Abstract
The precise mechanisms governing invasion at the leading edge of SCC and its subsequent metastasis are not fully understood. We aimed to define the cancer related molecular changes that distinguish non-invasive tumor from invasive SCC. To this end, we combined laser capture microdissection with cDNA microarray analysis. We defined invasion-associated genes as those differentially regulated only in invasive SCC nests, but not in actinic keratosis or in situ SCC, compared to normal epidermis. There were 383 up- and 354 down-regulated genes in the “invasion set.” SCC invasion was characterized by aberrant expression of various proteolytic molecules. We noted increased expression of MMP7 and IL-24 in invasive SCC. IL-24 induced the expression of MMP7 in SCC cells in culture. In addition, blocking of MMP7 by a specific antibody significantly delayed the migration of SCC cells in culture. These results suggest a possible contribution of IL-24 to SCC invasion via enhancing focal expression of MMP7, though IL-24 has been suggested to have anti-tumor growth effects in other cancer types. Identification of regional molecular changes that regulate cancer invasion may facilitate the development of new targeted treatments for aggressive cancer.
Introduction
Cutaneous squamous cell carcinoma (SCC), the second most frequent skin cancer, arises from interfollicular epidermal keratinocytes. Transformed malignant cells can proliferate in the epidermis as in situ SCC, eventually cross the basement membrane and enter the dermis to form invasive SCC. Invasion to the dermis is a critical event, since cancer cells are allowed to access lymphatic and to a lesser degree blood vessels, which may result in metastasis. The American Joint Committee on Cancer, in fact, added tumor depth (>2-mm thickness or Clark level ≥IV) as a high-risk feature of SCC (Farasat et al, 2011). Although usually curable by resection, SCC accounts for the majority of approximately 10,000 deaths from non-melanoma skin cancer in the United States each year (Weinberg et al, 2007).
Cancer invasion can be mediated by several processes including degradation of extracellular matrix (ECM) and growth and migration of tumor cells into surrounding stromal regions. Thus, studying the expression and regulation of tumor growth factors and proteolytic molecules within a tissue is important. Gene expression analysis, such as cDNA microarray analysis, has been applied to many cancer types including cutaneous SCC (Dooley et al, 2003; Nindl et al, 2006; Kathpalia et al, 2006; Haider et al, 2006; Hudson et al, 2010; Padilla et al, 2010). These prior studies identified numerous genes that might be involved in SCC pathogenesis. For example, we reported that matrix metallopeptidases (MMPs) including MMP1, MMP10, and MMP13 were selectively expressed in SCC but not in psoriasis, a benign inflammatory skin disease characterized by epidermal hyperproliferation, but without invasion into the dermis by keratinocytes (Haider et al, 2006). This suggests the potential importance of these MMPs in SCC progression.
Cytokines also have significant functions in tumor biology. Th1 cytokines, such as IFN-γ, have been thought to possess anti-tumor properties, whereas Th2 cytokines, such as IL-4, have primarily pro-tumor activity. Recent studies, however, reveal that the cytokines, such as IFN-γ, possess both anti- and pro-tumor activities depending on the tumor types as well as the tumor microenvironment (Zaidi and Merlino. 2011). IL-24, also known as melanoma differentiation associated gene-7 (mda-7), is a member of the IL-10 family of cytokines. It was initially found as a growth inhibitor of melanoma (Jiang et al. 1995). Mounting evidence established a role of IL-24 in inducing apoptosis and cell death in many solid cancers including melanoma as well as epithelial carcinomas of various organs (Dash et al. 2010). However, phase I clinical study using an adenovirus containing IL-24/mda-7 construct (Ad.IL24/mda-7, INGN241) was not able to control tumor growth of a penile, and a head and neck SCC (Cunningham et al, 2005). IL-24 is known to be highly produced in psoriasis lesional skin (Romer et al. 2003). In addition, IL-24 was as potent as IL-22 to induce keratinocytic hyperplasia in a reconstituted human epidermis culture model (Sa et al, 2007). It was thus speculated that IL-24 might have distinct functions in cutaneous SCC growth as opposed to anti-tumor functions established in other cancer types.
Cancer cells may change their phenotypes along with tumor progression, i.e. a switch of E-cadherin to N-cadherin expression (Hazan et al, 2004) and an epithelial-to-mesenchymal transition in invading tumor cells (Thiery et al, 2009). Cells at the invading front might, thus, be genomically distinct from bulk tumor cells. Therefore, we used laser capture microdissection (LCM) in order to collect subpopulations of SCC cells from human skin tissue. We generated the invasion signature gene set of cutaneous SCC, which is a set of genes that were differentially regulated in SCC invasive nests, but not in actinic keratosis (AK) or in situ SCC regions, compared to normal epidermis. We identified significant up-regulation of IL-24 and MMP7 mRNA in the invading front of cutaneous SCC. Molecular interaction of these two molecules and their potential role in SCC progression are discussed in this study.
Results
LCM combined with cDNA microarray analysis provides specific gene expression profiles for various stages of SCC progression
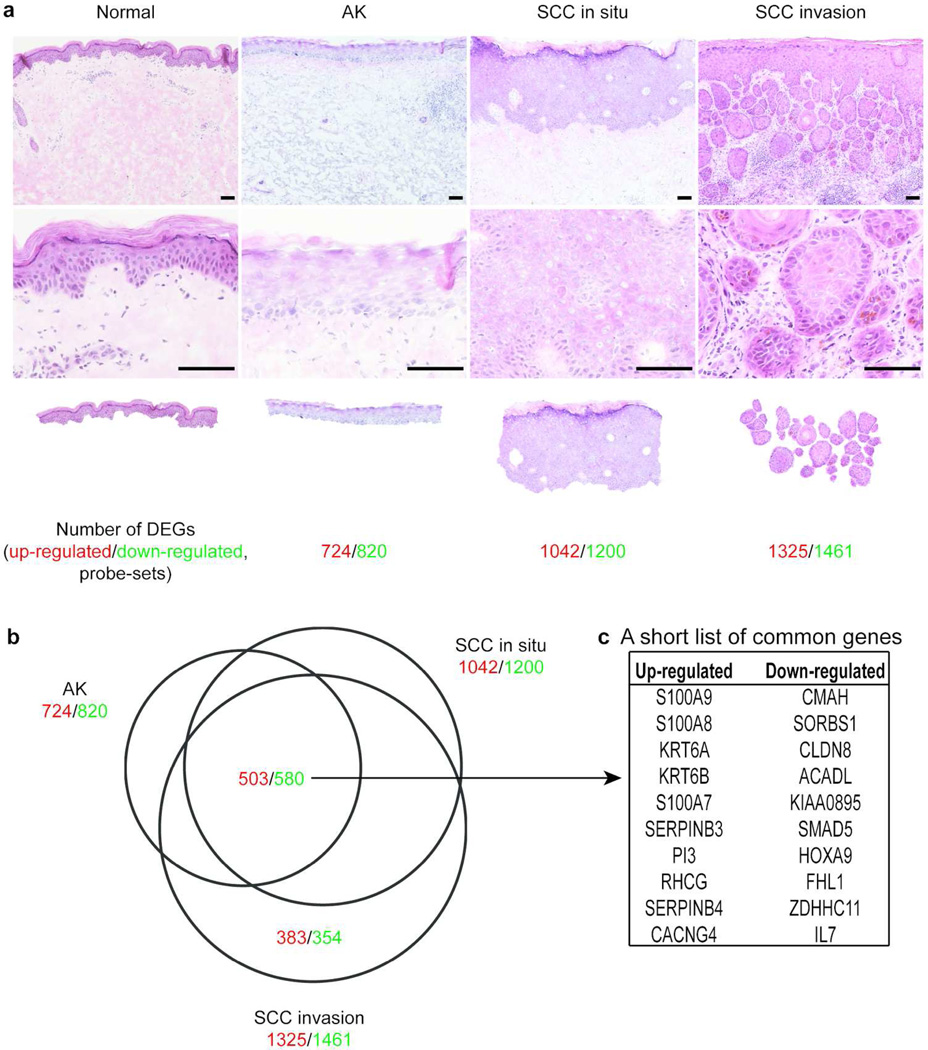
Tumor debulking samples were obtained during Mohs micrographic surgery for SCC. Three transformed epidermal regions in this study that represent the transition to invasive SCC were defined as follows: 1) actinic keratosis (AK atrophic type), regions of severe dysplasia at the basal layer of atrophic epidermis with solar elastosis in dermis, 2) in situ SCC, tumor regions with transformed keratinocytes throughout the entire epidermis that have not crossed the basement membrane, and 3) invasive SCC, tumor nests that have invaded the dermis and disconnected from the bulk tumor mass (Figure 1a). There were 724 up- and 820 down-regulated probe-sets in AK, 1042 up- and 1200 down-regulated probe-sets in in situ SCC, and 1325 up- and 1461 down-regulated probe-sets in invasive SCC compared to microdissected normal epidermis [fold change (FCH)>3.0 and false discovery rate (FDR)<0.05, Figure 1a]. A Venn-diagram demonstrated 1083 (503 up- and 580 down-regulated) commonly regulated probe-sets among the three regions, including S100As, KRT6, and serpins (Figure 1 b–c). A group of genes that was selectively regulated in invasive SCC, but not in dysplasia or in situ SCC, was of particular interest as these genes might have significant roles in SCC invasion to the dermis. This consists of 383 up- and 354 down-regulated probe-sets and these genes were designated as invasion signature genes (Table S1). The complete gene lists comparing each region to microdissected normal epidermis are found in Tables S2–S4.
Figure 1. Combined LCM and cDNA microarray analysis identified region specific gene expression changes in the SCC tissues.
(a) Images of H&E staining for each region and number of differentially expressed probe-sets identified in the corresponding regions compared to normal epidermis. Top panels; lower magnification, middle panels; higher magnifications, bottom panels; images of LCM. Scale bar=100µm. (b) A Venn-Diagram revealed the numbers of commonly regulated probe-sets among the dysplasia/cancer regions compared to normal epidermis as well as uniquely regulated probe-sets in the SCC invasion nests. (c) The top 10 up- and down-commonly regulated probe-sets among the three dysplasia/cancer regions compared to normal epidermis are listed. The numbers in red indicate up-regulated probe-sets whereas those in green indicate down-regulated probe-sets.
The invasion signature gene set characterized the tumor nests at the invasion front
Table 1 shows selected up- and down-regulated invasion signature genes. Genes encoding proteolytic molecules, such as MMPs and PLAU, were highly up-regulated. A cell adhesion molecule LAMC2 was also up-regulated. The expression of PDPN in cutaneous SCC was reported previously by qRT-PCR and by immunohistochemistry (Moussai et al, 2011; Schacht et al, 2005). In contrast, melanocyte-related genes such as TYRP1, DCT, and KIT, as well as keratinocyte differentiation markers including FLG, LOR, and LCE2B, were down-regulated. Ingenuity pathway analysis linked 28 canonical pathways to the SCC invasion signature gene set (Table S5, Fisher’s exact test, p<0.05). The most significant pathway was Leukocyte extravasation signaling (p=9.56×10−4), followed by Aryl hydrocarbon receptor signaling (p=1.46×10−3), and Hypoxia-inducible factor 1α signaling (p=1.54×10−3).
Table 1.
Selected up- and down-regulated SCC invasion signature genes
| a. Selected up-regulated SCC invasion signature genes | |||||||
|---|---|---|---|---|---|---|---|
| Symbol | AK | SCC in situ | SCC invasion | Description | |||
| FCH | FDR | FCH | FDR | FCH | FDR | ||
| MMP12 | 1.14 | 0.90 | 2.31 | 0.37 | 32.97 | < 0.01 | matrix metallopeptidase 12 |
| PTHLH | 0.76 | 0.67 | 3.09 | 0.05 | 17.89 | < 0.01 | parathyroid hormone-like hormone |
| MMP3 | 1.19 | 0.89 | 4.77 | 0.15 | 16.38 | < 0.01 | matrix metallopeptidase 3 |
| AIM2 | 1.80 | 0.34 | 2.72 | 0.08 | 14.61 | < 0.01 | absent in melanoma 2 |
| NEFL | 0.87 | 0.83 | 2.23 | 0.13 | 13.34 | < 0.01 | neurofilament, light polypeptide |
| MMP13 | 0.96 | 0.95 | 2.37 | 0.11 | 12.53 | < 0.01 | matrix metallopeptidase 13 |
| KRT19 | 1.17 | 0.78 | 2.80 | 0.02 | 11.33 | < 0.01 | keratin 19 |
| INHBA | 0.86 | 0.80 | 2.42 | 0.07 | 10.89 | < 0.01 | inhibin, beta A |
| IL7R | 1.62 | 0.36 | 2.86 | 0.03 | 9.45 | < 0.01 | interleukin 7 receptor |
| CXCL10 | 3.63 | 0.24 | 5.26 | 0.11 | 9.44 | 0.02 | chemokine (C-X-C motif) ligand 10 |
| SPP1 | 1.45 | 0.70 | 2.18 | 0.37 | 8.76 | < 0.01 | secreted phosphoprotein 1 |
| PLAU | 2.60 | < 0.01 | 2.73 | < 0.01 | 8.45 | < 0.01 | plasminogen activator, urokinase |
| PDPN | 0.41 | 0.01 | 2.11 | 0.03 | 8.04 | < 0.01 | podoplanin |
| KRT18 | 2.24 | 0.16 | 2.80 | 0.06 | 7.69 | < 0.01 | keratin 18 |
| FN1 | 0.67 | 0.61 | 3.30 | 0.09 | 7.06 | < 0.01 | fibronectin 1 |
| APOBEC3G | 2.45 | 0.03 | 2.55 | 0.02 | 6.96 | < 0.01 | apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G |
| LAMC2 | 0.79 | 0.69 | 1.17 | 0.77 | 6.77 | < 0.01 | laminin, gamma 2 |
| IL24 | 1.20 | 0.79 | 2.03 | 0.23 | 6.74 | < 0.01 | interleukin 24 |
| CXCR4 | 2.60 | 0.09 | 1.78 | 0.31 | 6.16 | < 0.01 | chemokine (C-X-C motif) receptor 4 |
| MMP7 | 1.09 | 0.89 | 1.16 | 0.77 | 5.43 | < 0.01 | matrix metallopeptidase 7 |
| b. Selected down-regulated SCC invasion signature genes | |||||||
|---|---|---|---|---|---|---|---|
| AK | SCC in situ | SCC invasion | |||||
| Symbol | FCH | FDR | FCH | FDR | FCH | FDR | Description |
| TYRP1 | 0.79 | 0.88 | 0.45 | 0.54 | 0.03 | < 0.01 | tyrosinase-related protein 1 |
| FLG | 0.60 | 0.70 | 0.14 | 0.09 | 0.04 | < 0.01 | filaggrin |
| LOR | 1.39 | 0.80 | 0.25 | 0.21 | 0.07 | 0.01 | loricrin |
| TFAP2B | 1.17 | 0.83 | 0.36 | 0.10 | 0.07 | < 0.01 | transcription factor AP-2 beta |
| HPGD | 0.41 | 0.18 | 0.29 | 0.05 | 0.08 | < 0.01 | hydroxyprostaglandin dehydrogenase |
| DCT | 1.24 | 0.81 | 0.31 | 0.11 | 0.08 | < 0.01 | dopachrome tautomerase |
| NFIB | 0.61 | 0.38 | 0.36 | 0.04 | 0.10 | < 0.01 | nuclear factor I/B |
| CYorf15B | 0.40 | 0.38 | 0.23 | 0.12 | 0.11 | 0.02 | chromosome Y open reading frame 15B |
| DDAH1 | 1.18 | 0.74 | 0.73 | 0.50 | 0.12 | < 0.01 | dimethylarginine dimethylaminohydrolase 1 |
| CHI3L1 | 0.76 | 0.73 | 0.33 | 0.11 | 0.12 | < 0.01 | chitinase 3-like 1 |
| LCE2B | 1.15 | 0.87 | 0.25 | 0.05 | 0.13 | < 0.01 | late cornified envelope 2B |
| ANXA9 | 1.17 | 0.81 | 0.54 | 0.26 | 0.13 | < 0.01 | annexin A9 |
| NEB | 1.06 | 0.90 | 0.41 | 0.02 | 0.13 | < 0.01 | nebulin |
| ALOX12 | 1.26 | 0.47 | 0.38 | < 0.01 | 0.14 | < 0.01 | arachidonate 12-lipoxygenase |
| CEP76 | 0.46 | 0.09 | 0.40 | 0.04 | 0.14 | < 0.01 | centrosomal protein 76kDa |
| ACSBG1 | 0.93 | 0.89 | 0.38 | 0.02 | 0.15 | < 0.01 | acyl-CoA synthetase bubblegum family member 1 |
| KIT | 0.64 | 0.43 | 0.42 | 0.09 | 0.16 | < 0.01 | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog |
| NFASC | 0.38 | < 0.01 | 0.36 | 0.00 | 0.16 | < 0.01 | neurofascin homolog |
| AADAC | 1.34 | 0.54 | 0.47 | 0.08 | 0.16 | < 0.01 | arylacetamide deacetylase |
| ATP6V0E2 | 0.80 | 0.55 | 0.38 | 0.00 | 0.17 | < 0.01 | ATPase, H+ transporting V0 subunit e2 |
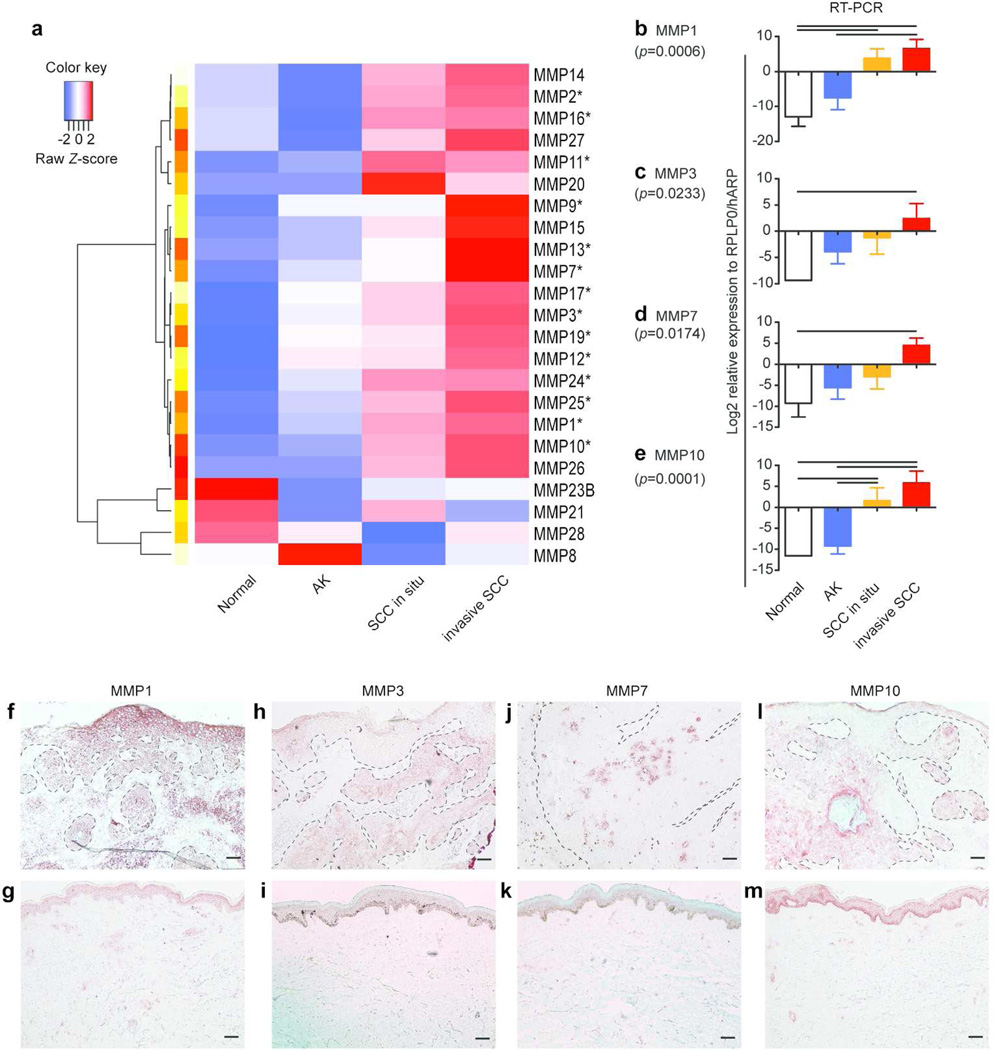
Regional expression of MMPs in the SCC tissue was mapped using pre-amplification quantitative RT-PCR
The expression of MMPs in SCC invasion nests was further investigated, as three of the top 6 genes in the invasion signature gene set were MMPs (MMP12, -3, and -13) and MMPs are critical for degrading the ECM surrounding tumor nests. Eleven MMPs were detected as differentially expressed probe-sets along with SCC progression by microarray analysis (Table 2). The expression of MMP9, -11, and -14 was increased even in AK. The expression of MMP1, -2, and -10 started to elevate in in situ SCC, and further increased in invasive SCC by approximately 2 to 7 fold compared to in situ SCC. MMP1 was the most abundant MMP in invasive SCC with a FCH=107.82, followed by MMP10 with a FCH=48.35. The expression of MMP3, -7, -12, -13, and -17 was selective for invasive SCC. The regional expression difference of all 23 known human MMPs was further tested using the same RNA used for microarray analysis by a more sensitive RT-PCR detection. A heat map clearly showed the increase of expression of multiple MMPs towards invasive SCC (Figure 2a). 12 out of 23 genes tested had significant difference among the four keratinocytic regions (p<0.05, Figure 2b–e and Figure S1). Eleven of those 12 genes were up-regulated in cancer regions compared to normal epidermis or AK. The expression of MMP28 was lower in in situ SCC than in normal epidermis. This was consistent with a previous report showing the specific expression of MMP28 in proliferating keratinocytes during wound healing, but not in SCC (Saarialho-Kere et al, 2002). mRNA of MMP8, -20, -23B, -26, and -27 was rarely detected in any keratinocytic regions tested (Figure S1). In addition, proportional odds model identified 14 MMPs as statistically significant (* in Figure 2a, p values in Table S6). This indicates that gene expression of those MMPs increases the odds of being in a phenotype of higher degree of malignancy. The results of cDNA microarray and pre-amplification RT-PCR were positively correlated as evidenced by Pearson’s r=0.788 (p<0.0001), when the 11 genes detected in both methods were compared (Figure S2).
Table 2.
MMP expression across regions measured by cDNA microarray analysis
| Symbol | AK | SCC in situ | SCC invasion | |||
|---|---|---|---|---|---|---|
| FCH | FDR | FCH | FDR | FCH | FDR | |
| MMP9 | 3.42 | 0.03 | 3.85 | 0.01 | 19.31 | <0.01 |
| MMP11 | 3.05 | <0.01 | 5.57 | <0.01 | 5.77 | <0.01 |
| MMP14 | 3.08 | <0.01 | 4.06 | <0.01 | 3.91 | <0.01 |
| MMP1 | 1.28 | 0.87 | 16.61 | 0.02 | 107.82 | <0.01 |
| MMP2 | 1.05 | 0.93 | 5.85 | <0.01 | 10.50 | <0.01 |
| MMP10 | 1.14 | 0.89 | 7.21 | <0.01 | 48.35 | <0.01 |
| MMP3 | 1.19 | 0.89 | 4.77 | 0.15 | 16.38 | <0.01 |
| MMP7 | 1.09 | 0.89 | 1.16 | 0.77 | 5.43 | <0.01 |
| MMP12 | 1.14 | 0.90 | 2.31 | 0.37 | 32.97 | <0.01 |
| MMP13 | −1.04 | 0.95 | 2.37 | 0.11 | 12.53 | <0.01 |
| MMP17 | 2.62 | <0.01 | 2.81 | <0.01 | 3.50 | <0.01 |
Figure 2. Mapping the regional expression differences of known MMPs was performed by RT-PCR and immunohistochemistry.
(a) A heat map of mean expression of all MMPs tested across the regions was shown. (b–e) RT-PCR was performed for (b) MMP1, (c) MMP3, (d) MMP7, and (e) MMP10 (n=5 for each region). Y-axis shows relative expression level of each gene compared to the housekeeping gene RPLP0/hARP in Log2 (mean ± SEM). A p-value was estimated among four keratinocytic regions (One-way ANOVA with Tukey’s correction). The resultant p values were shown in parentheses. A line between tow bars; statistical significance between two cell types. (f–m) Corresponding protein product was detected by IHC for (f–g) MMP1, (h–i) MMP3, (j–k) MMP7, (l–m) MMP10. Upper panels; normal skin, lower panels; SCC. Dotted lines; borders between epidermis/tumor nests and dermis. Scale bar=100µm.
Altered protein expression of the selected genes was confirmed by immunohistochemistry
Immunohistochemistry was performed in order to confirm the altered protein expression of the corresponding genes within tissue. MMP1, the most up-regulated MMP on our gene list, strongly stained both SCC tumor nests and the interstitial region surrounding tumor nests (Figure 2f–g). Staining of MMP3 and MMP7 was selective in the invading cancer cells, but not in the epidermis of normal skin or the epidermis above the tumor nests (Figure 2h–k), thus confirming the specificity and gradual increase of the mRNA expression in tumor nests. MMP10 strongly stained tumor nests in the dermis, but it also stained the basal layer of keratinocytes in normal skin (Figure 2l–m). These genomic and protein data provide the localization of MMPs within the cutaneous SCC tissues.
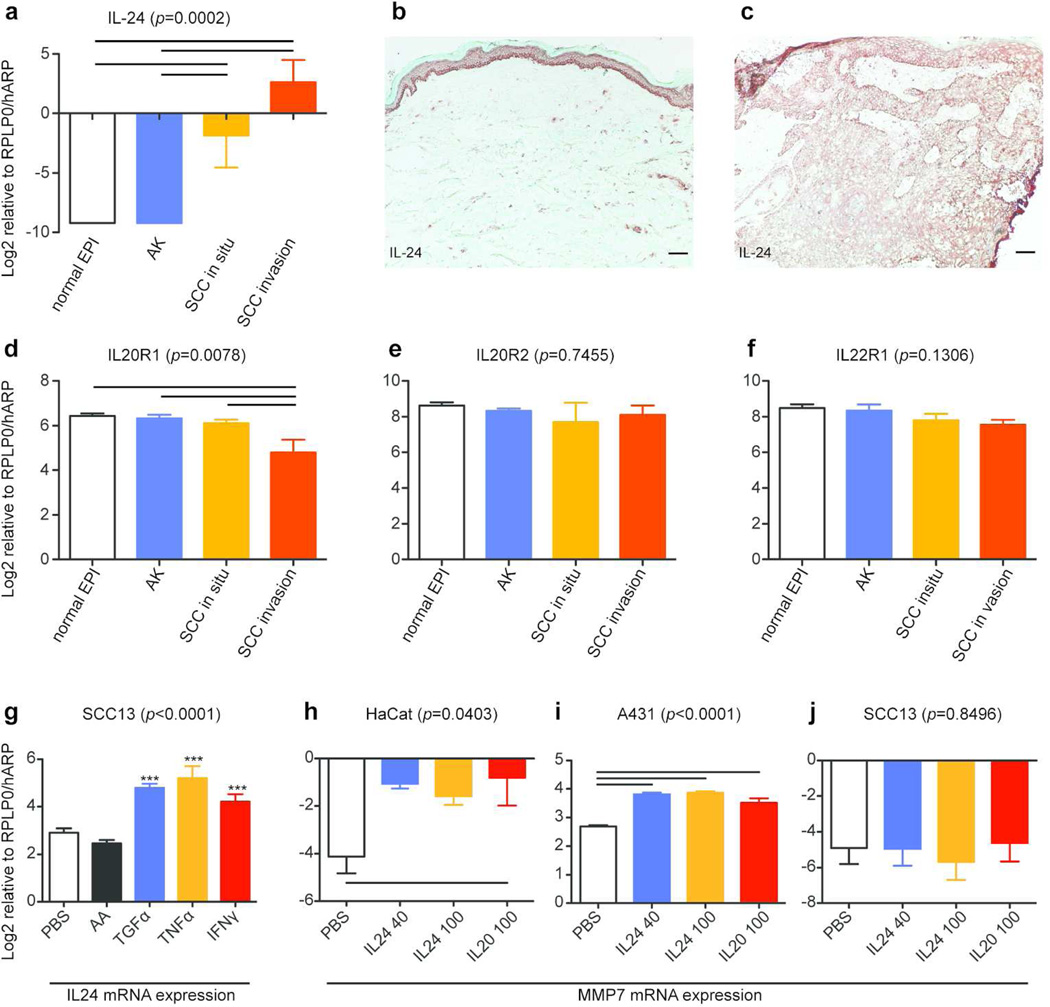
IL-24 was increased in SCC invasion front and up-regulated the expression of MMP7 in SCC cells in vitro
IL-24 was up-regulated in our SCC invasion signature gene set (Table 1a). mRNA and protein expression of IL-24 as well as its receptor subunits (IL-20R1, IL-20R2, and IL-22R1) within the tissues was confirmed. The expression of IL-24 mRNA was detected only in in situ SCC and invasive SCC, but not in AK (Figure 3a). An IL-24 antibody detected signals in SCC tumor nests, but not normal skin (Figure 3b–c). mRNA of the three receptor subunits was constitutively expressed in all regions, although IL20R1 was slightly decreased in invasive SCC (Figure 3d–f). It has been reported that some cytokines, including TGF-α, enhance the expression of IL-24 in HaCaT cells as well as normal human epidermal keratinocytes (Poindexter et al, 2010). The expression of IL-24 mRNA in two human cutaneous SCC cell lines was thus investigated [see detailed descriptions of these cells in the supplemental materials and methods (SMM)]. SCC13 cells (Rheinwald and Beckett, 1981) constitutively expressed IL-24 mRNA without addition of any cytokines. This expression was further increased by stimulation with TGF-α, TNF-α, and IFN-γ (Figure 3g). A431 cells, another SCC cell line (Giard et al, 1973; Price et al, 1988), did not express IL-24 regardless of the stimuli (data not shown). IL-24 utilizes the same receptors to activate cells as IL-20 (Sabat, 2010), and IL-20 has been reported to up-regulate the expression of MMP7 in HaCaT cells (Wang et al, 2006). Hence, we hypothesized that IL-24 might play a role in up-regulating the expression of MMP7 mRNA in SCC. MMP7 mRNA in A431 or HaCaT cells was increased when cultured with 40ng/ml or 100ng/ml of IL-24, or 100ng/ml of IL20 for 24 hours (Figure 3h–i). MMP7 mRNA was not increased in SCC13 cells with the addition of IL20 or IL24 (Figure 3j), although SCC13 cells expressed IL-24 mRNA constitutively. This might be in part explained by significantly lower expression of the IL-24 receptor subunits in SCC13 cells compared to others (Figure S3). Nevertheless, these data suggest a role of IL-24 in enhancing the expression of MMP7 in SCC cells, which may contribute to cancer progression.
Figure 3. IL-24 induced the expression of MMP7 in HaCaT and A431 cells.
(a–c) IL24 expression was examined at the mRNA level (a) and at the protein level in (b) normal skin and (c) SCC. Scale bar=100µm. (d-f) mRNA expression of (d) IL20R1, (e) IL20R2, and (f) IL22R1 was examined. (g) Expression levels of IL-24 mRNA in SCC13 cells after a twelve hour treatment with PBS or indicated cytokines are shown. AA; acidic acetate. (h-j) Expression of MMP7 mRNA in the three cell lines was evaluated after a 24 hour treatment with PBS, IL24 (40ng/ml and 100ng/ml), or IL20 (100ng/ml). Relative expression compared to RPLP0/hARP in Log2 is shown for all RT-PCR results (mean ± SEM). A line between two bars; statistical significance between the two conditions (one-way ANOVA with Tukey’s correction). ***; p<0.0001 against both PBS and AA (repeated measures ANOVA with Tukey’s correction).
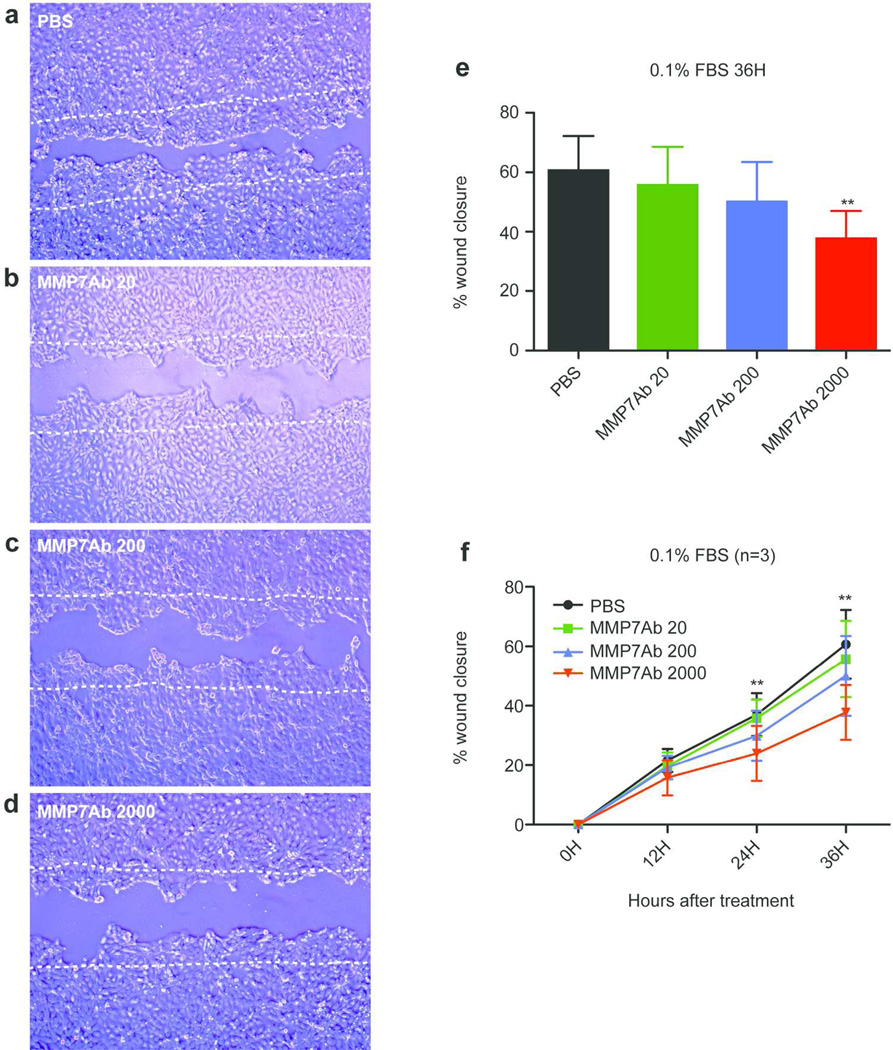
Blocking of MMP7 by a specific antibody significantly delayed the migration of A431 cells
Finally, the function of MMP7 in A431 cells was examined using a scratch assay. A431 cells were selected since this cell line expresses approximately 50-fold higher MMP7 mRNA than the other two cell lines tested (Figure 3h–j). A scratch was made when A431 cells reached 90% confluence, and they were maintained in the 0.1%FBS-containing media with or without an MMP7 antibody (MMP7Ab). The MMP7Ab used in this experiment was shown to block activity of MMP7 (Ito et al. 2007). Figure 4a–d show the gap between cells after a 36 hour treatment with PBS alone (a) or different concentrations of the MMP7Ab [20ng/ml (b), 200ng/ml (c), and 2000ng/ml (d)]. The effect of blocking MMP7 was evaluated by calculating a percent confluence for each condition at each time point. The MMP7Ab significantly delayed the migration of A431 cells at a concentration of 2000ng/ml compared to the other conditions (Figure 4e). This was also true after a 24 hour treatment with the MMP7Ab treatment at 2000ng/ml compared to PBS and 20ng/ml MMP7Ab (Figure 4f). A similar effect was observed when the A431 cells were maintained in media containing 10%FBS (Figure S4). These results suggest the involvement of MMP7 in the migration of cutaneous SCC.
Figure 4. Blocking of MMP7 delayed the migration of A431 cells.
Scratch was made on a culture of A431 cells at 90% confluence and cells were cultured in 0.1%FBS-containing media with or without indicated concentrations of anti-MMP7 antibody (MMP7Ab) for 36 hours. Cells were photographed every 12 hours. (a–d) Representative images of area after a 36 hour cultivation with (a) PBS, (b) 20ng/ml of MMP7Ab, (c) 200ng/ml of MMP7Ab, and (d) 2000ng/ml of MMP7Ab. Two white dotted lines depict the initial area. (e) The bar graph shows mean of percent wound closure for each treatment after 36 hours. (f) The graph depicts the chronological change of the percent wound closure for each condition. An error bar shows S.E.M. (n=3). **; p<0.01 between MMP7Ab2000 vs. PBS and MMP7Ab20 for a 24H treatment, and between MMP7Ab2000 vs. PBS, MMP7Ab20, and MMP7Ab200 for a 36H treatment (repeated measures ANOVA with Tukey’s correction).
Discussion
Previous studies including our own have examined gene expression profiling of cutaneous SCC using total RNA extracted from full thickness tumor tissues (Dooley et al, 2003; Nindl et al, 2006; Kathpalia et al, 2006; Haider et al, 2006; Hudson et al, 2010; Padilla et al, 2010). To our knowledge, no studies combining LCM and cDNA microarray analysis on cutaneous SCC have been published. Our current study confirmed many genes of previous findings, i.e., the enhanced expression of epidermal differentiation complex genes, such as S100As, SPRRs, IVL, and LCE3D. Hudson et al. proposed the expression of KRT13 as a marker of SCC. In this study, we found that KRT13 is specific to in situ and invasive SCC, but not AK. Thus, our study is unique in localizing hundreds of gene products to different stages of tumor progression in vivo by combining LCM and cDNA microarray analysis. This helps to identify molecular interactions that may happen in focal regions of SCC. One such example is the focal regulation of MMP7 by IL-24 cytokine.
IL-24 has been shown to induce apoptosis and cell death in many solid cancers (Dash et al. 2010). Its expression has been inversely correlated with lymph node metastasis or overall survival in some cancers (Ishikawa et al, 2005; Patani et al, 2010; Choi et al, 2011). However, a phase I clinical study of intra-tumoral injections of Ad.IL24/mda-7 resulted in progressive disease in two cases of SCC (Cunningham et al. 2005). New nodules arose around the periphery of the skin nodule of penile SCC after treatment. These results may thus suggest a putative pro-oncogenic function of IL-24 in SCC growth. Our findings may support this notion. We found that IL-24 was significantly overexpressed at the invasion nest of SCC. IL-24 mRNA was constitutively expressed in SCC13 cells and this was further enhanced by culturing with TNF-α, TGF-α, and IFN-γ. The cellular source of IL-24 may, however, be more complicated within the SCC microenvironment. Th2 cells and macrophages as well as melanocytes produce IL-24 (Poindexter et al, 2005), and those cells reside in the SCC microenvironment. In fact, IL-24 mRNA was also up-regulated in inflammatory infiltrating cell regions surrounding tumor nests compared to normal reticular dermis (Belkin et al, 2013). We thus concluded that IL-24 can be derived from multiple cell types including cancer cells within the SCC microenvironment. We have previously reported that IL-24 promoted neither proliferation nor apoptosis of A431 cells when cultured in a starvation condition (0.1%FBS) (Zhang et al, 2013). Instead, we report herein a function of IL-24 in increasing the expression of MMP7 in SCC cells.
mRNA of MMP7 was specifically up-regulated in the invasion nests. The expression of MMP7 was enhanced under a hypoxic condition in colon cancer cells (Remy et al, 2006). Ingenuity pathway analysis associated the genomic profile of invasive SCC with Hypoxia-inducible factor 1α signaling. Thus, the microenvironment of the invasive tumor front may be prone to induce MMP7 by multiple mechanisms. We confirmed the function of MMP7 in the migration of cutaneous SCC cells. The migration of A431 cells was significantly delayed by a specific MMP7Ab in a dose dependent fashion. MMP7 has been shown to be involved in the tumor migration of various cancers through multiple pathways. One such mechanism is proposed in colon cancer cells, where MMP7 cleaves β3 chain of laminin332, resulting in enhancing migration capacity (Remy et al, 2006). Laminin332 can be processed by various enzymes and its remodeling has been implicated in SCC migration (Marinkovich, 2007). All subunits of laminin332 were significantly up-regulated along with SCC progression (Table S4). Taken together, these results may suggest a function of MMP7 in SCC migration via accelerating laminin332 remodeling. In addition, it has been demonstrated that MMP7 shed E-cadherin and released an 80-kDa fragment of E-cadherin from A431 cells, which would facilitate an epithelial-to-mesenchymal transition like process (Shibata et al, 2009). MMP7 also sheds pro heparin binding epidermal growth factor (proHB-EGF) to yield mature HB-EGF (Hashimoto et al, 2002). Overall, MMP7 enhances proliferation, migration, and invasion of cancer cells (Ii et al, 2006). However, we acknowledge that the effect of blockade of MMP7 in 0.1%FBS-containing media was partially diminished in the 10%FBS-containing media, indicating the contribution of other factors to the migration of A431 cells. There was a discrepancy between the expression of IL-24 and MMP7 in the two SCC cell lines tested. It should be noted that strong expression of MMP7 was observed in the aggressive A431 cells. Whereas the less aggressive SCC13 cells and HaCaT cells, immortalized keratinocytes, had only weak or absent expression of MMP7. This observation may support an important role of MMP7 in SCC migration. The function of IL-24 or MMP7 needs to be conclusively demonstrated using appropriate in vivo models. However, current mouse models of SCC do not properly recapitulate human SCC, because mouse SCCs feature follicular differentiation. Therefore, development of the animal models that accurately represent invasion of human SCC is needed.
In summary, we characterized gene expression profiling of invasion front of SCC compared to laser captured AK and SCC that have not invaded the dermis. We suggest a pro-oncogenic role of IL-24 in cutaneous SCC, unlike other cancer types, via inducing MMP7 expression in cancer cells. Further elucidation of mechanisms governing regulation of invasion by SCC will allow for development of targeted therapy for aggressive disease not amenable to conventional intervention.
Materials and Methods
The detailed protocols and statistical analysis are described in the SMM.
Patients and Samples
Institutional review board (The Rockefeller University and Weill Cornell Medical College) approval and written informed consent were obtained before enrolling patients to participate in this study. The study was performed in adherence with the Declaration of Helsinki Principles. Cutaneous SCC samples were obtained from debulking during Mohs micrographic surgery. All tumors were obtained from sun-exposed skin.
LCM
Eight SCC tissues were subjected to LCM following the manufacturer’s protocol for CellCut system (Molecular Machines and Industries, Haslett, MI). Normal skin samples used in this study (n=10) as a reference came from our previous study (Kennedy-Crispin et al, 2012)
RNA extraction and quantification
RNA extraction was performed using RNeasy Kit (QIAGEN, Valencia, CA).
RNA amplification and hybridization
Total RNA was subjected to two-cycle cDNA synthesis (Affymetrix, Santa Clara, CA), with a slight modification described previously (Mitsui et al, 2012). Human Genome U133 A2.0 arrays (Affymetrix) were used. The data have been deposited to the Gene Expression Omnibus repository (GSE42677).
Cell culture
A431 cells were purchased from ATCC. SCC13 cells were kindly provided by professor Rheinwald (Harvard Medical Center, MA). Cells were grown in appropriate media. When they reached 80% confluence, the cells were starved in empty media for 24 hours, followed by stimulation with various cytokines at indicated concentrations for 24 hours.
Scratch assay
A431 cells were grown in a 10%FBS-containing media. When the cells reached 90% confluence, a wound was made by tip of a 200µl pipet tip and the media was replaced with either 0.1%- or 10%FBS-containing media. The MMP7Ab was purchased from R&D systems and added to the culture media at the indicated concentrations. The wounds were photographed every 12 hours up to 36 hours. Measurement of wound area and calculation of percent wound closure are described in the SMM.
Quantitative RT-PCR
Pre-amplification quantitative RT-PCR technique was used for measuring mRNA expression values in total RNA extracted from microdissected samples according to the company’s instruction (Applied Biosystems, Foster City, CA). Regular TaqMan RT-PCR method was used to detect the signals in total RNA extracted from cultured cells. All data were normalized to RPLP0/hARP. Primers and probes used in this experiment are listed in Table S7.
Immunohistochemistry
Frozen skin sections were prepared and standard procedures were used. Antibodies used in this experiment are listed in Table S8.
Statistical analysis
Microarray data were analyzed using R/Bioconductor packages (www.r-project.org). The Harshlight package (Suarez-Farinas et al, 2005) was used to scan Affymetrix chips for spatial artifacts. Expression values were obtained using gcrma algorithm. Expression values were linearly modeled in the limma package framework. For the comparison of interest, the moderated t-test was used to assess differential expression. P-values for each comparison were adjusted for multiple hypotheses using the Benjamini-Hochberg approach. Genes with FDR<0.05 and FCH>3.0 were considered as differentially expressed.
Statistical analysis of all RT-PCR data was performed using Graphpad Prism ver.5 (GraphPad Software, La Jolla, CA). A p<0.05 was considered as statistically significant.
Supplementary Material
Acknowledgement
We appreciate the assistance and advice from the Genomics Resource Center (Zhang, W. and Zhao, C.) and the Bioimaging Resource Center (Bhuvanendran, S. and North, A.) at The Rockefeller University. We thank Michelle A. Lowes for helpful comments and discussions on this manuscript. This research was supported by the Milstein Medical Program and in part by National Institutes of Health (NIH) grant 8 KL2 TR000151 (HM, MS-F, KRS, MVC, IC, and JGK). NG was supported by NIH MSTP grant GM07739. DF was supported by the Frederick J. and Theresa Dow Wallace Fund of the New York Community Trust. JAC was supported by Dana Foundation Human Immunology Consortium Grant.
Abbreviations
- SCC
squamous cell carcinoma
- AK
actinic keratosis
- LCM
laser capture microdissection
- MMP
matrix metallopeptidase
- ECM
extracellular matrix
- IL
interleukin
- H&E
hematoxylin and eosin
- RT-PCR
reverse transcribe polymerase chain reaction
- FCH
fold change
- FDR
false discovery rate
Footnotes
Conflict of interests: The authors declare no competing financial interests.
References
- Belkin DA, Mitsui H, Wang CQ, et al. CD200 upregulation in vascular endothelium surrounding cutaneous squamous cell carcinoma. JAMA Dermatol. 2013;149:178–186. doi: 10.1001/jamadermatol.2013.1609. [DOI] [PubMed] [Google Scholar]
- Choi Y, Roh MS, Hong YS, et al. Interleukin-24 is correlated with differentiation and lymph node numbers in rectal cancer. World J Gastroenterol. 2011;17:1167–1173. doi: 10.3748/wjg.v17.i9.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunningham CC, Chada S, Merritt JA, et al. Clinical and local biological effects of an intratumoral injection of mda-7 (IL24; INGN 241) in patients with advanced carcinoma: a phase I study. 2005. Mol Ther. 2005;11:149–159. doi: 10.1016/j.ymthe.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Dash R, Bhutia SK, Azab B, et al. mda-7/IL-24: a unique member of the IL-10 gene family promoting cancer-targeted toxicity. Cytokine Growth Factor Rev. 2010;21:381–391. doi: 10.1016/j.cytogfr.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley TP, Reddy SP, Wilborn TW, et al. Biomarkers of human cutaneous squamous cell carcinoma from tissues and cell lines identified by DNA microarrays and qRT-PCR. Biochem Biophys Res Commun. 2003;306:1026–1036. doi: 10.1016/s0006-291x(03)01099-4. [DOI] [PubMed] [Google Scholar]
- Farasat S, Yu SS, Neel VA, et al. A new American Joint Committee on Cancer staging system for cutaneous squamous cell carcinoma: Creation and rationale for inclusion of tumor (T) characteristics. J Am Acad Dermatol. 2011;64:1051–1059. doi: 10.1016/j.jaad.2010.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giald DJ, Aaronson SA, Todaro GJ, et al. In vitro cultivation of human tumors: establishment of cell lines derived from a series of solid tumors. J Natl Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- Giannelli G, Falk-Marzillier J, Schiraldi O, et al. Induction of cell migration by matrix metalloprotease-2 cleavage of laminin-5. Science. 1997;277:225–228. doi: 10.1126/science.277.5323.225. [DOI] [PubMed] [Google Scholar]
- Haider AS, Peters SB, Kaporis H, et al. Genomic analysis defines a cancer-specific gene expression signature for human squamous cell carcinoma and distinguishes malignant hyperproliferation from benign hyperplasia. J Invest Dermatol. 2006;126:869–881. doi: 10.1038/sj.jid.5700157. [DOI] [PubMed] [Google Scholar]
- Hashimoto G, Inoki I, Fujii Y, et al. Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. J Biol Chem. 2002;277:36288–36295. doi: 10.1074/jbc.M201674200. [DOI] [PubMed] [Google Scholar]
- Hazan RB, Qiao R, Keren R, et al. Cadherin switch in tumor progression. Ann N Y Acad Sci. 2004;1014:155–163. doi: 10.1196/annals.1294.016. [DOI] [PubMed] [Google Scholar]
- Hudson LG, Gale JM, Padilla RS, et al. Microarray analysis of cutaneous squamous cell carcinomas reveals enhanced expression of epidermal differentiation complex genes. Mol Carcinog. 2010;49:619–629. doi: 10.1002/mc.20636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ii M, Yamamoto H, Adachi Y, et al. Role of matrix metalloproteinase-7 (Matrilysin) in human cancer invasion, apoptosis, growth, and angiogenesis. Exp Biol Med (Maywood) 2006;231:20–27. doi: 10.1177/153537020623100103. [DOI] [PubMed] [Google Scholar]
- Ishikawa S, Nakagawa T, Miyahara R, et al. Expression of MDA-7/IL-24 and its clinical significance in resected non-small cell lung cancer. Clin Cancer Res. 2005;11:1198–1202. [PubMed] [Google Scholar]
- Ito T-K, Ishii G, Chiba H, et al. The VEGF angiogenic switch of fibroblasts is regulated by MMP-7 from cancer cells. Oncogene. 2007;26:7194–7203. doi: 10.1038/sj.onc.1210535. [DOI] [PubMed] [Google Scholar]
- Jiang H, Lin JJ, Su ZZ, Goldstein NI, et al. Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene. 1995;11:2477–2486. [PubMed] [Google Scholar]
- Kathpalia VP, Mussak EN, Chow SS, et al. Genome-Wide transcriptional profiling in human squamous cell carcinoma of the skin identifies unique tumor-associated signatures. J Dermatol. 2006;33:309–318. doi: 10.1111/j.1346-8138.2006.00075.x. [DOI] [PubMed] [Google Scholar]
- Kennedy-Crispin M, Billick E, Mitsui H, et al. Human keratinocytes have a response to injury that upregulates CCL20 and other genes linking innate and adaptive immunity. J Invest Dermatol. 2012;132:105–113. doi: 10.1038/jid.2011.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marinkovich MP. Laminin 332 in squamous-cell carcinoma. Nat Rev Cancer. 2007;7:370–380. doi: 10.1038/nrc2089. [DOI] [PubMed] [Google Scholar]
- Mitsui H, Suárez-Fariñas M, Belkin DA, et al. Combined use of laser capture microdissection and cDNA microarray analysis identifies locally expressed disease-related genes in focal regions of psoriasis vulgaris skin lesions. J Invest Dermatol. 2012;132:1615–1626. doi: 10.1038/jid.2012.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussai D, Mitsui H, Pettersen JS, et al. The human cutaneous squamous cell carcinoma microenvironment is characterized by increased lymphatic density and enhanced expression of macrophage-derived VEGF-C. J Invest Dermatol. 2011;131:229–236. doi: 10.1038/jid.2010.266. [DOI] [PubMed] [Google Scholar]
- Nindl I, Dang C, Forschner T, et al. Identification of differentially expressed genes in cutaneous squamous cell carcinoma by microarray expression profiling. Molecular Cancer. 2006;5:30. doi: 10.1186/1476-4598-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla RS, Sebastian S, Jiang Z, et al. Gene expression patterns of normal human skin, actinic keratosis, and squamous cell carcinoma: a spectrum of disease progression. Arch Dermatol. 2010;146:288–293. doi: 10.1001/archdermatol.2009.378. [DOI] [PubMed] [Google Scholar]
- Patani N, Douglas-Jones A, Mansel R, et al. Tumour suppressor function of MDA- 7/IL-24 in human breast cancer. Cancer Cell Int. 2010 doi: 10.1186/1475-2867-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poindexter NJ, Walch ET, Chada S, et al. Cytokine induction of interleukin-24 in human peripheral blood mononuclear cells. J Leukoc Biol. 2005;78:745–752. doi: 10.1189/jlb.0205116. [DOI] [PubMed] [Google Scholar]
- Poindexter NJ, Williams RR, Powis G, et al. IL-24 is expressed during wound repair and inhibits TGFalpha-induced migration and proliferation of keratinocytes. Exp Dermatol. 2010;19:714–722. doi: 10.1111/j.1600-0625.2010.01077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JE, Sauder DN, Fidler IJ. Tumorigenicity and metastatic behavior in nude mice of two human squamous cell carcinoma lines that differ in production of the cytokine ETAF/IL-1. J Invest Dermatol. 1988;91:258–262. doi: 10.1111/1523-1747.ep12470396. [DOI] [PubMed] [Google Scholar]
- Remy L, Trespeuch C, Bachy S, et al. Matrilysin 1 influences colon carcinoma cell migration by cleavage of the laminin-5 β3 chain. Cancer Res. 2006;66:11228–11237. doi: 10.1158/0008-5472.CAN-06-1187. [DOI] [PubMed] [Google Scholar]
- Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- Rømer J, Hasselager E, Norby PL, et al. Epidermal overexpression of interleukin-19 and-20 mRNA in psoriatic skin disappears after short-term treatment with cyclosporine a or calcipotriol. J Invest Dermatol. 2003;121:1306–1311. doi: 10.1111/j.1523-1747.2003.12626.x. [DOI] [PubMed] [Google Scholar]
- Sa SM, Valdez PA, Wu J, et al. The effects of IL-20 subfamily cytokines on reconstituted human epidermis suggest potential roles in cutaneous innate defense and pathogenic adaptive immunity in psoriasis. J Immunol. 2007;178:2229–2240. doi: 10.4049/jimmunol.178.4.2229. [DOI] [PubMed] [Google Scholar]
- Saarialho-Kere U, Kerkela E, Jahkola T, et al. Epilysin (MMP-28) Expression is associated with cell proliferation during epithelial repair. J Invest Dermatol. 2002;119:14–21. doi: 10.1046/j.1523-1747.2002.01790.x. [DOI] [PubMed] [Google Scholar]
- Sabat R. IL-10 family of cytokines. Cytokine Growth Factor Rev. 2010;21:315–324. doi: 10.1016/j.cytogfr.2010.11.001. [DOI] [PubMed] [Google Scholar]
- Schacht V, Dadras SS, Johnson LA, et al. Up-regulation of the lymphatic marker podoplanin, a mucin-type transmembrane glycoprotein, in human squamous cell carcinomas and germ cell tumors. Am J Pathol. 2005;166:913–921. doi: 10.1016/S0002-9440(10)62311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Marushima H, Asakura T, et al. Three-dimentional culture using radial flow bioreactor induces matrix metalloprotease 7-mediated EMT-like process in tumor cells via TGFβ1/Smad pathway. Int J Oncol. 2009;34:1433–1438. [PubMed] [Google Scholar]
- Suárez-Fariñas M, Pellegrino M, Wittkowski KM, et al. Harshlight: a “Corrective Make-Up” program for microarray chips. BMC Bioinformatics. 2005;6:294. doi: 10.1186/1471-2105-6-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, et al. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Wang F, Lee E, Lowes MA, et al. Prominent production of IL-20 by CD68/CD11c myeloid-derived cells in psoriasis: gene regulation and cellular effects. J Invest Dermatol. 2006;126:1590–1599. doi: 10.1038/sj.jid.5700310. [DOI] [PubMed] [Google Scholar]
- Weinberg AS, Ogle CA, Shim EK. Metastatic cutaneous squamous cell carcinoma: an update. Dermatol Surg. 2007;33:885–899. doi: 10.1111/j.1524-4725.2007.33190.x. [DOI] [PubMed] [Google Scholar]
- Zaidi MR, Merlino G. The two faces of interferon-γ in cancer. Clin Cancer Res. 2011;17:6118–6124. doi: 10.1158/1078-0432.CCR-11-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Fujita H, Mitsui H, et al. Increased Tc22 and T-reg/Tc ratio accelerate progression of transplant-associated SCC. PLoS ONE. 2013;8:e62154. doi: 10.1371/journal.pone.0062154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.