Abstract
TIG3 is a tumor suppressor protein that plays a key role in controlling cell proliferation. TIG3 is observed at reduced levels in epidermal squamous cell carcinoma, and restoration of expression in skin cancer cells reduces cell survival. TIG3 suppresses cell survival via mechanisms that involve localization at the plasma membrane and at the centrosome. TIG3 interacts at the plasma membrane to activate enzymes involved in keratinocyte terminal differentiation, and at the centrosome to inhibit daughter centrosome separation during mitosis leading to cessation of cell proliferation and induction of apoptosis. An important goal is identifying the motifs required for TIG3 localization at these intracellular sites as a method to understand the function of TIG3 at each location. TIG3 encodes an N-terminal hydrophilic region (amino acids 1–135) and a C-terminal membrane anchoring domain (amino acids 135–164). We show that the C-terminal hydrophobic domain targets intact TIG3 to the plasma membrane, but when isolated as an independent element localizes at the mitochondria. We further demonstrate that a segment of the N-terminal hydrophilic region targets the centrosome. These studies provide important insights regarding the mechanisms that guide subcellular localization of this keratinocyte survival regulator.
Keywords: Keratinocyte differentiation, tumor suppressor, cell cycle, centrosome, microtubules, mitochondria, apoptosis, TIG3
Introduction
The TIG3 (Tazarotene-induced gene 3) tumor suppressor protein was originally discovered as a cell survival regulator in human keratinocytes (DiSepio et al., 1998) that has important biological actions in epidermis. TIG3 localizes to the plasma membrane and at the centrosome in normal keratinocytes and skin cancer cells (Sturniolo et al., 2005; Sturniolo et al., 2003; Scharadin et al., 2011; Scharadin et al., 2012). The C-terminal hydrophobic region serves to anchor TIG3 to the cell membrane (Deucher et al., 2000; Sturniolo et al., 2005; Sturniolo et al., 2003) where it activates transglutaminase. TIG3 localization at the centrosome (Scharadin et al., 2011; Scharadin et al., 2012) is associated with inhibition of daughter centrosome separation during mitosis, altered microtubule and organelle distribution and cessation of cell proliferation (Scharadin et al., 2011; Scharadin et al., 2012).
TIG3 expression is confined to cells that are undergoing differentiation in epidermis, and appears to be involved in this process. Thus, TIG3 mRNA and protein levels are reduced in hyperproliferative epidermal diseases including psoriasis and skin cancer (Duvic et al., 2000; Duvic et al., 2003; Duvic et al., 1997), and treating psoriatic lesions with retinoid increases TIG3 level to decrease cell proliferation and activate differentiation (DiSepio et al., 1998; Sturniolo et al., 2003). In cultured keratinocytes, TIG3 reduces cell proliferation and increases cornified envelope formation (Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003). Moreover, TIG3 is expressed at low levels in keratinocyte monolayers but at much higher levels in differentiated keratinocytes grown as epidermal equivalents (Jans et al., 2008). TIG3 also suppresses survival of transformed keratinocytes via a process that involves activation of caspase-associated cell death (Scharadin et al., 2011).
TIG3 displays significant homology to the H-rev107 family of class II tumor suppressors (DiSepio et al., 1998; Ou et al., 2008; Tsai et al., 2007; Jiang et al., 2005; Huang et al., 2000). These proteins encode an N-terminal hydrophilic region and a C-terminal membrane-anchoring domain (DiSepio et al., 1998). The N-terminal domain encodes several motifs which are conserved among the H-rev107 family members, including the NCEHFV and LRYG regions (Deucher et al., 2000). From a functional perspective, the relative quantity of TIG3 that distributes to the plasma membrane versus the pericentrosomal location may have a significant impact on biological outcome. Thus, an important goal is identifying the mechanisms that control TIG3 intracellular distribution. This requires identification of motifs that target TIG3 to specific subcellular compartments. In the present study we identify a centrosome-localizing motif in the N-terminal hydrophilic region and suggest a molecular mechanism that may control subcellular distribution.
Results
C-terminal hydrophobic domain
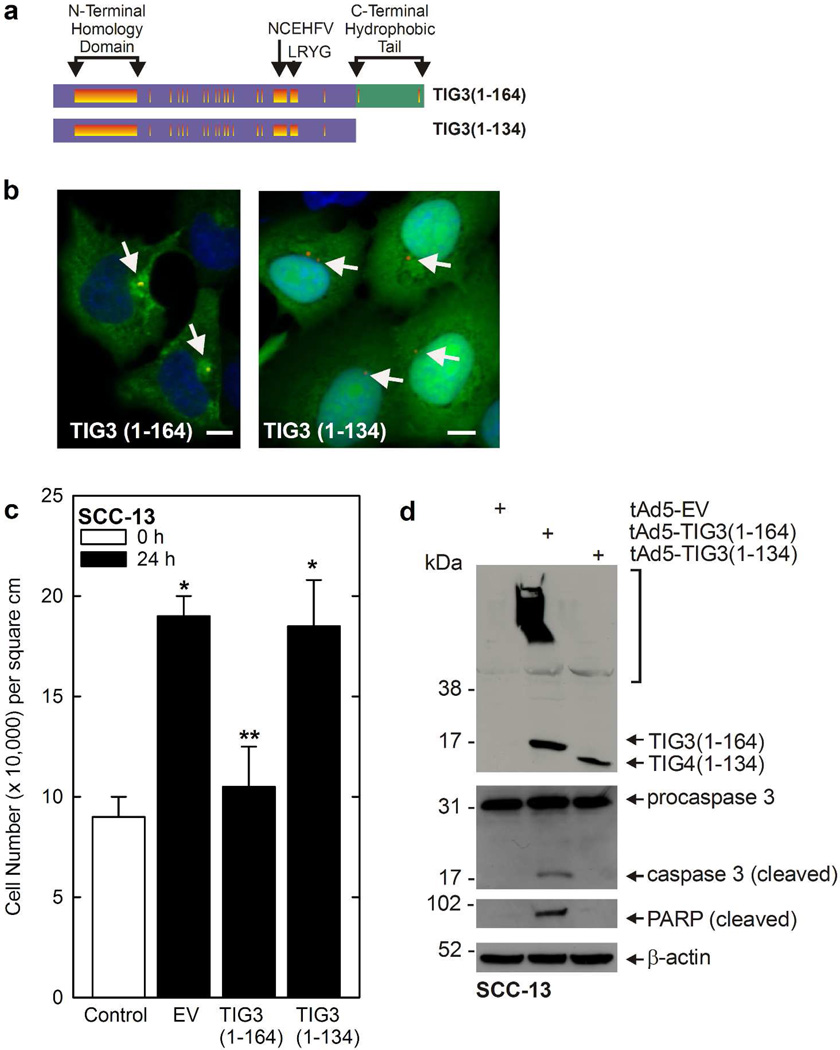
The TIG3 protein encodes a 134 amino acid N-terminal hydrophilic region and a thirty amino acid C-terminal hydrophobic domain (DiSepio et al., 1998; Deucher et al., 2000). The C-terminal hydrophobic domain is conserved among TIG3-related tumor suppressors (DiSepio et al., 1998). To assess the function of this domain, adenoviruses were constructed that encode full-length TIG3 and a mutant lacking the C-terminal 30 amino acids (Fig. 1A). TIG3 was then expressed in epidermis-derived SCC-13 skin cancer cells. Consistent with an absence of endogenous TIG3 in cancer cells, these cells do not express TIG3 (Scharadin et al., 2011; Scharadin et al., 2012). Fig. 1B shows that full-length TIG3 (TIG31–164) distributes in a punctate manner with a particular accumulation around the centrosome where it co-localizes over pericentrin (yellow signal). Recent co-staining studies show that centrosomal markers co-localize with TIG3, and that other organelle markers co-localize to a lesser extent (Scharadin et al., 2011; Scharadin et al., 2012). In contrast, TIG31–134 distributes diffusely throughout the cytoplasm and does not localize with pericentrin (Fig. 1B).
Fig. 1.
TIG3 C-terminal domain is required for appropriate intracellular localization and function. A TIG3 protein domain structure. Purple indicates the N-terminal hydrophilic region and green the C-terminal hydrophobic region. Regions of the protein that are conserved in other H-rev107 family members are shown in orange. The N-terminal Homology Domain, and the NCEHFV and LRYG motifs are shown (Deucher et al., 2000). B Subcellular localization of TIG31–164 and TIG1–134. SCC-13 cells were infected with 10 MOI TIG31–164 or 50 MOI TIG31–134 adenovirus and after 24 h the cells were fixed and stained with anti-TIG3 and appropriate secondary antibody (green). The sections were also stained with anti-pericentrin and appropriate secondary (red). The yellow signal indicates co-localization of TIG3 and pericentrin, red indicates absence of co-localization. Arrows indicate the centrosomes. Bars = 10 µm. C Impact of full-length TIG31–164 and TIG31–134 on cell survival. Cells were infected with 10 MOI of empty (EV) and TIG31–164 and TIG31–134 encoding adenoviruses and at 24 h post-infection the cells were harvested for cell count. The open bar indicates cell number at the time of virus delivery (t = 0) and the shaded bars indicated cell number at 24 h later. The values are mean ± SEM. Single asterisks indicate a significant increase in cell number (n = 3, p < 0.05) as compared to the control, while the double asterisks indicate reduced cell number compared to the other 24 h data points (n = 3, p < 0.05) as determined using the Student’s t-test. D Impact of TIG31–164 and TIG31–134 on SCC-13 cell apoptosis. SCC-13 cells were infected with the indicated adenovirus and after 24 h the cells were harvested for assay of caspase and PARP status. The arrows indicate TIG3 monomers and the bracket indicated high molecular weight crosslinked TIG3 as previously described (Eckert et al., 2009; Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003).
The diffuse cytoplasmic localization corresponds with the loss of TIG3 biological activity. SCC-13 cells were infected with EV, TIG31–164, or TIG31–134 encoding adenovirus and after 24 h cell number was determined (black bars). Fig. 1C shows that cell number doubles in 24 h in EV-infected cultures, but that cell number does not increase in TIG31–164 expressing cells. In contrast, TIG31–134 does not reduce cell survival. To assay biological endpoints, cell lysate was collected from cells to measure caspase activity. Fig. 1D shows that full-length TIG3 induces caspase and PARP cleavage, but that this is not observed in cells expressing TIG31–134. These findings suggest that the C-terminal hydrophobic region of TIG3 is important for proper TIG3 subcellular localization and function.
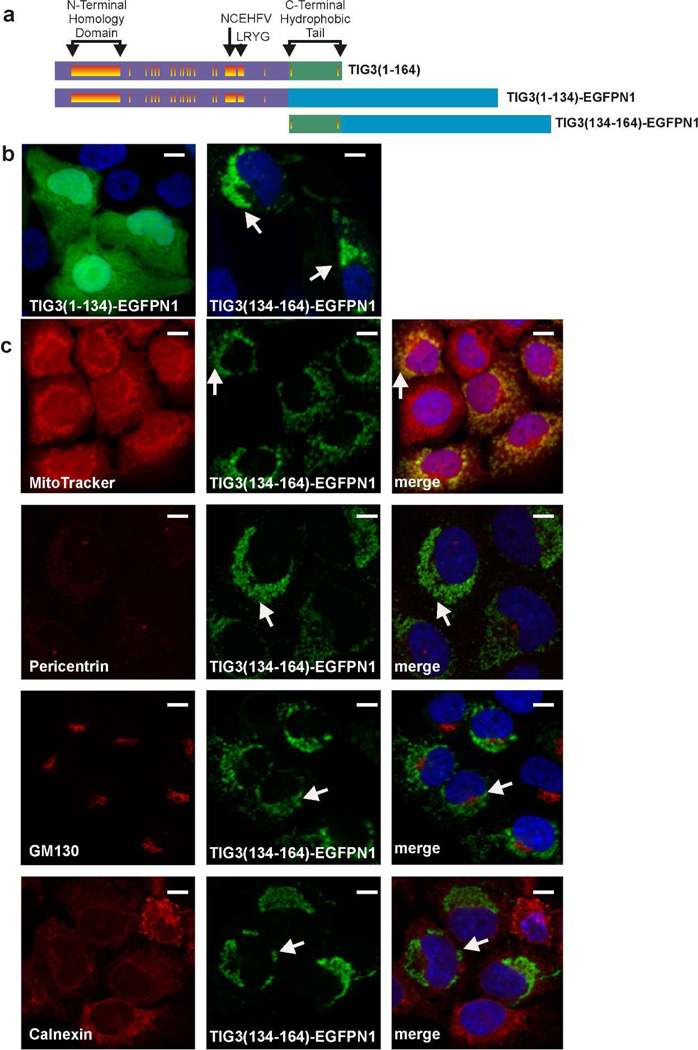
We next created GFP fusion proteins encoding the N-terminal or C-terminal regions of TIG3 (Fig. 2A), and examined the distribution in SCC-13 cells. Fig. 2B shows that TIG31–134-EGFPN1 assumes a cytoplasmic distribution similar to TIG31–134. Interestingly, TIG3134–164-EGFPN1 distributes to a unique intracellular location. To identify this location, TIG31–134-EGFPN1 expressing cells were stained to detect the mitochondria (MitoTracker), the centrosome (pericentrin), the Golgi (GM130) and the endoplasmic reticulum (calnexin). Fig. 2C shows that TIG31–134-EGFPN1 co-localizes with mitochondria (MitoTracker) as indicated by the yellow signal in the merged image.
Fig. 2.
Role of the C- and N-terminal domains in guiding subcellular localization. A Schematic of TIG3-EGFPN1 fusion proteins. The proteins are as described in Fig. 1A, and the blue rectangle designates EGFPN1. B Subcellular localization of TIG31–134-EGFPN1 and TIG3134–164-EGFPN1 in SCC-13 cells. SCC-13 cells were electroporated with 3 µg of the indicated plasmid and after 24 h EGFPN1 fluorescence was detected by confocal microscopy. The arrows indicate the novel subcellular distribution of TIG3134–164-EGPN1. No signal was observed in non-electroporated cells (not shown). C TIG3134–164-EGFPN1 localizes at the mitochondria. Cells were electroporated with 3 µg of TIG3134–164-EGFPN1 and after 24 h the cells were fixed and stained with anti-pericentrin (centrosome), GM130 (Golgi), or calnexin (ER), or incubated with MitoTracker (mitochondria) (red). The images were then visualized by confocal microscopy. The merged images are the composite of the green and red channels and yellow indicates co-localization. Arrows indicate perinuclear TIG3 localization. Red, green (EGFPN1) and merged channels are shown. Bars = 10 µm.
N-terminal hydrophilic region
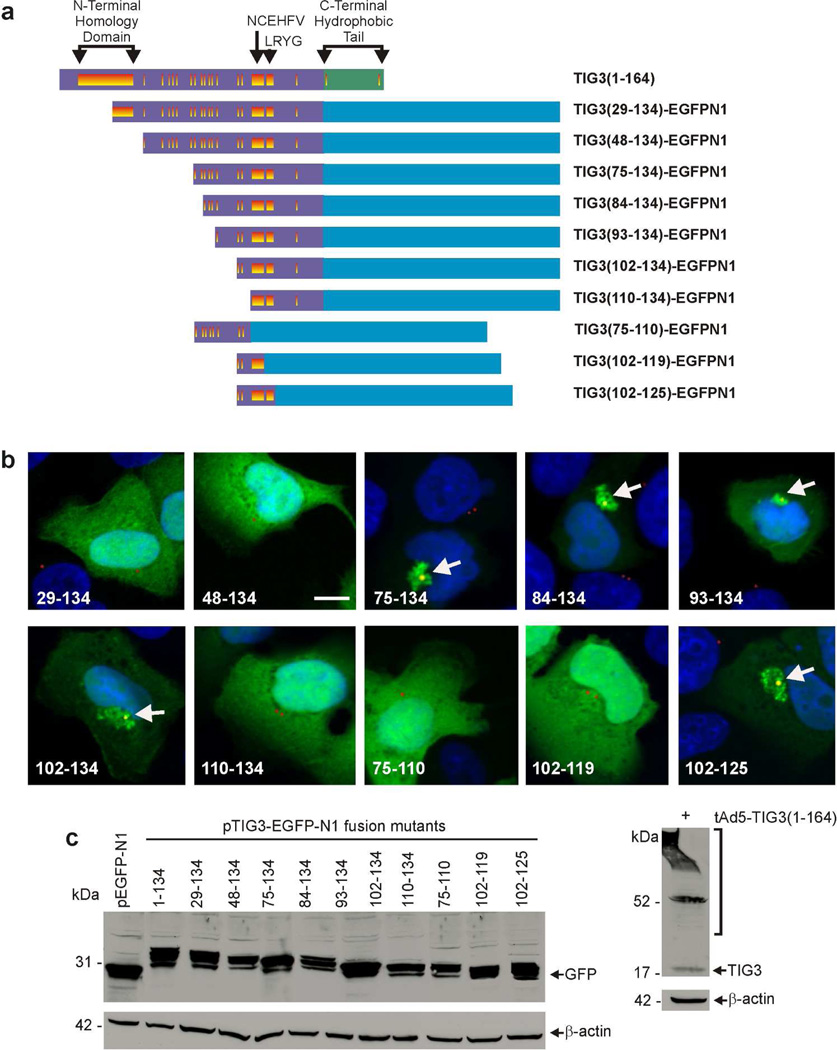
The above studies indicate that neither the N- or C-terminal regions mediate TIG3 pericentrosomal localization, suggesting that the localizing domain may be masked. We therefore generated a series of N-terminal hydrophilic domain-EGFPN1 fusion mutants to identify the pericentrosomal localization motif (Fig. 3A). The TIG329–134, TIG348–134, and TIG3110–134, mutants distribute in the cytoplasm (Fig. 3B), similar to the distribution of TIG31–134 (Fig. 1B). However, like full-length TIG3 (Fig. 1B), TIG375–134, TIG384–134, TIG393–134, TIG3102–134 and TIG3102–125 concentrate at a perinuclear location, suggesting that a centrosome localization signal exists within TIG3 amino acids 102 to 125 (Fig. 3B). This is supported by the finding that truncation of the N-terminus (TIG3110–134) or C-terminus (TIG375–110, TIG3102–119) results in loss of pericentrosomal localization (Fig. 3B). TIG3-GFP fusion mutant-expressing cells were co-stained to detect pericentrin, a centrosome protein (Scharadin et al., 2011; Scharadin et al., 2012). The TIG375–134, TIG384–134, TIG393–134, TIG3102–134 and TIG3102–125 fusion proteins co-localize with the pericentrin signal, but the other mutants do not (Fig. 3B). As previously reported (Scharadin et al., 2011; Scharadin et al., 2012) co-staining with anti-GM130, a Golgi marker, revealed minimal TIG3 co-localization with TIG3 (not shown). Fig. 3C shows that the TIG3 mutant proteins are expressed and that there is no correlation between expression level and centrosome localization, suggesting that the localization pattern is not an artifact related to expression level.
Fig. 3.
Presence of putative centrosome localization signal in the TIG3 N-terminal hydrophilic region. A Schematic showing the TIG3 fusion proteins in which segments of the TIG3 N-terminal hydrophilic region are fused to EGFPN1 (blue). The TIG3 domains are as described in Fig. 1A. B Subcellular distribution of fusion proteins. SCC-13 cells were electroporated with 3 µg of each indicated plasmid and after 24 h the EGFPN1 signal was detected by confocal microscopy (green) and pericentrin was detected with anti-pericentrin and appropriate secondary antibody (red). Co-localization of TIG3 mutant and pericentrin is indicated by the yellow signal (arrows). TIG375–134, TIG384–134, TIG393–134, TIG3102–134, and TIG3102–125 co-localize with pericentrin at a pericentrosomal location (arrows). Bars = 10 µm. C Expression level of TIG3-EGFPN1 fusion proteins. SCC-13 cells were transfected with 3 µg of each construct and after 24 h the cells were harvested and cell extract was prepared and incubated with anti-GFP and appropriate secondary antibody (left panel). As a comparison SCC-13 cells were infected with 10 MOI of tAd5-TIG3 adenovirus and after 24 h extracts were prepared for electrophoresis and detection with anti-TIG3 (right panel). An arrow indicates the TIG3 monomer and the brackets indicate crosslinked forms of TIG3 (Eckert et al., 2009; Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003).
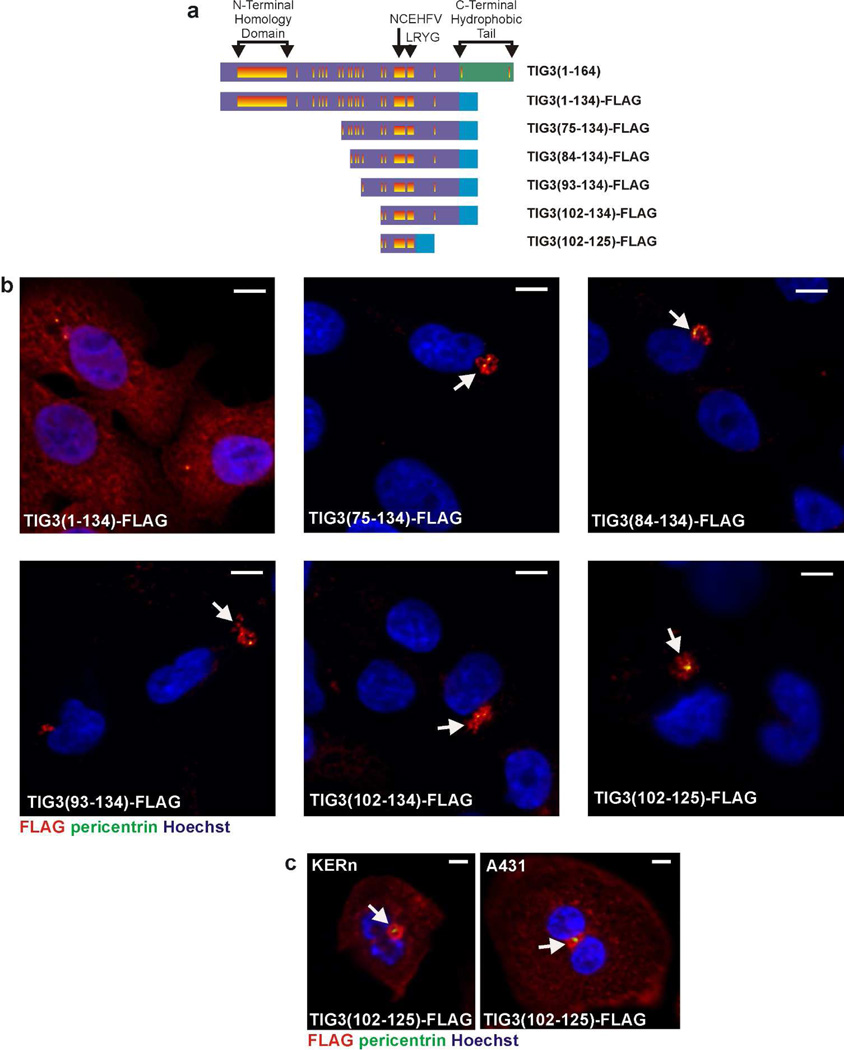
The adjacent panel in Fig. 3C shows wild-type TIG3 expression in cells, illustrating the fact that wild-type TIG3 is much smaller than the TIG3-EGFPN1 fusion proteins used in this experiment due to the size (27 kDa) of EGFPN1. We were concerned that the fusion of EGFPN-1 to the TIG3 mutants may influence subcellular distribution. We therefore examined the location of selected TIG3-FLAG fusion proteins (Fig. 4A). The FLAG tag adds only a small number of amino acids. Analysis of the TIG375–134, TIG384–134, TIG393–134, and TIG3102–134 FLAG-fusion proteins in SCC-13 cells reveals a perinuclear distribution, as evidence by co-localization with pericentrin (yellow), consistent with that observed for the EGFPN1 fusion proteins (Fig. 4B). These findings indicate that a specific TIG3 motif targets pericentrosomal localization of TIG3. We also confirmed that the centrosome localizing motif operates in other epidermis-derived cells by showing that the TIG3102–125 FLAG-fusion protein localizes at the centrosome in A431 cells and normal human foreskin keratinocytes (Fig. 4C).
Fig. 4.
Subcellular localization of TIG3-FLAG fusion proteins. A Schematic of the fusion proteins in which the segments of the N-terminal hydrophilic region are fused to FLAG epitope. The TIG3 domains are described in the legend to Fig. 1A. B SCC-13 cells were electroporated with 3 µg of each TIG3-FLAG fusion protein and after 24 h the cells were fixed and co-stained with the centrosome marker, pericentrin (green), and Cy3-conjugated anti-FLAG (red). Each of these fusion proteins localize at the centrosome (yellow, arrows). Bars = 10 µm. C TIG3(102–125)-FLAG accumulates at the centrosome in A431 cells and normal foreskin keratinocytes (KERn). Cells were electroporated with plasmid encoding TIG3(102–125)-FLAG and then stained with antibodies as indicated above. Bars = 10 µM.
We also assessed whether these constructs are functionally active. We transfected SCC-13 cells with each FLAG-tagged TIG3-mutant shown in Fig. 4A/B and after 48 h co-stained the cells with anti-FLAG and anti-cleaved PARP. PARP cleavage is an endpoint of TIG3 action (Scharadin et al., 2011; Scharadin et al., 2012). Only one mutant, TIG375–134-FLAG retained residual ability to stimulate PARP cleavage (an indicator of apoptosis) (Scharadin et al., 2011; Scharadin et al., 2012). In each of three experiments 10, 12 and 14% of TIG375–134-FLAG-positive cells were PARP cleavage-positive (not shown). Based on these findings, we conclude that the TIG3 region responsible for centrosome localization, amino acids 102 – 125, does not includes the TIG3 domains required to activate cell death.
Conserved elements required for TIG3 centrosomal localization
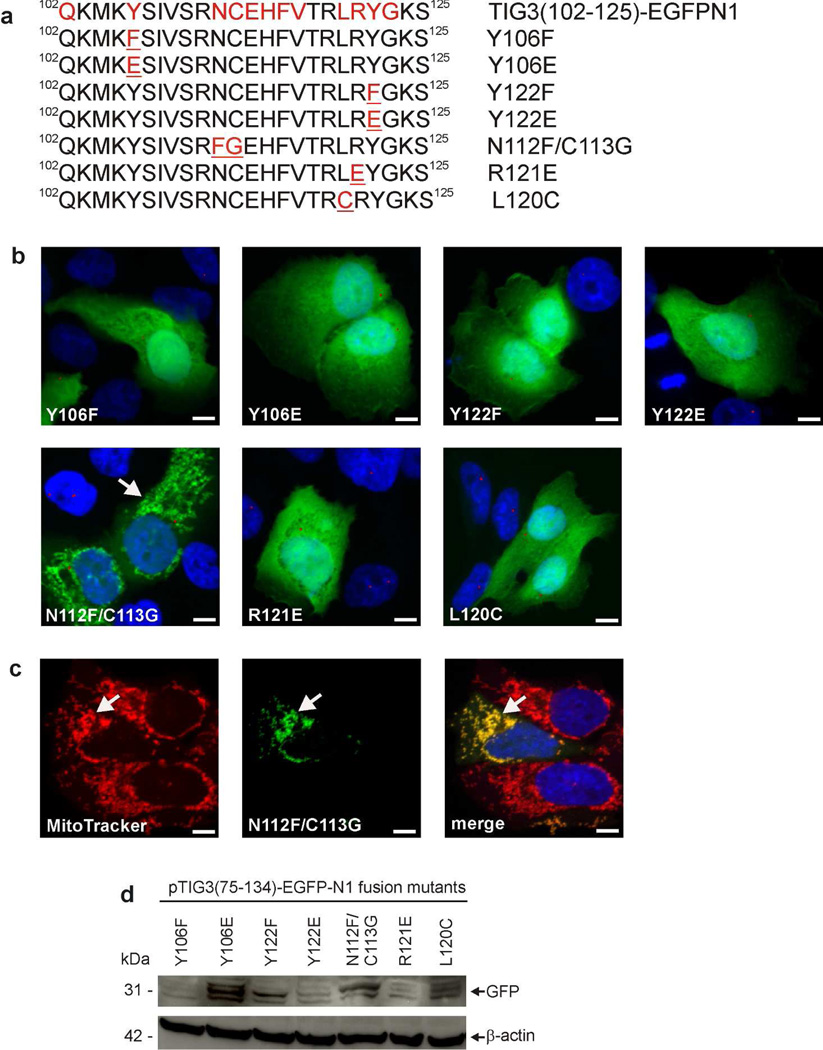
These findings suggest that TIG3 amino acids 102 and 125 are sufficient for TIG3 localization at the centrosome. Within this region are three conserved elements including the NCEHFV and LRYG motifs and a conserved tyrosine (DiSepio et al., 1998) (Fig. 5A). To determine the role of these motifs in TIG3 localization to the centrosome, we tested the impact of specific mutations in each motif. Fig. 5A shows the authentic sequence (top) and each mutated sequence. Expression studies reveal that many of these mutants localize in the cytoplasm (Fig. 5B). While none of the mutants distribute at the centrosome, one mutant, N112F/C113G, localizes at the mitochondria (Fig. 5C). Fig. 5D confirms that each mutant is expressed. These findings indicate that all conserved motifs, present between amino acids 102 and 125, are required for pericentrosomal TIG3 localization.
Fig. 5.
Detailed analysis of the centrosome localization motif. A Role of highly-conserved amino acids in the centrosome-localization motif. pTIG375–134-EGFPN1 encodes amino acids 75–134 of TIG3 fused to the N-terminus of EGFPN1. This plasmid was used as a substrate to generate plasmids encoding the indicated mutated sequences. Conserved sequences are indicated in red and mutated sequences are underlined in red. The distribution of TIG3102–125-EGFPN1 is shown in Fig. 3B. B SCC-13 cells were electroporated with 3 µg of each plasmid and after 24 h the cells were fixed and imaged by confocal microscopy. One of the mutants, N112F/C113G, localizes with punctate structures (arrow), while the others distribute throughout the cytosol. TIG3 mutant staining (anti-FLAG) is green and anti-pericentrin staining is red. Bars = 10 µm. C The N112F/C113G mutant distributes to the mitochondria. SCC-13 cells, expressing the N112F/C113G mutant (green), were incubated with MitoTracker dye to stain mitochondria (red) and imaged by confocal microscopy. The arrows show localization of the N112F/C113G mutant and the yellow color in the merged image indicates co-localization of this mutant with MitoTracker. Bars = 10 µm. D Expression of the mutant proteins. Cells were transfected with 3 µg of each plasmid and after 24 h lysates were prepared for immunoblot detection of each mutant using anti-GFP antibody. Similar results were observed in each of three experiments.
Discussion
TIG3 (Tazarotene-induced gene 3) was originally identified as expressed in keratinocytes treated with the anti-psoriasis drug, tazarotene (DiSepio et al., 1998), and subsequently identified in other tissues (Huang et al., 2000; Uyama et al., 2009b; Uyama et al., 2009a; Ou et al., 2008; Shyu et al., 2005). TIG3 belongs to the NIpC/P60 superfamily of proteins that share homology with lecithin:retinol acyltransferase (LRAT) and the H-rev107 tumor suppressor family (Anantharaman and Aravind, 2003; Husmann et al., 1998; Hajnal et al., 1994; Jahng et al., 2003). The 164 amino acid TIG3 protein includes N-terminal hydrophilic and C-terminal hydrophobic regions. The C-terminal domain, amino acids 135–164, acts as a membrane anchor (Deucher et al., 2000; DiSepio et al., 1998). The N-terminal domain, amino acids 1–134, encodes a hydrophilic domain that includes conserved motifs (Han et al., 2010; Uyama et al., 2009b; Uyama et al., 2009a).
TIG3 subcellular distribution
TIG3 accumulates at the cell membrane and at the centrosome in SCC-13 cells and normal keratinocytes. Membrane association permits TIG3 interaction with type I transglutaminase (TG1) (Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003) which leads to increased TG1 expression and activity resulting in enhanced cornified envelope formation (Sturniolo et al., 2005; Sturniolo et al., 2003). The C-terminal membrane-anchoring domain of TIG3 is required for this activity, and C-terminus-deleted TIG3 redistributes to the cytoplasm and is inactive (Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003).
TIG3 also distributes to a perinuclear localization in normal and transformed keratinocytes (Scharadin et al., 2011), and staining with organelle markers indicates that the centrosome and surrounding area is the major site of localization (Scharadin et al., 2011; Scharadin et al., 2012). The centrosome is a key controller of cell structure, mitosis and intracellular transport (Soldati and Schliwa, 2006; Lim et al., 2009; Doxsey et al., 2005), and serves to structure microtubules in interphase cells during cell division (Doxsey et al., 2005; Schliwa and Woehlke, 2003). Centrosome accumulation of TIG3 causes redistribution of the microtubules, increased microtubule stability, altered microtubule growth, and inhibition of daughter centrosome separation during cell division (Scharadin et al., 2011; Scharadin et al., 2012).
Identifying motifs that control TIG3 subcellular location
Our goal in this study is to identify motifs that drive organelle-specific TIG3 localization. We show that deletion of the C-terminal hydrophobic region results in TIG3 localization in the cytoplasm. This is consistent with our previous reports (Eckert et al., 2009; Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003; Deucher et al., 2000) and the reports of others (Sers et al., 1997; Akiyama et al., 1999; Scharadin et al., 2011). We also show that the isolated TIG3 hydrophobic region targets the mitochondria. This is an unexpected, but potentially important observation. Full-length TIG3 drives differentiation of normal human keratinocytes (Eckert et al., 2009; Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003), but TIG3 mutants lacking the amino terminus are pro-apoptotic. Moreover, the apoptotic activity increases as the TIG3 N-terminus is shortened (Jans et al., 2008). This would predict that the c-terminus may be involved in apoptosis. Our present study, showing that this region targets the mitochondria, is consistent with a pro-apoptotic role of the c-terminus. Understanding the implications of this interesting observation will require additional study.
The TIG3 N-terminal region
The function of the TIG3 N-terminal hydrophilic region is not well understood. It has been reported to encode phospholipase A2 activity (Jahng et al., 2003; Han et al., 2010); however, it is not likely that this activity is required for TIG3 anti-cancer function, as we have shown that mutant encoding only the N-terminal hydrophilic region of TIG3 accumulate in the cytoplasm and have no biological activity. The fact that the TIG3 c-terminal domain localizes at the mitochondria, suggesting that the centrosome localization domain must reside within the N-terminus. To assess this possibility, we constructed a series of TIG3-GFP fusion proteins in which various lengths of the N-terminal hydrophilic region was linked to EGFPN1. Truncation experiments identify a region spanning amino acids 102 to 125 that is a minimal region required for centrosome localization. Centrosome localization of this region was confirmed using TIG3102–125-EGFPN1 and TIG3102–125-FLAG fusion proteins. This region also prompted pericentrosomal localization in A431 skin cancer cells and normal human keratinocytes, indicating that the centrosome-localizing function operates in multiple cell types. This region is conserved among TIG3 and other related members of the H-rev107 protein family (Deucher et al., 2000). There is a conserved motif at amino acid at position 102 (Q102) that is part of the centrosome localization motif (Fig. 5A). The highly conserved NCEHFV and LRYG motifs are also contained within this region (Fig. 5A) (Deucher et al., 2000). Additionally, two conserved tyrosine residues (amino acids 106 and 122) are present (Deucher et al., 2000). Systematic mutation of these amino acids suggests that each of these conserved elements is required for TIG3 targeting to the centrosome.
We also examined the ability of the TIG3 centrosome-localizing motif mutants to activate apoptosis. This analysis revealed that TIG384–134-FLAG, TIG393–134-FLAG, TIG3(102–134)-FLAG and TIG3(102–125)-FLAG are inactive, but that TIG3(75–134)-FLAG retains minimal activity. Thus, amino acids 102–125 of TIG3 specify localization to the centrosome, but lack the protein domains required to drive cell death or differentiation. This indicates that other elements in the protein encode the death domains (Scharadin et al., 2011; Scharadin et al., 2012).
Mechanism of TIG3 intracellular targeting
The 3D structure of the N-terminal hydrophilic region of H-rev107, which shares homology with TIG3, has been determined (Ren et al., 2010b; Ren et al., 2010a; DiSepio et al., 1998). Assuming similar structures, the TIG3 centrosome localization motif corresponds to the β6 sheet and α3 helix of H-rev107 (Ren et al., 2010b; Ren et al., 2010a). A key question is whether this structure can help explain our localization results. The amino acid 102–125 motif that we show is required for TIG3 localization to the centrosome spans the β-sheet and α- helix segments at the N-terminal hydrophilic region immediately upstream of the hydrophobic tail. We suggest that the TIG3 subcellular distribution depends upon protein conformation and that the TIG3 C-terminal hydrophobic domain may cover the β6-sheet/α3-helix region to reduce TIG3 interaction at the centrosome. We further propose that moving this domain aside, via an unknown mechanism, may uncover the β6-sheet/α3-helix region (amino acids 102 – 125) and thereby permit centrosome localization.
In summary, these findings suggest that TIG3 accesses various cellular locations using a variety of targeting motifs, including a C-terminal hydrophobic region that targets membranes and the mitochondria, and a centrosome localizing motif located in the N-terminal hydrophilic domain. It appears possible that the ultimate intracellular destination of TIG3 is determined by change in TIG3 structure that either hides or exposes the centrosome-localizing domain.
Materials and Methods
Cell culture, cloning, adenovirus infection, and electroporation
SCC-13 cells were obtained from American Type Culture Collection (Rockville, MD) and were maintained in high glucose DMEM (Gibco, 11960-044) supplemented with 2 mM L-glutamine, 1 mM sodium pyruvate, 100 U/ml penicillin, 100 µg/ml streptomycin and 5% fetal bovine serum (Sigma, St. Louis, MO). TIG3 constructs were produced by amplifying the desired sequence using primers and then cloning into pcDNA3 or pEGFP-N1 plasmids (Clontech, Mountain View, CA) using NEB Quick Ligase. The sequence of each plasmid was confirmed by DNA sequencing. Adenoviruses were produced as previously described (Sturniolo et al., 2003). tAd5-TIG31–164, is a tetracycline-inducible virus that encodes the full-length TIG3 protein and a tetracycline-responsive enhancer element (Sturniolo et al., 2005). The Ad5-TA virus encodes the tetracycline transactivator (TA). The tAd5-TIG31–134 virus encodes TIG3 protein lacking the C-terminal hydrophobic region, and tAd5-EV is an empty virus used as a control (Sturniolo et al., 2005; Sturniolo et al., 2003). For infection, cells are washed with phosphate-buffered saline, incubated with 10 MOI of TIG3-encoding or empty virus in the presence of 5 MOI of Ad5-TA in Dulbecco’s Modified Eagle’s medium (DMEM) containing 6 µg polybrene/ml (Sigma, H9268). After 5 h, the medium is replaced with virus-free medium and incubation is continued for an additional 24 h prior to preparation of cells and extracts for cell count, immunocytochemistry and immunoblot. For delivery of DNA by electroporation, cells were treated with 3 µg of plasmid using a Lonza Nucleofector II device and human keratinocyte nucleofector kit. Cells were incubated for 24 – 48 h post-electroporation prior to fixation and immunostaining.
Immunological methods
Extracts for immunoblot were prepared in 20 mM Tris- HCl, pH 7.5 lysis buffer containing 150 mM NaCl, 1 mM EGTA, 1 mM EDTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate, 1 mM sodium vanadate, 1 µg/ml leupeptin (Cell Signaling, 9803, Danvers, MA) and 1 mM phenylmethylsulfonyl fluoride. Equal amounts of protein were electrophoresed on Ready Gels (Bio-Rad, Hercules, CA) and transferred to nitrocellulose membranes for immunoblot. For immunofluorescence, cells maintained on coverslips, were infected with adenoviruses or electroporated with plasmid. After 24 – 48 h, the cells were fixed in 4% paraformaldehyde, permeabilized with methanol, and incubated with organelle marker-specific antibody followed by Alexa Fluor 488-conjugated goat anti-rabbit IgG (1:1000) or Alexa Fluor 555-conjugated goat anti-mouse IgG (1:1000) secondary antibody. Cells were then incubated with Hoechst 33258 (1:2000), washed thoroughly and mounted on slides using Fluoromount (Sigma, F4680). Confocal cell images were generated using an Olympus IX81 spinning disk confocal microscope.
Antibodies and Reagents
Polyclonal rabbit anti-TIG3 production was as previously described (Jans et al., 2008; Sturniolo et al., 2005; Sturniolo et al., 2003). β-actin (A1978) and FLAG-M2-Cy3 (A9594) antibodies were purchased from Sigma (St Louis, MO). Caspase 3 (9665), Caspase 9 (9502), and cleaved PARP (9541) antibodies were from Cell Signaling (Danvers, MA). GM130 (610822) and calnexin (610524) antibodies were purchased from BD Transduction Laboratories (San Jose, CA). Pericentrin (ab28144) antibody was from Abcam (Cambridge, MA). GFP antibody (sc-8334) was purchased from Santa Cruz Biotechnology (Santa Cruz, Ca). Alexa Fluor 488-conjugated goat anti-rabbit (A-11034), Alexa Fluor 555-conjugated goat anti-mouse (A-21424), Hoechst 33258 (H3569), and MitoTracker Red CMXRos (M7512) were from Invitrogen (Carlsbad, CA). Rabbit HRP-conjugated IgG (NA934) and mouse HRP-conjugated IgG (NXA931) were from GE Healthcare (Piscataway, NJ). Cells were incubated with 200 nM MitoTracker Red CMXRos for 30 min prior to fixation.
FLAG immunohistochemistry
To monitor FLAG-tagged TIG3 mutant expression, SCC-13 cells, grown on coverslips, were washed twice with TBS then fixed in 3 % paraformaldehyde + 0.5 % Triton X-100. The cells were washed in TBS four times, blocked in 10 % goat serum, and incubated with the Cy3-conjugated FLAG antibody for 1 h.
Apoptosis assay - detection of cleaved PARP
SCC-13 cells (1.2 million) were transfected with 3 µg of plasmid encoding FLAG-tagged TIG3 proteins and plated on glass coverslips. After 48 h the cells were fixed and co-stained with anti-FLAG and anti-cleaved PARP. The cells were imaged using a confocal microscopy and the percentage of FLAG-positive cells that were cleaved PARP-positive was determined as an index of ability of each protein to induce apoptosis.
Acknowledgements
This work was supported by a grant from the National Institutes of Health to Richard L. Eckert (NIH R01 AR49713).
Abbreviations
- TIG3
tazarotene-induced gene 3
- RIG1
retinoid-inducible gene 1
- RARRES3
retinoic acid receptor responder 3
- LRAT
lecithin:retinol acyltransferase
Footnotes
The authors declare no conflict of interest.
References
- Akiyama H, Hiraki Y, Noda M, et al. Molecular cloning and biological activity of a novel Ha-Ras suppressor gene predominantly expressed in skeletal muscle, heart, brain, and bone marrow by differential display using clonal mouse EC cells, ATDC5. J Biol Chem. 1999;274:32192–32197. doi: 10.1074/jbc.274.45.32192. [DOI] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L, et al. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deucher A, Nagpal S, Chandraratna RA, et al. The carboxy-terminal hydrophobic domain of TIG3, a class II tumor suppressor protein, is required for appropriate cellular localization and optimal biological activity. Int J Oncol. 2000;17:1195–1203. doi: 10.3892/ijo.17.6.1195. [DOI] [PubMed] [Google Scholar]
- DiSepio D, Ghosn C, Eckert RL, et al. Identification and characterization of a retinoid-induced class II tumor suppressor/growth regulatory gene. Proc Natl Acad Sci U S A. 1998;95:14811–14815. doi: 10.1073/pnas.95.25.14811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S, Zimmerman W, Mikule K, et al. Centrosome control of the cell cycle. Trends Cell Biol. 2005;15:303–311. doi: 10.1016/j.tcb.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Duvic M, Helekar B, Schulz C, et al. Expression of a retinoid-inducible tumor suppressor, Tazarotene-inducible gene-3, is decreased in psoriasis and skin cancer. Clin Cancer Res. 2000;6:3249–3259. [PubMed] [Google Scholar]
- Duvic M, Nagpal S, Asano AT, et al. Molecular mechanisms of tazarotene action in psoriasis. J Am Acad Dermatol. 1997;37:S18–S24. [PubMed] [Google Scholar]
- Duvic M, Ni X, Talpur R, et al. Tazarotene-induced gene 3 is suppressed in basal cell carcinomas and reversed in vivo by tazarotene application. J Invest Dermatol. 2003;121:902–909. doi: 10.1046/j.1523-1747.2003.12488.x. [DOI] [PubMed] [Google Scholar]
- Eckert RL, Sturniolo MT, Jans R, et al. TIG3: a regulator of type I transglutaminase activity in epidermis. Amino Acids. 2009;36:739–746. doi: 10.1007/s00726-008-0123-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajnal A, Klemenz R, Schafer R, et al. Subtraction cloning of H-rev107, a gene specifically expressed in H-ras resistant fibroblasts. Oncogene. 1994;9:479–490. [PubMed] [Google Scholar]
- Han BG, Cho JW, Cho YD, et al. Expression, purification and biochemical characterization of the N-terminal regions of human TIG3 and HRASLS3 proteins. Protein Expr Purif. 2010;71:103–107. doi: 10.1016/j.pep.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Huang SL, Shyu RY, Yeh MY, et al. Cloning and characterization of a novel retinoid-inducible gene 1(RIG1) deriving from human gastric cancer cells. Mol Cell Endocrinol. 2000;159:15–24. doi: 10.1016/s0303-7207(99)00207-5. [DOI] [PubMed] [Google Scholar]
- Husmann K, Sers C, Fietze E, et al. Transcriptional and translational downregulation of H-REV107, a class II tumour suppressor gene located on human chromosome 11q11-12. Oncogene. 1998;17:1305–1312. doi: 10.1038/sj.onc.1202060. [DOI] [PubMed] [Google Scholar]
- Jahng WJ, Xue L, Rando RR, et al. Lecithin retinol acyltransferase is a founder member of a novel family of enzymes. Biochemistry. 2003;42:12805–12812. doi: 10.1021/bi035370p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans R, Sturniolo MT, Eckert RL, et al. Localization of the TIG3 transglutaminase interaction domain and demonstration that the amino-terminal region is required for TIG3 function as a keratinocyte differentiation regulator. J Invest Dermatol. 2008;128:517–529. doi: 10.1038/sj.jid.5701035. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Wu MS, Chen LM, et al. Identification and characterization of the retinoic acid response elements in the human RIG1 gene promoter. Biochem Biophys Res Commun. 2005;331:630–639. doi: 10.1016/j.bbrc.2005.03.214. [DOI] [PubMed] [Google Scholar]
- Lim HH, Zhang T, Surana U, et al. Regulation of centrosome separation in yeast and vertebrates: common threads. Trends Cell Biol. 2009;19:325–333. doi: 10.1016/j.tcb.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Ou CC, Hsu SC, Hsieh YH, et al. Downregulation of HER2 by RIG1 involves the PI3K/Akt pathway in ovarian cancer cells. Carcinogenesis. 2008;29:299–306. doi: 10.1093/carcin/bgm263. [DOI] [PubMed] [Google Scholar]
- Ren X, Lin J, Jin C, et al. 1H, 13C and 15N resonance assignments of human H-REV107 N-terminal domain. Biomol NMR Assign. 2010a;4:175–178. doi: 10.1007/s12104-010-9238-5. [DOI] [PubMed] [Google Scholar]
- Ren X, Lin J, Jin C, et al. Solution structure of the N-terminal catalytic domain of human H-REV107--a novel circular permutated NlpC/P60 domain. FEBS Lett. 2010b;584:4222–4226. doi: 10.1016/j.febslet.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Scharadin TM, Jiang H, Jans R, et al. TIG3 Tumor Suppressor-Dependent Organelle Redistribution and Apoptosis in Skin Cancer Cells. PLoS One. 2011;6:e23230. doi: 10.1371/journal.pone.0023230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharadin TM, Jiang H, Martin S, et al. TIG3 interaction at the centrosome alters microtubule distribution and centrosome function. J Cell Sci. 2012;125:2604–2614. doi: 10.1242/jcs.096495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, Woehlke G, et al. Molecular motors. Nature. 2003;422:759–765. doi: 10.1038/nature01601. [DOI] [PubMed] [Google Scholar]
- Sers C, Emmenegger U, Husmann K, et al. Growth-inhibitory activity and downregulation of the class II tumor-suppressor gene H-rev107 in tumor cell lines and experimental tumors. J Cell Biol. 1997;136:935–944. doi: 10.1083/jcb.136.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu RY, Chang SC, Yu JC, et al. Expression and regulation of retinoid-inducible gene 1 (RIG1) in breast cancer. Anticancer Res. 2005;25:2453–2460. [PubMed] [Google Scholar]
- Soldati T, Schliwa M, et al. Powering membrane traffic in endocytosis and recycling. Nat Rev Mol Cell Biol. 2006;7:897–908. doi: 10.1038/nrm2060. [DOI] [PubMed] [Google Scholar]
- Sturniolo MT, Chandraratna RA, Eckert RL, et al. A novel transglutaminase activator forms a complex with type 1 transglutaminase. Oncogene. 2005;24:2963–2972. doi: 10.1038/sj.onc.1208392. [DOI] [PubMed] [Google Scholar]
- Sturniolo MT, Dashti SR, Deucher A, et al. A novel tumor suppressor protein promotes keratinocyte terminal differentiation via activation of type I transglutaminase. J Biol Chem. 2003;278:48066–48073. doi: 10.1074/jbc.M307215200. [DOI] [PubMed] [Google Scholar]
- Tsai FM, Shyu RY, Jiang SY, et al. RIG1 suppresses Ras activation and induces cellular apoptosis at the Golgi apparatus. Cell Signal. 2007;19:989–999. doi: 10.1016/j.cellsig.2006.11.005. [DOI] [PubMed] [Google Scholar]
- Uyama T, Jin XH, Tsuboi K, et al. Characterization of the human tumor suppressors TIG3 and HRASLS2 as phospholipid-metabolizing enzymes. Biochim Biophys Acta. 2009a;1791:1114–1124. doi: 10.1016/j.bbalip.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Uyama T, Morishita J, Jin XH, et al. The tumor suppressor gene H-Rev107 functions as a novel Ca2+-independent cytosolic phospholipase A1/2 of the thiol hydrolase type. J Lipid Res. 2009b;50:685–693. doi: 10.1194/jlr.M800453-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]