR-ABL1, consistent with insensitivity of the BCR-ABL1T315I mutant to nilotinib. Similar findings were also observed for dasatinib and ponatinib (Fig. S1C,E). Interestingly, OP449 was effective at inhibiting growth of nilotinib and ponatinib resistant primary CML cells (Fig 2D, S1D) by increasing apoptosis (Fig S1F). Taken together, our data suggest the use of SET antagonists such as OP449 in combination with ABL1 tyrosine kinase inhibitors may represent a novel and more efficient therapeutic strategy for the treatment of CML. Furthermore, OP449 alone may be therapeutically beneficial for patients harboring tyrosine kinase inhibitor-resistant BCR-ABL1 mutations.
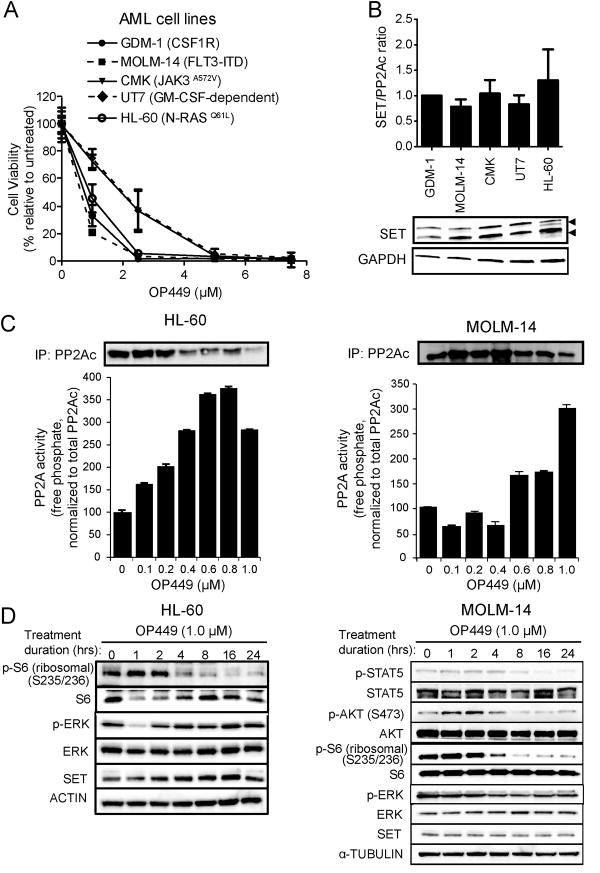
OP449 inhibits the growth of AML cells harboring various genetic lesions
PP2A activity has been previously shown to be significantly reduced in AML cells, and pharmacological restoration of PP2A activity causes growth inhibition of leukemic cells (24, 28). Further, elevated SET levels are significantly correlated with poor disease outcome in AML (33).To evaluate the efficacy of OP449 in AML cells, we first tested a panel of five AML cell lines harboring different genetic lesions (Fig. 3A). MOLM-14 cells (which harbor a FLT3-ITD mutation; IC50: 0.59 μM), GDM-1 (over-expressing CSF1R; IC50: 0.82 μM), and HL-60 (NRASQ61L; IC50: 0.94 μM) were the most sensitive to OP449. Other cell lines demonstrated comparable intermediate sensitivity, including CMK cells (JAK3A572V; IC50: 1.73 μM) and GM-CSF-dependent UT7 cells (IC50: 1.78 μM; Fig. 3A). Each of these cell lines exhibited similar levels of SET expression (Fig. 3B). Similar results were obtained when SET levels were normalized with total PP2Ac levels (Fig. S2A). While the ratio of the SETα and SETβ (lower band) proteins were demonstrated to be prognostic for more aggressive disease in chronic lymphocytic leukemia (CLL) (29), no significant differences in band intensities were noted in the AML cell lines. We also confirmed that treatment with OP449 resulted in a dose-dependent increase in PP2A activity in HL-60 and MOLM-14 cell lines (normalized to total PP2Ac, a subunit of PP2A protein levels; Fig. 3C). Further, we demonstrated time-dependent reduction in levels of phosphorylated S6 ribosomal protein and ERK in HL-60 cells and phosphorylated STAT5, AKT and S6 ribosomal protein in MOLM-14 cells, suggesting efficacy of OP449 at inhibiting downstream signaling in AML cells (Fig. 3D). Of note, the basal phosphorylation of STAT5 and AKT was very low in HL-60 cells, precluding identification of any significant differences over time (data not shown). These results show that OP449 is cytotoxic to AML cells driven by diverse oncogenic genetic lesions.
Figure 3. OP449 inhibits growth and downstream kinase signaling in AML cell lines.
(A) Inhibition of growth of AML cell lines by OP449. Cells were cultured in the presence of graded concentrations of OP449 for 72 hrs and cell viability was analyzed by standard MTS assay. Results are graphed as the mean percent viability relative to untreated controls ± standard deviation. (B) SET expression levels in AML cells lines tested. SET/ACTIN ratios were quantified from densitometric analysis of protein expression in each of the indicated AML cell lines. (C) Reactivation of PP2A in AML cells following OP449 treatment. HL-60 and MOLM-14 cells were exposed to graded concentrations of OP449 for 24 hrs and free phosphate levels were measured. Results are normalized to the total amount of immunoprecipitated PP2Ac protein as detected by western blot analysis ± standard deviation. (D) Inhibition of downstream kinase signaling in AML cells by OP449. HL-60 and MOLM-14 cells were incubated for the indicated durations in the presence of OP449, harvested and lysed, and immunoblotted with the indicated antibodies. All experiments shown are representative of three independent experiments.
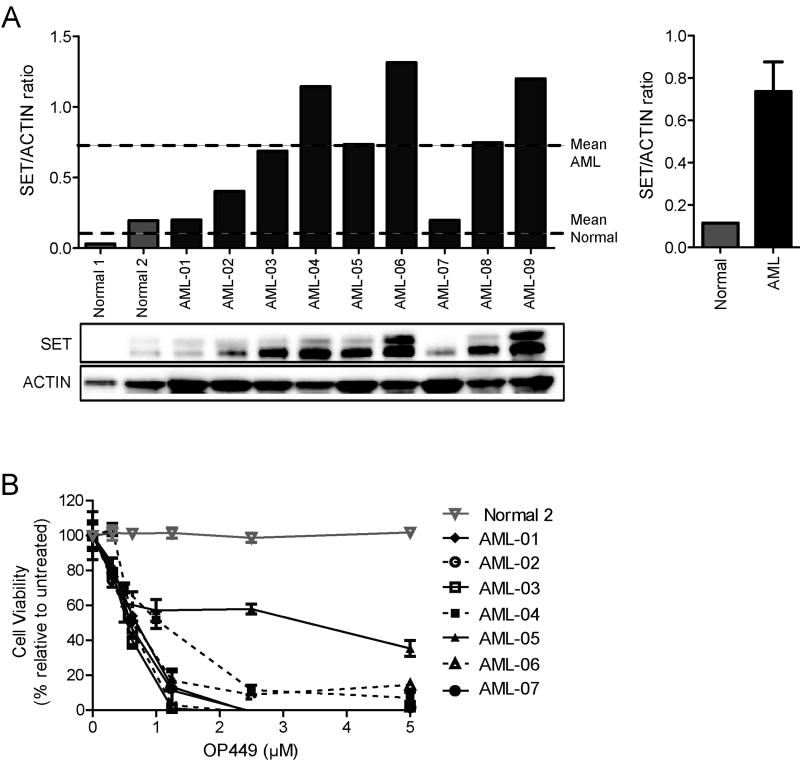
AML patient samples show overexpression of SET and are sensitive to SET inhibition by OP449
A recent study reported that SET is overexpressed in 28% of AML samples and its expression is associated with poor prognosis (33). To determine whether the efficacy of OP449 extends to primary AML samples, we screened a small cohort of nine AML patient samples for expression of SET as compared to normal CD34+ cells. We observed that seven out of nine patient samples show significantly increased SET expression as compared to normal CD34+ cells (Fig. 4A). Similar results were obtained when SET levels were normalized with total PP2Ac levels (Fig. S2B). Additionally, consistent with previous studies (33), we found SET protein levels were significantly increased in a subset of AML specimens (3/9; 30%) when normalized within the disease. No discernible disease subtype appeared overrepresented among those samples with SET overexpression (Table S2). However, examination of a larger patient cohort would be required for more conclusive analysis of disease subtype and to establish correlation between SET expression levels and OP449 sensitivity. Next, we tested the efficacy of OP449 in primary AML samples as compared to normal CD34+ cells and observed that OP449 significantly reduced the viability of AML cells (with IC50 ranges from 0.6 – 0.9 μM for most of the samples except one sample (AML-05) which was not very responsive to OP449 treatment) (Fig. 4B). Overall, similar to our findings in CML cells, OP449 inhibited growth of primary AML patient cells with variety of genetic lesions, including cells from a patient positive for the FLT3-ITD mutation.
Figure 4. SET is overexpressed in primary AML cells and OP449 inhibits growth of primary AML patient cell.
(A) SET expression levels in primary AML samples and normal CD34+ cells tested. SET/ACTIN ratios were quantified from densitometric analysis of protein expression in each of the indicated samples. Dashed lines represent mean SET/ACTIN ratios for normal CD34+ and AML cells, respectively. Bar graph shows mean SET/ACTIN ratios for normal CD34+ and AML cells ± standard error. (B) Inhibition of primary AML cell viability by OP449. Primary AML and normal CD34+ cells were cultured in the presence of graded concentrations of OP449 for 72 hrs and cell viability was analyzed by standard MTS assay. Results are graphed as the mean percent viability relative to untreated controls ± standard deviation.
OP449 inhibits AML tumor growth in a murine leukemia model
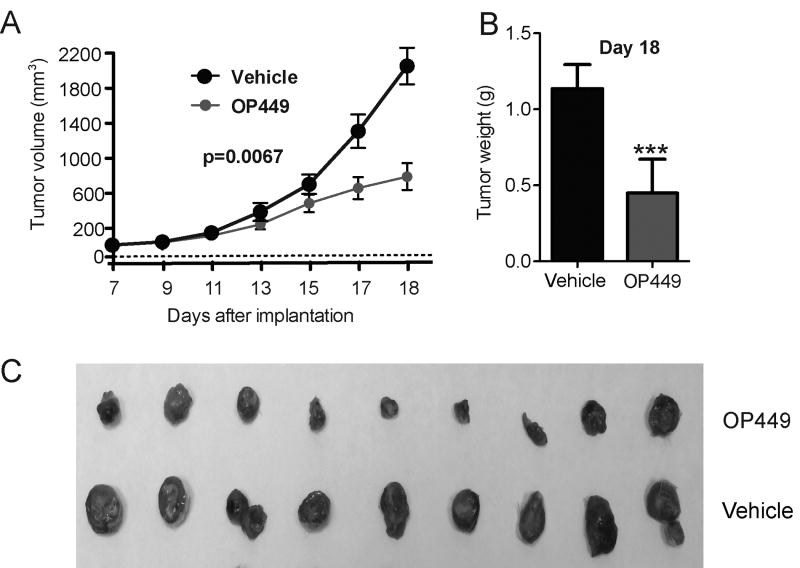
Previous studies have reported pharmacological activation of PP2A reduces leukemia burden in CML cells (21-23). We have also shown that OP449 is effective in reducing tumor growth in a Burkitt's lymphoma model (29). To evaluate this compound's potential antitumor efficacy against AML cells in vivo, we tested efficacy of OP449 in RAG2-/-γc-/- xenograft mice bearing human leukemia HL-60 cell-derived tumors. Animals were treated with 5mg/kg OP449 or vehicle by intraperitoneal injection every 3 days. We observed that treatment with OP449 significantly inhibited tumor growth measured over time (p=0.0067; Fig. 5A) and resulted in a more than two-fold reduction in tumor burden measured at the end of the experiment as compared to vehicle-treated controls (Day 18: 1.14±0.06 g vs. 0.45±0.08 g, respectively; p<0.001; Fig. 5B,C). These results demonstrate the in vivo efficacy of OP449 in a murine leukemia model.
Figure 5. OP449 inhibits AML tumor growth in vivo.
Mice were treated every 3 days with either 5mg/kg OP449 or vehicle control beginning at 7 days post-implantation of HL-60 cells. (A) Inhibition of tumor growth in vivo by OP449. Mean tumor volumes ± standard deviation over the course of treatment are shown. (B) Comparisons of tumor burden between treatments. Final tumor mass for OP449- and vehicle-treated HL-60 tumors harvested on day 18 after implantation was compared. *** denotes p<0.001. (C) Visual representation of extracted tumors is shown.
OP449 synergistically reduces growth of AML cells in combination with relevant kinase inhibitors and chemotherapy
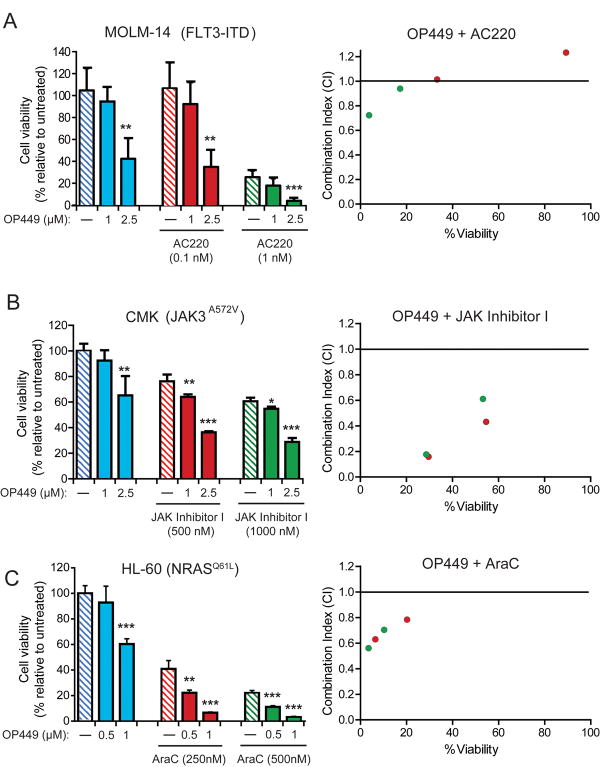
To determine if combined inhibition of relevant tyrosine kinases and SET antagonism is more effective for inhibiting the growth of AML cells, we assessed the effects of combinations of OP449 with either FLT3 or JAK kinase inhibitors in MOLM-14 and CMK cells, respectively. Treatment of MOLM-14 cells with 2.5 μM OP449 or 1 nM AC220 alone resulted in reduced cell viability by 58% and 75%, respectively; combined treatment significantly and synergistically reduced cell growth by close to 96% (CI value: 0.723; Fig. 6A, Table S3). Similarly, for CMK cells we observed ∼40% reduction in viability with 2.5 μM OP449 and ∼22% with 500 nM JAK inhibitor I (a pan-JAK family kinase inhibitor) alone, while the combination reduced viability by approximately 65% (p<0.001 as compared to each drug alone) and was highly synergistic (CI value: 0.158; Fig. 6B, Table S3). Additionally, HL-60 cells showed ∼40% reduced growth with 1 μM OP449 and ∼60% reduced growth with 250 nM cytarabine (AraC), while combination treatment led to a 94% reduction in viability (CI value 0.630; p<0.001 as compared to each drug alone; Fig 6C, Table S3). These results suggest that select combinations of OP449 with relevant tyrosine kinase inhibitors or chemotherapy are capable of improved, synergistic suppression of growth of AML cells harboring various mutationally-activated kinase pathways.
Figure 6. Combination of OP449 with tyrosine kinase inhibitors or chemotherapy enhances inhibition of AML cell growth.
(A) Combined treatment of FLT3-ITD positive AML cells with OP449 and a FLT3 inhibitor. MOLM-14 cells (FLT3-ITD) were incubated for 72 hrs with OP449 and AC220 alone or in combination and cell viability was measured by standard MTS assay. (B) Combined treatment of JAK3-mutant AML cells with OP449 and a pan-JAK inhibitor. CMK cells (JAK3A572V) were incubated for 72 hrs with OP449 and JAK Inhibitor I alone or in combination and cell viability was measured by standard MTS assay. (C) Combined treatment of NRAS-mutant AML cells with OP449 and cytarabine (AraC). HL-60 cells (NRASQ61L) were incubated for 72 hrs with OP449 and AraC alone or in combination and cell viability was measured by standard MTS assay. Results are graphed as the mean percent proliferation relative to untreated controls ± standard deviation. * denotes p<0.05, **denotes p<0.001 and *** denotes p<0.001, where treatment with OP449 alone was compared to untreated or combination treatment was compared to the respective tyrosine kinase inhibitors or chemotherapy alone. Right panels show combination indices (CI) for all drug combinations, where a CI value less than 1.0 is considered synergistic.
Discussion
Although great strides have been made in the treatment of the patients with CML and AML harboring well-characterized genetic lesions, intrinsic and acquired drug resistance is a persistent clinical problem. Therefore the development of novel therapeutic approaches is urgently required to treat these patients. Recent studies have proposed SET oncoprotein as a novel therapeutic target for the treatment of leukemia (26, 29). Perrotti and colleagues reported that SET is overexpressed in CML cells, resulting in decreased PP2A activity (22, 23, 36). Similarly, SET overexpression in CLL (29) and AML patients is associated with poor prognosis (33). Further, the pharmacological activation of PP2A by FTY720 reduced viability of CML cells by inhibiting BCR-ABL1 kinase signaling (22, 23). However, given potential toxicity concerns encountered for FTY720 in clinical evaluation in multiple sclerosis (38, 39), evaluation of this compound in CML blastic phase patients has not been pursued. Therefore, investigation of alternative SET-targeted agents for reactivation of PP2A is warranted.
Recently, we reported that OP449, a novel compound antagonized SET's inhibition of PP2A in CLL and non-Hodgkin's lymphoma (29). In this report, we evaluated the efficacy of OP449 for inhibiting the growth of CML and AML cells. We show that SET antagonism leads to growth suppression, enhanced apoptosis, and impaired clonogenicity of tyrosine kinase inhibitor-sensitive and -resistant CML and AML cell lines and primary patient cells. We also demonstrated the in vivo efficacy of OP449 in an acute myeloid leukemia xenograft model. Since a fine balance between kinase and phosphatase activity is critical for normal cell growth, we tested the efficacy and specificity of the combination of SET antagonism using OP449 with specific tyrosine kinase inhibitors or chemotherapy in various CML and AML cell lines and primary patient samples. We showed that combined targeting of specific tyrosine kinases or chemotherapy and SET is both synergistic and selective for inhibiting leukemia cell growth both in CML and AML patients as compared to monotherapy. Notably, these findings extended to difficult to treat myeloid disease, including blastic phase CML patients harboring highly drug-resistant BCR-ABL1 mutations and AML patients with FLT3-ITD, JAK3 mutant or RAS mutant-positive cells.
Mechanistically, we demonstrated that the treatment of CML and AML cells with OP449 increased PP2A activity in a dose-dependent manner and OP449-mediated cytotoxicity could be partially neutralized by inhibition of PP2A with okadaic acid. Together these results validate that OP449 inhibits leukemia cell growth through a PP2A-dependent mechanism as would be expected by a SET-targeting agent. Furthermore, we showed that treatment of CML and AML cells with OP449 decreased phosphorylation of STAT5, AKT and ERK. Importantly, in CML cells OP449 not only decreased tyrosine phosphorylation but also reduced protein levels of BCR-ABL1. It has been suggested that increased PP2A activity is associated with increased tyrosine phosphatase activity of SHP-1 in CML cells, which in turn dephosphorylates BCR-ABL1 and leads to proteasome-dependent degradation of BCR-ABL1 (22, 23). Previous studies have shown that overexpression or amplification of BCR-ABL1 is associated with decreased imatinib sensitivity in CML patients (40). Our results suggest that SET antagonism using OP449 in combination with standard therapy may offer additional therapeutic advantage to those patients having disease persistence and tyrosine kinase inhibitor resistance by reducing oncoprotein levels.
With the recent addition of ponatinib to the pharmacopeia of clinically approved ABL1 inhibitors for CML, control of resistance due to point mutations in the kinase domain of BCR-ABL1, including the T315I mutant, appears to be largely tenable. However, BCR-ABL1 compound mutations confers high-level resistance to multiple drugs including ponatinib (16, 18, 19). Our data suggest that even cells that express highly recalcitrant BCR-ABL1 mutations such T315I and the E255V/T315I compound mutant remain sensitive to OP449, warranting further exploration of SET antagonism as a therapeutic strategy in these patients.
The treatment of AML remains challenging because the molecular abnormalities in AML cells are more complex and more heterogeneous than those found in CML cells. Recently, selective inhibitors have been developed for many genes and pathways that are altered in AML. However, a successful implementation of treatment with these agents has been impeded by an incomplete understanding of the genetic changes that drive the disease process. Given the efficacy we demonstrated with OP449 for a variety of mutationally and functionally-activated targets in AML cell lines and primary patient cells, we believe SET antagonism represents a promising paradigm for the treatment of genetically heterogeneous AML cells.
The therapeutic strategy of combined targeting of SET and kinase pathways has broader application to various cancers. For instance, Piazza et al. recently reported that mutations in SET binding protein 1 (SETBP1) in atypical CML (aCML) lead to elevated SET protein levels and reduced PP2A activity (41). SETBP1 deregulation has also been reported in AML (42, 43), myelodysplastic syndromes (MDS), myelofibrosis (MF), myeloproliferative neoplasms (MPN) (44-49) and T cell-precursor ALL patients (50). While the precise mechanistic signaling consequences of such variants have not yet been fully characterized, our findings warrant investigation of the efficacy of SET antagonism in cells harboring such mutations, as well as in tandem with inhibitors of potentially simultaneously dysregulated kinase signaling pathways.
Taken together, our findings demonstrate the potential for development of new therapeutics such as OP449 for chronic and acute leukemias that act by antagonizing SET and increasing the activity of PP2A. Importantly, we show for the first time that combined targeting of SET and relevant oncogenic kinase pathways not only increases efficacy but also maintains selective toxicity for leukemia progenitor cells, which suggests that this approach may also offer a new treatment strategy for targeting residual disease in patients in remission on kinase inhibitor therapy. Therefore our data advocate for the further investigation of such agents in combination with approved kinase inhibitors. Moreover, further preclinical testing of OP449 for the efficacy and toxicity might advance it into clinical trials. Overall, this and future studies will help guide the establishment of a novel paradigm for combined targeting of phosphatase and tyrosine kinase signaling pathways to offer improved therapeutic options in patients with treatment-refractory malignancies.
Supplementary Material
Translational Relevance.
Molecularly targeted therapy has achieved remarkable success in leukemia patients with defined oncogenic lesions. However, drug resistance and relapse is common in large number of cases. The SET oncoprotein is known to be overexpressed in myeloid leukemias including chronic myeloid leukemia (CML) and acute myeloid leukemia (AML). Here, we show that combined targeting of SET and tyrosine kinases provides more efficient and selective inhibition of CML and AML leukemia cell growth from specimens that collectively harbor a broad range of oncogenic lesions. This approach of combined targeting of SET and relevant oncogenic kinase pathways may offer new treatment strategies for treatment-refractory malignancies. To our knowledge, this is the first report showing that SET antagonism in combination with standard targeted therapies may provide an improved treatment option for these patients.
Acknowledgments
We thank Sarah Bowden for administrative support. We thank Dr. Robert H. Collins (University of Texas Southwestern Medical Center, Dallas) and Dr. Jorge E. Cortes (M. D. Anderson Cancer Center, Houston) for providing valuable reagents.
Financial support: This study is supported by an NIH/NCI-STTR grant (10843287, B.J.D., D.C.), Oncotide Pharmaceuticals and in part by The Leukemia & Lymphoma Society. A.A. is supported by National Cancer Institute K99 Career Development Award (5 K99 CA151670 02), Collins foundation, Knight Pilot Project, and Friends of Doernbecher grants. J.W.T. is supported by grants from the V Foundation for Cancer Research, the William Lawrence and Blanche Hughes Fund, and the National Cancer Institute (4 R00CA151457-03). R.S. is supported by Leukemia and Lymphoma Society Scholar Award. B.J.D. is an investigator of Howard Hughes Medical Institute. R.P. and M.D.O. is supported by Ministerio de Ciencia e Innovación (PI11/02443), Departamento Salud del Gobierno de Navarra (78/2012), ISCIII-RTICC (RD12/0036/0063), and Fundación para la Investigación Médica Aplicada y UTE (Spain).
Footnotes
Disclosure of potential conflicts of interest: B.J.D serves as a consultant to MolecularMD, Blueprint Medicines, Gilead Sciences, Cell Therapeutics, Inc., AstraZeneca, Cylene Pharmaceuticals and Lorus Therapeutics. B.J.D. has a subcontract from Oncotide's NIH STTR 1R41CA165570-01. OHSU has clinical trial contracts with Novartis, Bristol-Myers Squibb and ARIAD to pay for patient costs, nurse and data manager salaries, and institutional overhead. B.J.D. does not derive salary, nor does his laboratory receive funds, from these contracts. OHSU and B.J.D. have a financial interest in Molecular MD. Technology used in some of these studies has been licensed to MolecularMD by OHSU. This potential individual and institutional conflict of interest has been reviewed and managed by OHSU. M.V is an Associate Professor at Duke and his potential individual and institutional conflict of interest has been reviewed and is managed by the DUMC Conflict of Interest Committee. R.P. and M.D.O. declare no competing financial interests. D.J.C. and M.P.V. are shareholders (>5%) and employees of Oncotide Pharmaceuticals.
Authorship Contribution: A.A., D.J.C., and M.D.O. designed the research, performed experiments, analyzed results and wrote the paper. R.J.M., R.P., and J.O. performed experiments and analyzed results. C.A.E. provided assistance in analyzing data and writing the paper. J.W.T., M.P.V., R.S., and B.J.D. provided critical feedback, and reviewed of the paper. B.J.D. also provided important resources and samples.
References
- 1.Agarwal A, Byrd J, Deininger MW. The Molecular Biology of the Chronic Leukemias; DeVita, Hellman, and Rosenberg's Cancer: Principles and Practice of Oncology. 2011 [Google Scholar]
- 2.Birkenkamp KU, Geugien M, Lemmink HH, Kruijer W, Vellenga E. Regulation of constitutive STAT5 phosphorylation in acute myeloid leukemia blasts. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2001;15:1923–31. doi: 10.1038/sj.leu.2402317. [DOI] [PubMed] [Google Scholar]
- 3.Druker BJ, Guilhot F, O'Brien SG, Gathmann I, Kantarjian H, Gattermann N, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. The New England journal of medicine. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 4.Chu S, Xu H, Shah NP, Snyder DS, Forman SJ, Sawyers CL, et al. Detection of BCR-ABL kinase mutations in CD34+ cells from chronic myelogenous leukemia patients in complete cytogenetic remission on imatinib mesylate treatment. Blood. 2005;105:2093–8. doi: 10.1182/blood-2004-03-1114. [DOI] [PubMed] [Google Scholar]
- 5.Traer E, MacKenzie R, Snead J, Agarwal A, Eiring AM, O'Hare T, et al. Blockade of JAK2-mediated extrinsic survival signals restores sensitivity of CML cells to ABL inhibitors. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012;26:1140–3. doi: 10.1038/leu.2011.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agarwal A, Fleischman AG, Petersen CL, Mackenzie R, Luty S, Loriaux M, et al. Effects of plerixafor in combination with BCR-ABL kinase inhibition in a murine model of CML. Blood. 2012;120:2658–68. doi: 10.1182/blood-2011-05-355396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101:690–8. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 8.Agarwal A, Eide CA, Harlow A, Corbin AS, Mauro MJ, Druker BJ, et al. An activating KRAS mutation in imatinib-resistant chronic myeloid leukemia. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22:2269–72. doi: 10.1038/leu.2008.124. [DOI] [PubMed] [Google Scholar]
- 9.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. The Journal of clinical investigation. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 11.Kantarjian H, Shah NP, Hochhaus A, Cortes J, Shah S, Ayala M, et al. Dasatinib versus imatinib in newly diagnosed chronic-phase chronic myeloid leukemia. The New England journal of medicine. 2010;362:2260–70. doi: 10.1056/NEJMoa1002315. [DOI] [PubMed] [Google Scholar]
- 12.O'Hare T, Walters DK, Stoffregen EP, Jia T, Manley PW, Mestan J, et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65:4500–5. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 13.Weisberg E, Manley PW, Breitenstein W, Bruggen J, Cowan-Jacob SW, Ray A, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer cell. 2005;7:129–41. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Saglio G, Kim DW, Issaragrisil S, le Coutre P, Etienne G, Lobo C, et al. Nilotinib versus imatinib for newly diagnosed chronic myeloid leukemia. The New England journal of medicine. 2010;362:2251–9. doi: 10.1056/NEJMoa0912614. [DOI] [PubMed] [Google Scholar]
- 15.Jabbour E, Hochhaus A, Cortes J, La Rosee P, Kantarjian HM. Choosing the best treatment strategy for chronic myeloid leukemia patients resistant to imatinib: weighing the efficacy and safety of individual drugs with BCR-ABL mutations and patient history. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2010;24:6–12. doi: 10.1038/leu.2009.193. [DOI] [PubMed] [Google Scholar]
- 16.O'Hare T, Shakespeare WC, Zhu X, Eide CA, Rivera VM, Wang F, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer cell. 2009;16:401–12. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cortes JE, Kantarjian H, Shah NP, Bixby D, Mauro MJ, Flinn I, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. The New England journal of medicine. 2012;367:2075–88. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khorashad JS, Kelley TW, Szankasi P, Mason CC, Soverini S, Adrian LT, et al. BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships. Blood. 2013;121:489–98. doi: 10.1182/blood-2012-05-431379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eide CA, Zabriskie MS, Adrian LT, Lange T, Deininger MW, Druker BJ, et al. Resistance Profiling of BCR-ABL Compound Mutations Linked to Tyrosine Kinase Inhibitor Therapy Failure in Chronic Myeloid Leukemia. American Society of Hematology: Blood (ASH Annual Meeting Abstracts) 2011 [Google Scholar]
- 20.Estey E, Dohner H. Acute myeloid leukaemia. Lancet. 2006;368:1894–907. doi: 10.1016/S0140-6736(06)69780-8. [DOI] [PubMed] [Google Scholar]
- 21.Perrotti D, Neviani P. ReSETting PP2A tumour suppressor activity in blast crisis and imatinib-resistant chronic myelogenous leukaemia. British journal of cancer. 2006;95:775–81. doi: 10.1038/sj.bjc.6603317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neviani P, Santhanam R, Oaks JJ, Eiring AM, Notari M, Blaser BW, et al. FTY720, a new alternative for treating blast crisis chronic myelogenous leukemia and Philadelphia chromosome-positive acute lymphocytic leukemia. The Journal of clinical investigation. 2007;117:2408–21. doi: 10.1172/JCI31095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neviani P, Santhanam R, Trotta R, Notari M, Blaser BW, Liu S, et al. The tumor suppressor PP2A is functionally inactivated in blast crisis CML through the inhibitory activity of the BCR/ABL-regulated SET protein. Cancer cell. 2005;8:355–68. doi: 10.1016/j.ccr.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 24.Cristobal I, Garcia-Orti L, Cirauqui C, Alonso MM, Calasanz MJ, Odero MD. PP2A impaired activity is a common event in acute myeloid leukemia and its activation by forskolin has a potent anti-leukemic effect. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2011;25:606–14. doi: 10.1038/leu.2010.294. [DOI] [PubMed] [Google Scholar]
- 25.Roberts KG, Smith AM, McDougall F, Carpenter H, Horan M, Neviani P, et al. Essential requirement for PP2A inhibition by the oncogenic receptor c-KIT suggests PP2A reactivation as a strategy to treat c-KIT+ cancers. Cancer Res. 2010;70:5438–47. doi: 10.1158/0008-5472.CAN-09-2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Switzer CH, Cheng RY, Vitek TM, Christensen DJ, Wink DA, Vitek MP. Targeting SET/I(2)PP2A oncoprotein functions as a multi-pathway strategy for cancer therapy. Oncogene. 2011;30:2504–13. doi: 10.1038/onc.2010.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Switzer CH, Glynn SA, Ridnour LA, Cheng RY, Vitek MP, Ambs S, et al. Nitric oxide and protein phosphatase 2A provide novel therapeutic opportunities in ER-negative breast cancer. Trends in pharmacological sciences. 2011;32:644–51. doi: 10.1016/j.tips.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang Y, Huang Q, Lu Y, Li X, Huang S. Reactivating PP2A by FTY720 as a novel therapy for AML with C-KIT tyrosine kinase domain mutation. Journal of cellular biochemistry. 2012;113:1314–22. doi: 10.1002/jcb.24003. [DOI] [PubMed] [Google Scholar]
- 29.Christensen DJ, Chen Y, Oddo J, Matta KM, Neil J, Davis ED, et al. SET oncoprotein overexpression in B-cell chronic lymphocytic leukemia and non-Hodgkin lymphoma: a predictor of aggressive disease and a new treatment target. Blood. 2011;118:4150–8. doi: 10.1182/blood-2011-04-351072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perrotti D, Neviani P. Protein phosphatase 2A (PP2A), a drugable tumor suppressor in Ph1(+) leukemias. Cancer metastasis reviews. 2008;27:159–68. doi: 10.1007/s10555-008-9119-x. [DOI] [PubMed] [Google Scholar]
- 31.Christensen DJ, Ohkubo N, Oddo J, Van Kanegan MJ, Neil J, Li F, et al. Apolipoprotein E and peptide mimetics modulate inflammation by binding the SET protein and activating protein phosphatase 2A. J Immunol. 2011;186:2535–42. doi: 10.4049/jimmunol.1002847. [DOI] [PubMed] [Google Scholar]
- 32.Agarwal A, MacKenzie R, Oddo J, Vitek MP, Christensen DJ, Druker BJ. A Novel SET Antagonist (OP449) Is Cytotoxic to CML Cells, Including the Highly-Resistant BCR-ABLT315I Mutant, and Demonstrates Enhanced Efficacy in Combination with ABL Tyrosine Kinase Inhibitors. American Society of Hematology: Blood (ASH Annual Meeting Abstracts) 2011 [Google Scholar]
- 33.Cristobal I, Garcia-Orti L, Cirauqui C, Cortes-Lavaud X, Garcia-Sanchez MA, Calasanz MJ, et al. Overexpression of SET is a recurrent event associated with poor outcome and contributes to protein phosphatase 2A inhibition in acute myeloid leukemia. Haematologica. 2012;97:543–50. doi: 10.3324/haematol.2011.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsuo Y, MacLeod RA, Uphoff CC, Drexler HG, Nishizaki C, Katayama Y, et al. Two acute monocytic leukemia (AML-M5a) cell lines (MOLM-13 and MOLM-14) with interclonal phenotypic heterogeneity showing MLL-AF9 fusion resulting from an occult chromosome insertion, ins(11;9)(q23;p22p23) Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 1997;11:1469–77. doi: 10.1038/sj.leu.2400768. [DOI] [PubMed] [Google Scholar]
- 35.Agarwal A, Bumm TG, Corbin AS, O'Hare T, Loriaux M, VanDyke J, et al. Absence of SKP2 expression attenuates BCR-ABL-induced myeloproliferative disease. Blood. 2008;112:1960–70. doi: 10.1182/blood-2007-09-113860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Neviani P, Harb JG, Oaks JJ, Santhanam R, Walker CJ, Ellis JJ, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. The Journal of clinical investigation. 2013;123:4144–57. doi: 10.1172/JCI68951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Garcia L, Garcia F, Llorens F, Unzeta M, Itarte E, Gomez N. PP1/PP2A phosphatases inhibitors okadaic acid and calyculin A block ERK5 activation by growth factors and oxidative stress. FEBS letters. 2002;523:90–4. doi: 10.1016/s0014-5793(02)02950-2. [DOI] [PubMed] [Google Scholar]
- 38.Sharma S, Mathur AG, Pradhan S, Singh DB, Gupta S. Fingolimod (FTY720): First approved oral therapy for multiple sclerosis. Journal of pharmacology & pharmacotherapeutics. 2011;2:49–51. doi: 10.4103/0976-500X.77118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kappos L, Antel J, Comi G, Montalban X, O'Connor P, Polman CH, et al. Oral fingolimod (FTY720) for relapsing multiple sclerosis. The New England journal of medicine. 2006;355:1124–40. doi: 10.1056/NEJMoa052643. [DOI] [PubMed] [Google Scholar]
- 40.Jiang X, Zhao Y, Smith C, Gasparetto M, Turhan A, Eaves A, et al. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2007;21:926–35. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 41.Piazza R, Valletta S, Winkelmann N, Redaelli S, Spinelli R, Pirola A, et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nature genetics. 2012;45:18–24. doi: 10.1038/ng.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Albano F, Anelli L, Zagaria A, Coccaro N, Casieri P, Minervini A, et al. SETBP1 and miR_4319 dysregulation in primary myelofibrosis progression to acute myeloid leukemia. Journal of hematology & oncology. 2012;5:48. doi: 10.1186/1756-8722-5-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cristobal I, Blanco FJ, Garcia-Orti L, Marcotegui N, Vicente C, Rifon J, et al. SETBP1 overexpression is a novel leukemogenic mechanism that predicts adverse outcome in elderly patients with acute myeloid leukemia. Blood. 2010;115:615–25. doi: 10.1182/blood-2009-06-227363. [DOI] [PubMed] [Google Scholar]
- 44.Damm F, Itzykson R, Kosmider O, Droin N, Renneville A, Chesnais V, et al. SETBP1 mutations in 658 patients with myelodysplastic syndromes, chronic myelomonocytic leukemia and secondary acute myeloid leukemias. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013;27:1401–3. doi: 10.1038/leu.2013.35. [DOI] [PubMed] [Google Scholar]
- 45.Laborde RR, Patnaik MM, Lasho TL, Finke CM, Hanson CA, Knudson RA, et al. SETBP1 mutations in 415 patients with primary myelofibrosis or chronic myelomonocytic leukemia: independent prognostic impact in CMML. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013 doi: 10.1038/leu.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meggendorfer M, Bacher U, Alpermann T, Haferlach C, Kern W, Gambacorti-Passerini C, et al. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013 doi: 10.1038/leu.2013.133. [DOI] [PubMed] [Google Scholar]
- 47.Sakaguchi H, Okuno Y, Muramatsu H, Yoshida K, Shiraishi Y, Takahashi M, et al. Exome sequencing identifies secondary mutations of SETBP1 and JAK3 in juvenile myelomonocytic leukemia. Nature genetics. 2013 doi: 10.1038/ng.2698. [DOI] [PubMed] [Google Scholar]
- 48.Thol F, Suchanek KJ, Koenecke C, Stadler M, Platzbecker U, Thiede C, et al. SETBP1 mutation analysis in 944 patients with MDS and AML. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2013 doi: 10.1038/leu.2013.145. [DOI] [PubMed] [Google Scholar]
- 49.Makishima H, Yoshida K, Nguyen N, Przychodzen B, Sanada M, Okuno Y, et al. Somatic SETBP1 mutations in myeloid malignancies. Nature genetics. 2013 doi: 10.1038/ng.2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Panagopoulos I, Kerndrup G, Carlsen N, Strombeck B, Isaksson M, Johansson B. Fusion of NUP98 and the SET binding protein 1 (SETBP1) gene in a paediatric acute T cell lymphoblastic leukaemia with t(11;18)(p15;q12) British journal of haematology. 2007;136:294–6. doi: 10.1111/j.1365-2141.2006.06410.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.