Abstract
Background
Episodic memory (EM) is known to decline with age and the rate of decline is variable across individuals. A single nucleotide polymorphism (rs17070145) in the WWC1 gene that encodes the KIBRA protein critical for long term potentiation (LTP) and memory consolidation has previously been associated with EM performance as well as differences in hippocampal engagement during EM tasks using fMRI. In the current study, we explore the effect of this polymorphism on EM related activity and cognitive performance across the adult lifespan using fMRI.
Methods
Two hundred thirty-two healthy, Caucasian subjects (age range 18–89 years) completed a battery of cognitive tests as well as an EM task during an fMRI scan.
Results
WWC1 T carriers had significantly better delayed recall performance than CC individuals (p=0.006). The relationship between increasing age and recall scores, both immediate and delayed, was also significantly different between WWC1 genotype groups (p=0.01). In addition to the age-related decline in hippocampal formation (HF) activation bilaterally during encoding and retrieval (p<0.05; FDRSVC-HF-ROI), we observed an age by WWC1 genotype interaction on activation in the HF during encoding and retrieval. The CC group showed a significant negative association between HF activity and increasing age while no such association was observed in the T carrier group (left HF p=0.04; R-Z correlation difference during encoding and retrieval; right HF p=0.0008; R-Z correlation difference during retrieval).
Conclusion
Our results provide evidence of a dynamic relationship between rs17070145 polymorphism and increasing age on neuronal activity in the hippocampal region.
Keywords: Episodic Memory (EM), Aging, WWC1, KIBRA, Long term potentiation (LTP), single nucleotide polymorphism (SNP), hippocampus
Age-related changes in declarative memory vary greatly from individual to individual. Converging evidence suggests that both environmental and inherited factors play a substantial role in an individual’s declarative memory abilities throughout his or her, lifespan (1–3). Heritability estimates derived from twin and familial studies suggest that approximately 50% of the variance observed between younger individuals in declarative memory performance can be attributed to genetic variation. This remains the case for middle-aged as well as elderly individuals (4, 5). Additionally, some studies have shown that the impact of genes on individual components of declarative memory performance is not necessarily equal, and some processes, such as acquisition, may have their own unique genetic component (6). Genetic association studies have been used to uncover the molecular architecture that supports cognitive processes, including declarative memory. In the context of aging, these studies can be helpful in uncovering the molecular mechanisms underlying individual variation in alteration of declarative memory with advancing age by identifying genetic variations that place individuals at an increased risk for decline or those conferring resilience against decline. To date, the candidate gene approach, in which potential genes are selected based on a priori knowledge of their biological importance, function, and/or association with disease, has produced a substantial number of reports showing how common human variation in memory-related genes such as APOE, BDNF, and COMT account for some of the individual differences in cognitive ability and the way it changes with increasing age (2).
The protein KIBRA has been the subject of much investigation of its role in the molecular basis of human memory. This followed a 2006 genome wide association study (GWAS) of memory that reported a significant association between performance on an episodic memory task and allelic variation in the gene that encodes KIBRA, WWC1, in two independent samples (7). Specifically, the authors reported that carriers of a single nucleotide polymorphism (SNP) resulting in a C→ T substitution within the ninth intron of the WWC1 gene show significantly better delayed word recall performance than C homozygote individuals. This association was found for both the 5 minute and 24 hour portions of a free recall task, but not for performance on the immediate recall section of the same task or on tasks of executive function, working memory, or attention. Furthermore, the authors found that individuals lacking the T allele had increased neuronal activity in the medial temporal lobe (MTL) during the retrieval phase of an episodic memory fMRI task when compared to T allele carriers. The authors interpreted this finding as the C homozygotes having neuronal inefficiency in the MTL as the genotype groups were matched for both fMRI task performance as well as performance on 5 minute delayed recall.
Subsequent studies using older individuals have confirmed the initial finding of T allele carriers exhibiting better performance on episodic memory tasks. Studies have even reported that the effect of this polymorphism on episodic memory scores becomes greater with increasing age (8). Other studies, however, have reported no effect (9–11) as well as the opposite effect on memory performance (12). In addition, some previous reports link this polymorphism to risk for Alzheimer’s disease, although the findings have been inconsistent (13–15). Recently, a meta-analysis found that T allele carriers perform significantly better on tests of episodic memory and that the rs17070145 SNP accounted for 0.5% of the variance in performance on episodic memory tasks (16). To date, only one additional study has investigated the impact of this SNP on neuronal activity during an episodic memory task. In this study, Kauppi et al. (17) reported an effect of this gene on neuronal activity during the retrieval phase of an fMRI episodic memory task such that T allele carriers of the rs17070145 polymorphism had increased hippocampal activity when compared to C homozygotes. This is in the opposite direction as what would be predicted by the original report (7). While the authors of the more recent report suggest that the contradictory findings between studies may be due to differences in sample size and/or the particular details of the episodic memory tasks used, it may also be possible that the differences in findings are the products of differences in the age range of the subjects between studies. The mean age of participants in the sample with fMRI data in the initial report by Papassotiropoulos et al. (7) was 22 years, and the mean age of participants in the sample with fMRI data in the recent report by Kauppi et al. (17) was 57 years. It could be the case that this polymorphism exerts differential effects on memory related MTL activity across the adult life span. This notion is supported by a recent study which reported that KIBRA knockout mice exhibited an age dependent impairment in the molecular formation of memory (LTP and LDP), as well as age dependent deficits in learning and the retention of previously learned information (18).
To further explore the possible interaction between genetic variation in the WWC1 gene and the normal aging process on neuronal activity during an episodic memory task, we conducted an fMRI study during an incidental encoding and retrieval task in healthy individuals across the adult lifespan. Given the results of previous studies, we hypothesized that T allele status would be associated with superior performance on cognitive tasks assessing free recall, that this association would become greater with increasing age, and that the effect of this polymorphism would not be associated with performance on tasks involving other cognitive abilities. Furthermore, we speculated that in a sample of performance matched individuals across genotype groups there would be WWC1 genotype based group differences in the association between increasing age and neuronal activity during a declarative memory task. In particular based on prior evidence on the differential effect of this gene on hippocampal engagement at younger (7) and older (17) ages as well as the effect of normal increasing age on hippocampal function (19), we hypothesized that C homozygotes would have increased MTL activity compared to T allele carriers during young adulthood reflecting inefficient memory related brain function needed to maintain memory, which then declines with increasing age as hippocampal function declines. Individuals carrying the T allele on the other hand may show a relatively slower decline in MTL function with increasing age.
Methods
Subjects
Two hundred thirty-two healthy Caucasian volunteers (113 carriers of the rs17070145 T allele and 120 non-carriers) were selected for the current study. A detailed description of the participants is reported in Supplement 1. All study participants under 55 years of age were recruited as part of the CBDB/NIMH Sibling Study (under NIH Protocol Number: 95-M-0150, PI: Daniel Weinberger); those of age 55 and older were recruited through the Elderly Cohort Study (under NIH Protocol Number: 00-M-0085, PI: Venkata Mattay). Subjects from both protocols underwent similar testing procedures.
Genotyping
The WWC1 SNP rs17070145 genotyping analysis was performed using the Taqman 5′-exonuclease allelic discrimination assay. The primer-probe assay set was obtained from Assays by Design from Applied Biosystems (Foster City, CA). Individuals were divided into two groups, carriers of the T allele substitution (CT/TT) and non-carriers (CC), for subsequent analysis based on the observations of Papassotiropoulos et al. (7). The two genotype groups were matched for sex (p=0.6), years of completed education (p=0.7), chronological age (p=0.9), handedness (p=0.7) and Wechsler Adult Intelligence Scale (WAIS-R) IQ(p=0.65) (Table S1 in Supplement 1). The groups were also matched for polymorphisms in four known memory-related genes: BDNF (p=0.12), SERT (p=0.12), COMT (p=0.87), and APOE (p= 0.87).
Neuropsychological Testing
Subjects underwent the following battery of neuropsychological testing to assess the effect of the WWC1 polymorphism on a wide variety of cognitive abilities: Wechsler Memory Scale (WMS) Logical Memory (LM-I and LM-II), Trails A/B, WAIS-R IQ, Letter-Number Sequencing (LNS), and Category and Letter (FAS) Fluency. All scores were standardized according to conventional measures. All non-imaging statistical analysis was done using R 2.15.0 software (http://www.R-project.org/). Prior to group comparisons, all scores were tested for normality using the Lilliefors (Kolmogorov-Smirnov) test in the R package nortest (http://CRAN.R-project.org/package=nortest). Genotype groups were compared using Welch two-sample t-test (normal distribution) or two-sample Mann-Whitney U-test (non-normal distribution).
For the WMS LM task, a direct comparison of scaled results was not possible as the young and elderly subjects in the current sample were administered different versions of the WMS LM task (WMS-R and WMS-III, respectively). For this reason, the analysis utilized unscaled scores from the first story (Story A) that was similar in both versions of the task, for each participant. WWC1 group differences in the relationship between increasing age and WMS LM performance was tested using a two-tailed Fisher’s r-to-Z transform test (p<0.05) in the R package psych (http://personality-project.org/r/psych.manual.pdf).
fMRI Experimental Design
BOLD fMRI was collected on a GE Signa (Milwaukee, WI, USA) 3 Tesla scanner during the encoding and subsequent retrieval of semantically unrelated aversive and neutral complex scenes selected from the International Affective Picture System (20). For both the encoding and retrieval sessions, the scenes were presented in a blocked fashion with two blocks of aversive or neutral scenes alternating with blocks of resting state. Detailed description of the experimental paradigm and fMRI data acquisition are provided in Supplement 1.
Image Analysis
Images were processed using SPM5 (http://www.fil.ion.ucl.ac.uk/spm). In the first level analyses, linear contrasts were computed for each subject producing t-statistical parameter maps at each voxel for encoding and retrieval of neutral visual stimulus relative to fixation. A detailed description of the preprocessing methods and first level processing is included in Supplement 1. Since we were primarily interested in exploring the effect of the WWC1 polymorphism on the association between increasing age and physiologic activity underlying episodic memory without the influence of the stimulus emotional content (e.g. negative valence in the aversive stimuli) and since this task has shown a significant effect of valence on age related differences in episodic memory function (19), we limited the analysis in the current study only to the neutral stimuli.
Individual subject first-level contrast images related to the encoding and retrieval sessions of neutral scenes, were entered into second level multiple regression analyses to explore the main effects of age, the WWC1 polymorphism, as well as their interaction on the regions of the brain that were activated in each task. Since our primary goal was to explore the potential way in which age-related changes in neuronal activity are affected by the WWC1 gene which is expressed in memory-related brain structures (7, 21) and important for hippocampal dependent memory (18), these analyses were limited to neural activation within the hippocampal formation (HF) which included the hippocampal (HC) and parahippocampal (PHC) regions using the WFU Pickatlas toolbox (version 1.04, Functional MRI Laboratory at the Wake Forest University School of Medicine, http://fmri.wfubmc.edu/). For the purpose of this study we aimed to test whether the relationship between increasing age and physiologic activity in the HF differed between WWC1 groups by using a two-tailed Fisher’s r-to-Z transform test. To this end, a significance threshold (p<0.005, cluster size (k)>10, uncorrected) within HF was used on the age-by-WWC1 contrast to identify potential clusters of activity that fit our a priori hypothesis of WWC1 group differences in the relationship between age and hippocampal activity. We used a statistical threshold that is higher than that used in a recent study reporting significant WWC1 group differences in hippocampal activity (p<0.01; uncorrected) (17). For each significant interaction cluster, the median value from each individuals 1st level contrast image was extracted. Prior to WWC1 group correlation testing, the extracted data was tested for outliers using the outlier function in the R package outliers (http://CRAN.R-project.org/package=outliers).
Briefly, the outlier function identifies the value within a vector that is furthest from the sample mean. After identifying potential outliers, WWC1 genotype dependent group differences in the correlation between increasing age and HF activation were tested using a two-tailed Fisher’s r-to-Z transform test (p<0.05) in the R package psych (http://personality-project.org/r/psych.manual.pdf). These tests were then repeated with the potential outliers removed from the analysis. If the removal of the single data point resulted in a change in the significance of the test, either from non-significant to significant or vice versa, we concluded that the value was indeed an outlier and was not included in subsequent tests. To ensure that the removal of outliers did not significantly change our results in image space, we reanalyzed the data in SPM without the outliers. Sensitivity analysis revealed that with our sample size (113 T carriers +120 CC homozygotes) and an alpha level of 0.05, we had sufficient power (β=0.80) to detect WWC1 group differences in correlation coefficients similar in effect size to previous reports with this SNP. Significance for the pure effects of both age and WWC1 genotypes inside the HF was set at an alpha of 0.05. The threshold for significance outside the HF (whole brain) was set at an alpha of 0.05. False discovery rate (FDR) for multiple comparisons was used in both of these analyses.
Results
Neuropsychological Testing
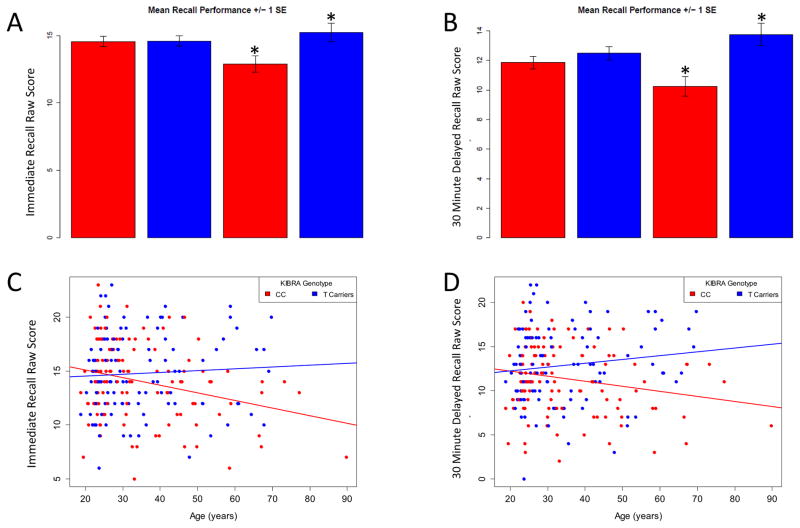
In the full sample, there were trends for age and genotype effects on story A immediate recall (WMS LM1 task; p=0.08, p=0.1 respectively). A main effect of genotype was observed for thirty-minute recall (p=0.006) in the full sample, with T carriers performing better than individuals homozygous for the C allele (Table 2). Additionally, we observed a greater genotype effect on both immediate as well as delayed recall performance when the analysis was restricted to subjects over the age of 40 (n= 66). T carriers (n=32, 13 males, age (mean)=52.5) performed significantly better than did non-carriers (n=34, 17 male, age(mean)=53) on both immediate (p=0.01) and thirty-minute (p=0.0008) recall (Fig. 1a, Table 2). Correlation analysis investigating potential WWC1 genotype differences in the relationship between age and story A recall performance for both immediate and delayed recall scores, showed significant group differences in the correlation between increasing age and performance on immediate (Z-score=2.59, Cohen’s d=0.34, p=0.01), as well as delayed recall scores (Z-score= 2.62, Cohen’s d=0.35, p=0.01)(Fig.1b). For both tests, the CC group had a negative relationship between increasing age and recall performance whereas no such relationship was observed in the T carrier group (Figure 1c, 1d). Aside from a trend in phonetic fluency (p=0.07) in which T carriers slightly outperformed CC individuals, the genotype groups did not differ on any other measures tested (Tables 1 & 2).
Table 2.
Neuropsychological Test Score Comparisons for WWC1 SNP rs17070145 Genotype Groups (subjects over 40 years old)
| CC | CT/TT | Total Sample | (p-value) | |
|---|---|---|---|---|
| Trail Making Test Trails B Score | 54.03 (9.45) | 52.16 (11.94) | 53.53 (10.5) | (0.4) |
| Letter-Number Sequencing | 11.55 (2.5) | 12.38 (83) | 12 (2.97) | (0.2) |
| DSST (WAIS subtest) | 12.74 (2.14) | 12.91 (1.78) | 13 (2.97) | (0.7) |
| Semantic Fluency | 52.69 (9.58) | 56.28 (9.83) | 54.39 (10.2) | (0.1) |
| Phonetic Fluency | 45.36 (9.6) | 50.59 (12.97) | 45.25 (10.25) | (0.07) |
| Story A Immediate Recall | 12.88 (3.48) | 15.22 (3.92) | 14.41 (3.61) | (0.01) |
| Story A 30 Minute Delayed Recall | 10.24 (3.81) | 13.75 (4.31) | 12.09 (4.1) | (0.0008) |
Values reported as mean (standard deviation); Welch two-sample t-test used for group comparisons
Bold indicates a significant difference between groups
Figure 1.
WWC1 SNP rs17070145 genotype group differences in free recall performance. (A) In subjects over 40 years of age, T-allele carriers have better immediate free recall performance when compared to CC individuals (p=0.01). (B) T-allele carriers have better 30 minute delayed free recall performance when compared to CC individuals (entire sample, p=0.006). Group differences remain significant when restricted to individuals over 40 years of age (p=0.0008). WWC1 genotype groups have significantly different correlations between increasing age and performance on immediate (C) and delayed recall (D) (p=0.01 for both immediate and delayed recall). Raw scores were computed by summing the number of correct responses for each individual. Adult C= CC homozygotes under 40 years old, Adult T= T-allele carriers under 40 years old, Older C= CC homozygotes over 40 years old, Older T= T-allele carriers over 40 years old. * = p<0.05, age> 40
Table 1.
Neuropsychological Test Score Comparisons for WWC1 SNP rs17070145 Genotype Groups (entire sample)
| CC | CT/TT | Total Sample | (p-value) | |
|---|---|---|---|---|
| Trail Making Test Trails B Score | 53.63 (10.03) | 53.42 (11.02) | 53.53 (10.5) | (0.9) |
| Letter-Number Sequencing | 12 (2.97) | 12 (2.97) | 12 (2.97) | (0.7)‡ |
| DSST (WAIS subtest) | 13 (2.97) | 13 (2.97) | 13 (2.97) | (0.9)‡ |
| Semantic Fluency | 54.25 (9.98) | 54.53 (10.46) | 54.39 (10.2) | (0.8) |
| Phonetic Fluency | 44.84 (9.81) | 45.69 (11.89) | 45.25 (10.25) | (0.6) |
| Story A Immediate Recall | 14.07 (3.56) | 14.77 (3.65) | 14.41 (3.61) | (0.1) |
| Story A 30 Minute Delayed Recall | 11.39 (3.86) | 12.84 (4.22) | 12.09 (4.1) | (0.006) |
Values reported as mean (standard deviation); Welch two-sample t-test used for group comparisons unless noted with ‡
Groups compared using two-sample Mann-Whitney U-test; Bold indicates a significant difference between groups
Episodic Memory task during fMRI
Task accuracy was recorded during the fMRI paradigm as the percentage of correct responses during the retrieval session. There were no significant effects of age, genotype, or their interaction found on task accuracy. There was no effect of the WWC1 genotype on encoding (p=0.6) or retrieval (p=0.9) response times. There was however a significant correlation between age and retrieval response time (p=0.03) whereby older individuals exhibited increased response times. There was a trend for a positive association between response time and age during encoding (p=0.1).
Episodic Memory Related Activity during fMRI
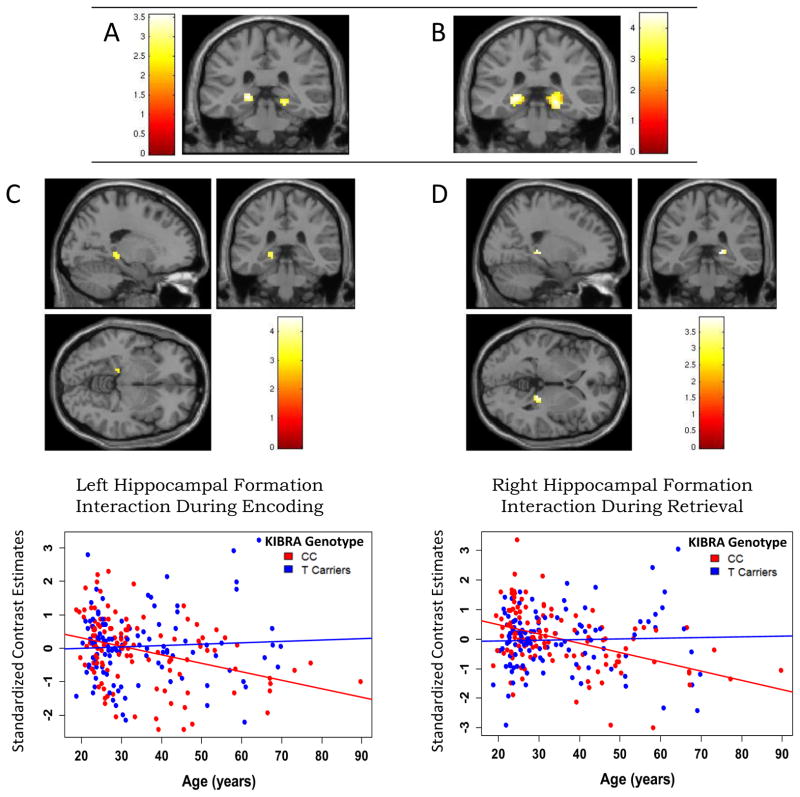
During the encoding session we observed significant bilateral age-related decline in activity within the hippocampal formation ROI (L: k=20, xyz=−18, −33, −3; R: k=8, xyz=21, −33, −9; p<0.05 FDR; Fig. 2.a, Table S2 in Supplement 1). Outside of the hippocampal formation we found decreasing activity with increasing age in the left enthorinal region (Table S2 in Supplement 1). While there was no significant main effect of WWC1 genotype (either CC> T Carriers or T Carriers> CC) during encoding, there was a significant WWC1 genotype-by-age interaction in the left hippocampal formation (k=24, xyz=−27, −24, −9, p<0.003 uncorrected). The results were similar on reanalysis after removing outliers as described above (k=27, xyz=−27, −33, −3, p=0.003 uncorrected)
Figure 2.
A & B Effect of age on hippocampal formation (HF) neuronal activity during an episodic memory task. (A) Bilateral age-related decline in HF activity during the incidental encoding session (p<0.05, k>5, hippocampal formation ROI, FDR corrected for multiple comparaisons). (B) Bilateral age-related decline in HF activity during the retrieval session (p<0.05, k>5, hippocampal formation ROI, FDR corrected for multiple comparaisons). C&D WWC1 SNP rs17070145 genotype group differences in the correlation between age and activation during an episodic memory task. (C) The WWC1 CC group (red) exhibits a negative correlation between age and left hippocampal formation neuronal activity during the encoding session, which is not observed for the T-allele carrier group (blue). Age-Activation correlation coefficient (pearsons r): CC r= −0.36, CT/TT r= −0.01; Fishers r-z: z-score=2.76, p=0.04). (D) The WWC1 CC group exhibits a negative correlation between age and right hippocampal formation neuronal activity during the retrieval session, which is not observed for the T-allele carrier group. Age-Activation correlation coefficient (pearsons r): CC r= −0.43, CT/TT r= 0.03; Fishers r-z: z-score=3.66, p=0.0008). Similar differences between WWC1 groups in the relationship between increasing age and episodic memory related activity during retrieval session were also observed in the left hippocampal formation (Age-Activation correlation coefficient (pearsons r): CC r= −0.37, CT/TT r= −0.11; Fishers r-z: z-score=2.09, p=0.04. Data not shown.)
During retrieval we found increasing age to be associated with a decline in activity in the hippocampal formation ROI (L: k=119, xyz= −24, −33, −6; R: k=97, xyz= 21, −33, −9; p<0.05 FDR)(Fig. 2.b; Table S3 in Supplement 1). Additionally, age-related decline in activity was found in the left occipital, left posterior fusiform, bilateral inferior frontal, right posterior cingulate, bilateral thalamus, and right superior parietal regions (Table S3 in Supplement 1). As was the case in during the encoding portion of the task, there was no significant main effect of WWC1 genotype on activity within or outside of the hippocampal formation during the retrieval session. There was a WWC1 genotype-by-age interaction observed in three clusters within the hippocampal formation (L: k=14, xyz=−18, −36, 0 and k=12, xyz= −24, −27, −15; R: k=13, xyz=21, −33, 3 p<0.005 uncorrected). However, reanalysis after removal of outliers as described above showed that only two of these three clusters remained significant (L: k=12, xyz=−18, −36, 0, p<0.005 uncorrected; R: k=15, xyz=21, −33,3; p<0.005 uncorrected, p<0.007 FDRSVC-HF-ROI) with an interaction pattern similar to that observed in the encoding session.
Further analysis using a two-tailed Fisher’s r-to-Z transform test on these clusters showed significant WWC1 genotype based group differences in the correlation between age and neuronal activity ( Fishers r-z p=0.0008 in the right HF and p=0.04 in the left HF during the retrieval session; Fishers r-z p=0.04 in the left HF during encoding) (Table S4 in Supplement 1). Specifically, we found that within the CC group, age was negatively correlated with hippocampal activity and that this strong negative association was largely absent in the T carrier group. Importantly, there was a significant difference between the group correlation coefficients, suggesting WWC1 group differences in the association between hippocampal activity and increasing age during both the encoding and retrieval portions of our task (Fig. 2 C&D; Table S4 in Supplement 1).
Discussion
This study adds to evidence for effects of WWC1 on human episodic memory at both the level of cognitive performance and hippocampal function. Our results replicate previous reports linking allelic differences in the WWC1 gene with human episodic memory performance and provide evidence of a dynamic relationship between WWC1 genotype and increasing age on neuronal activity in the hippocampal region. We found that increasing age was associated with an exaggerated effect of this polymorphism on measures of recollection memory. Interestingly, we found no evidence for significant effects of WWC1 genotype or its interaction with age on performance during the fMRI episodic memory task even though we found strong genotype differences in other measures of episodic memory. One possible explanation for this may be that while we observed differences in measures of recollection based memory, the performance collected during our episodic memory task is a measure of recognition memory, which may further add evidence to the specificity of this polymorphism to measures of recollection memory. Moreover, the task performed during the fMRI procedure is a much simpler task and the information encoded is much more easily recognized than the neuropsychological test performed as part of our clinical test battery.
More importantly, our fMRI results suggest that as a group, CC individuals show a clear negative association between hippocampal engagement during both the encoding and retrieval and increasing age. This association is significantly different than the association between hippocampal engagement and age that was observed in the T allele carrier group. To date, only two studies have reported WWC1 genotype differences in hippocampal function while performing an episodic memory task, but the results were in the opposite direction of one another (7, 17). Although we did not find any WWC1 group differences in mean hippocampal function, the results of our current study may in fact unify these previous two seemingly contradictory results by showing age-related differences in the effect of this polymorphism on hippocampal engagement during an episodic memory task - the WWC1 group differences in mean hippocampal function at different ages effectively canceled out a main effect of genotype when examined across the whole group including subjects at younger and older ages. Interestingly, similar age-related differences were reported with another gene that has been found to exhibit age-dependent effects on memory function. Filippini et al. (22) reported similar age-related differences in genotype effects on brain function when comparing subjects carrying the Alzheimer’s disease risk associated APOE E4 allele to those without it (22). The study reported that in young adults, E4 carriers had increased activation when compared to non-E4 carriers and that this association was switched in older adults (non-E4>E4). The authors suggest that while no clinical differences were observable between the genotype groups, the lower activation in the older E4 carriers may be indicative of changes at the level of brain function prior to the onset of observable cognitive deficits. This interpretation may also fit well for our findings given that increased risk for developing AD has been linked to WWC1 CC status, although we should mention these findings have not been conclusive.
Unlike the previous two studies of hippocampal physiology associated with WWC1 genotype, we observed a significant age-by-genotype interaction during the encoding session of our task which followed the same pattern that we found during the retrieval session. The nature of our task may account for this novel finding. The previous two studies (7, 17) used an fMRI paradigm in which the encoding session was comprised of individuals learning associations which were subsequently tested for during the retrieval session. Subjects in the current study underwent an incidental encoding session which did not include the learning of new associations. In fact, our paradigm requires subjects to make a decision as to whether the images presented to them are of an indoor scene or an outdoor scene which requires the use of previously learned associations. In light of this fact, it could be the case that our novel finding of group genotype differences in the relationship between age and encoding related hippocampal engagement may provide additional evidence for the impact of WWC1 on the retrieval of previously learned knowledge. This notion is further supported by the results of a study using adult KIBRA knockout (KO) mice which found that while KO mice could reach a level of association based memory performance equal to wild type (WT) mice, they required a longer period of time (delayed learning) to reach an equal performance level and KO mice failed to retain the previously learned association as evidenced by performance deficits after 24 hours (18). The authors additionally reported that adult KO mice had deficits in synaptic plasticity at Schaffer collateral-CA1 synapses which were not evident in juvenile KO mice and that all KO mice had dysfunctional activity dependent AMPAR membrane trafficking. While the functional nature of WWC1 SNP rs17070145 remains unknown, future studies should investigate the electrophysiological and behavioral properties of hippocampal neurons from animals transfected with this human polymorphism to assess if its effects are asserted though its interaction with other molecules rather than through expression differences.
Of course our current study has limitations. For one, our fMRI paradigm, which is a blocked design, may not be optimal for assessing episodic memory function. In particular, during the retrieval session subjects were shown old as well as new images resulting in a combination of brain activity related to retrieval of previously encoded material in addition to the encoding of new information. Nonetheless, activation invoked by our blocked design may represent the general state of encoding and retrieving information rather than discrete episodic events which could be assessed better using an event-related design. The cross sectional nature of our study could also be viewed as a limitation. While longitudinal studies are useful in understanding the relationship between point in time measures and future changes (e.g. subsequent declines in function), these studies are difficult to conduct due to practical reasons such as the rapidly changing nature of neuroimaging technology and participant dropout rates. It should be emphasized that our results are not evidence of genotype differences in the rate at which individual hippocampal function changes over time. Rather, our results provide evidence for genotype group differences in the correlation between hippocampal function and increasing chronological age. Lastly, as we touched upon earlier, the functional nature of this intronic SNP remains unknown which limits the power to make truly meaningful interpretations of the underlying molecular mechanisms driving these genetic differences in episodic memory performance. Unraveling this mystery should be a goal for researchers, one that will yield the information much needed to make biologically plausible interpretations of the results from studies investigating the impact of genetic variation in WWC1 and episodic memory function.
Supplementary Material
Acknowledgments
This work was supported by the Division of Intramural Research Programs, National Institute of Mental Health, National Institutes of Health, Bethesda, Maryland and The Lieber institute of Brain Development, Baltimore, Maryland.
Footnotes
Financial Disclosures
All authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cabeza R, Daselaar SM, Dolcos F, Prince SE, Budde M, Nyberg L. Task-independent and Task-specific Age Effects on Brain Activity during Working Memory, Visual Attention and Episodic Retrieval. Cerebral Cortex. 2004;14:364–375. doi: 10.1093/cercor/bhg133. [DOI] [PubMed] [Google Scholar]
- 2.Mattay VS, Goldberg TE, Sambataro F, Weinberger DR. Neurobiology of cognitive aging: Insights from imaging genetics. Biological Psychology. 2008;79:9–22. doi: 10.1016/j.biopsycho.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rasch B, Papassotiropoulos A, de Quervain DF. Imaging genetics of cognitive functions: focus on episodic memory. NeuroImage. 2010;53:870–877. doi: 10.1016/j.neuroimage.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 4.Bearden CE, Karlsgodt KH, Bachman P, van Erp TG, Winkler AM, Glahn DC. Genetic Architecture of Declarative Memory: Implications for Complex Illnesses. Neuroscientist. 2012;18(5):516–32. doi: 10.1177/1073858411415113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deary IJ, Wright AF, Harris SE, Whalley LJ, Starr JM. Searching for genetic influences on normal cognitive ageing. Trends in Cognitive Sciences. 2004;8:178–184. doi: 10.1016/j.tics.2004.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Panizzon MS, Lyons MJ, Jacobson KC, Franz CE, Grant MD, Eisen SA, et al. Genetic architecture of learning and delayed recall: A twin study of episodic memory. Neuropsychology. 2011;25:488–498. doi: 10.1037/a0022569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papassotiropoulos A, Stephan DA, Huentelman MJ, Hoerndli FJ, Craig DW, Pearson JV. Common Kibra alleles are associated with human memory performance. Science. 2006;314:475–478. doi: 10.1126/science.1129837. [DOI] [PubMed] [Google Scholar]
- 8.Schaper K, Kolsch H, Popp J, Wagner M, Jessen F. KIBRA gene variants are associated with episodic memory in healthy elderly. Neurobiol Aging. 2008;29:1123–1125. doi: 10.1016/j.neurobiolaging.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Need AC, Attix DK, McEvoy JM, Cirulli ET, Linney KN, Wagoner AP. Failure to replicate effect of Kibra on human memory in two large cohorts of European origin. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:667–668. doi: 10.1002/ajmg.b.30658. [DOI] [PubMed] [Google Scholar]
- 10.Sédille-Mostafaie N, Sebesta C, Huber K, Zehetmayer S, Jungwirth S, Tragl KH, et al. The role of memory-related gene polymorphisms, KIBRA and CLSTN2, on replicate memory assessment in the elderly. Journal of Neural Transmission. 2012;119:77–80. doi: 10.1007/s00702-011-0667-9. [DOI] [PubMed] [Google Scholar]
- 11.Wersching H, Guske K, Hasenkamp S, Hagedorn C, Schiwek S, Jansen S, et al. Impact of Common KIBRA Allele on Human Cognitive Functions. Neuropsychopharmacology. 2011;36:1296–1304. doi: 10.1038/npp.2011.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nacmias B, Bessi V, Bagnoli S, Tedde A, Cellini E, Piccini C, et al. KIBRA gene variants are associated with episodic memory performance in subjective memory complaints. Neuroscience Letters. 2008;436:145–147. doi: 10.1016/j.neulet.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Burgess JD, Pedraza O, Graff-Radford NR, Hirpa M, Zou F, Miles R. Association of common KIBRA variants with episodic memory and AD risk. Neurobiol Aging. 2011;32(3):557.e1–9. doi: 10.1016/j.neurobiolaging.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Corneveaux JJ, Liang WS, Reiman EM, Webster JA, Myers AJ, Zismann VL. Evidence for an association between KIBRA and late-onset Alzheimer’s disease. Neurobiol Aging. 2008;31:901–909. doi: 10.1016/j.neurobiolaging.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodríguez-Rodríguez E, Infante J, Llorca J, Mateo I, Sánchez-Quintana C, García-Gorostiaga I, et al. Age-dependent association of KIBRA genetic variation and Alzheimer’s disease risk. Neurobiology of Aging. 2009;30:322–324. doi: 10.1016/j.neurobiolaging.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 16.Milnik A, Heck A, Volger C, Heinze H-J, de Quervain DF-F, Papassotiropoulos A. Association of KIBRA with Episodic and Working Memory: A Meta-Analysis. Am J Med Genet Part B. 2011;159B:958–969. doi: 10.1002/ajmg.b.32101. [DOI] [PubMed] [Google Scholar]
- 17.Kauppi K, Nilsson L-G, Adolfsson R, Eriksson E, Nyberg L. KIBRA Polymorphism Is Related to Enhanced Memory and Elevated Hippocampal Processing. The Journal of Neuroscience. 2011;31:14218–14222. doi: 10.1523/JNEUROSCI.3292-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Makuch L, Volk L, Anggono V, Johnson RC, Yu Y, Duning K, et al. Regulation of AMPA Receptor Function by the Human Memory-Associated Gene KIBRA. Neuron. 2011;71:1022–1029. doi: 10.1016/j.neuron.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murty VP, Sambataro F, Das S, Tan HY, Callicott JH, Goldberg TE, et al. Age-related alterations in simple declarative memory and the effect of negative stimulus valence. J Cogn Neurosci. 2009;21(10):1920–33. doi: 10.1162/jocn.2009.21130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lang PJ, Bradley MM, Cuthbert B International Affective Picture System (IAPS) Technical Manual and Affective Ratings. The Center for Research in Psychophysiology, University of Florida; Gainesville, FL: 1999. [Google Scholar]
- 21.Johannsen S, Duning K, Pavenstadt H, Kremerskothen J, Boeckers TM. Temporal-spatial expression and novel biochemical properties of the memory-related protein KIBRA. Neuroscience. 2008;155:1165–1173. doi: 10.1016/j.neuroscience.2008.06.054. [DOI] [PubMed] [Google Scholar]
- 22.Filippini N, Ebmeier KP, MacIntosh BJ, Trachtenberg AJ, Frisoni GB, Wilcock GK, et al. Differential effects of the APOE genotype on brain function across the lifespan. NeuroImage. 2011;54:602–610. doi: 10.1016/j.neuroimage.2010.08.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.