Abstract
Metabolism is the process by which cells convert relatively simple extracellular nutrients into energy and building blocks necessary for their growth and survival. In cancer cells, metabolism is dramatically altered compared with normal cells. These alterations are known as the Warburg effect. One consequence of these changes is cellular addiction to glutamine. Because of this, in recent years the enzyme glutaminase has become a key target for small molecule therapeutic intervention. Like many oncotargets, however, glutaminase has a number of upstream partners that might offer additional druggable targets. This review summarizes the work from the current decade surrounding glutaminase and its regulation, and suggests strategies for therapeutic intervention in relevant cases.
Keywords: Glutaminase, inhibitors, Warburg effect, glutamine addiction
Introduction
During the past decade, cancer metabolism has drawn a significant amount of attention as a target for therapeutic intervention. This is because most cancer cells experience the Warburg effect, described in 1924 by the German biochemist Otto Warburg who observed that cancer cells undergo a high rate of glycolysis and lactic acid fermentation, even under normoxic conditions [1]. The Warburg effect has since been intensively studied, with numerous reports characterizing various aspects of the altered metabolism that it describes. One interesting facet, from a chemotherapeutic point of view, is that, in a large number of cases, cells undergoing a Warburg effect exhibit a marked dependence upon glutamine, to the extent that these cells are referred to as being ‘glutamine addicted’ [2]. Glutamine addiction arises from the need for extracellular glutamine to be consumed for anaplerotic input in[SB1] the citric acid cycle, which accounts for the majority of the bioenergetic needs of normal (nontransformed) cells. The key gatekeeper of this input is the enzyme glutaminase.
Glutaminase exists as two forms in humans: one that was originally named kidney-type glutaminase (KGA) derived from the GLS1 gene and a second called liver-type glutaminase (LGA) derived from the GLS2 gene [3]. Whereas LGA is expressed primarily in the liver, KGA has been found to be ubiquitously distributed [4]. We will thus focus on KGA, because its ubiquitous distribution renders it more likely to be relevant to a number of cancer types. Indeed, KGA has been shown to be upregulated in tumors from diverse systems such breast, lung, cervix, brain and B cells, with glutaminase inhibition having slowed the proliferation of these cancer cell lines [2,5–8]. KGA exists as two splice variants that differ only in their C-terminal regions, with the longer form retaining the acronym KGA and the shorter form being called glutaminase C (GAC) [9]. GAC has been detected in a wide variety of cancer cell lines in culture [2,10].
KGA and GAC are generally believed to localize to the mitochondria, although the exact intramitochondrial localization is still under debate [11,12]. The primary function of the glutaminase enzymes is to catalyze the hydrolysis of l-glutamine to l-glutamate, the latter being generally unable to enter the mitochondria directly. As l-glutamate is formed it is converted to α-ketoglutarate by the enzyme glutamate dehydrogenase (GDH). This product can then be utilized directly in the citric acid cycle, leading to energy and building block production. One other important function of glutamine metabolism is to provide precursors for glutathione production, which helps to maintain the oxidative status of cells. Indeed, glutaminase has been directly linked to redox balance in cancer cells [13–15].
In their inactive states, KGA and GAC exist primarily as dimeric species. In vitro, KGA or GAC can be activated via the addition of inorganic phosphate, which is thought to stimulate the formation of an active tetramer [16,17]. Little is currently known about the regulation of either enzyme in cells, but research performed during the past several years has begun to uncover a number of pathways that might lead to their expression and activation. Additionally, a minimum of ~40 mM phosphate is required to activate glutaminase in vitro, whereas serum phosphate levels tend to be ~1 mM, further suggesting that, in cells, glutaminase is activated by mechanisms involving something other than inorganic phosphate [18,19]. As is the case for many signaling proteins implicated in oncogenic transformation, the upstream regulators of glutaminase activity could serve as excellent drug targets, with some in fact already being the focus of drug discovery efforts.
This review summarizes several recently investigated mechanisms by which glutaminase activity can be modulated via pharmacological agents and examines the most recently developed small molecule inhibitors against KGA and/or [SB2]GAC and their regulators. Specifically, we examine two recently discovered glutaminase inhibitors [968 and bis-2-(5-phenylacetamido-1,2[SB3],4-thiadiazol-2-yl)ethyl sulfide (BPTES)], proceed to a discussion about upregulators of glutaminase [c-Myc, nuclear factor κB (NF-κB), signal transducer and activator of transcription 1 (STAT1) and the rapidly accelerated fibrosarcoma-mitogen-activated protein kinase kinase-extracellular signal-regulated[SB4] kinase (Raf-MEK-ERK) pathway], examine some potential negative regulators [anaphase-promoting complex/cyclosome (APC/C) and LON[SB5]] and finally discuss several recent discoveries in the brain, where glutamine metabolism serves the additional purpose of operating as a neurotransmitter. The scope of this review does not permit a similar in-depth analysis of metabolic pathways downstream of KGA and GAC. However, glutamine and glutamate metabolism downstream of KGA and GAC in cancer is extensively reviewed elsewhere, and the reader is invited to consult these sources for such information [20–22].
Direct inhibition of KGA and GAC[SB6]
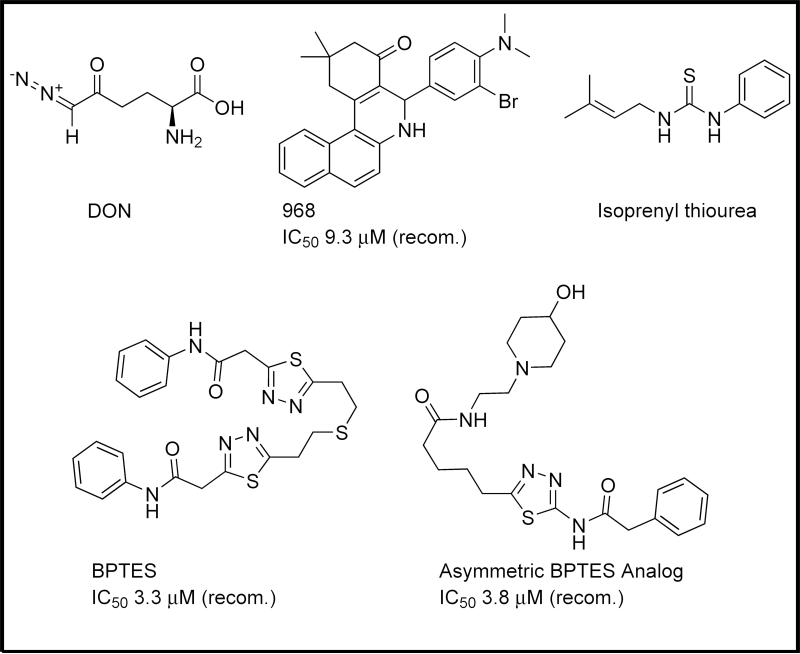
For many years, the predominant drug used to target KGA directly was 6-diazo-5-oxy-l-norleucine (DON; Figure 1). DON acts as an irreversible glutamine-competitive inhibitor. Although effective against glutaminase, DON is not selective and has several verified targets [23].
Figure 1.
Select inhibitors of kidney-type glutaminases. A number of inhibitors of glutaminase are reported in the literature, from the unselective 6-diazo-5-oxy-l-norleucine (DON) to the minimally potent thiourea scaffold and the recently reported 968 and bis-2-(5-phenylacetamido-1,2[SB17],4-thiadiazol-2-yl)ethyl sulfide (BPTES), and their analogs. IC50 data are presented for each inhibitor versus recombinant (recom.) glutaminase or cell systems as available.
During the past decade, several new small molecules have been discovered that inhibit KGA and its splice variant GAC. One of these molecules is 968 (Figure 1), which was discovered by our laboratory and was shown to be an allosteric regulator of GAC [2,24]. The inhibitory potential of 968 has been described in a number of cancer cell lines in culture, as well as in a mouse xenograft model [2]. Owing to the hydrophobic nature of the molecule, it has been difficult to use in animal models and almost all studies to date have been in cell culture. Our most recent report has described the SAR surrounding the ‘hot-spot’ region of the molecule: the halo-benzene ring, which was initially determined to be crucially important for inhibitory potency. We have determined that the electronic nature of the substituents on the ring is relatively unimportant but that they must impart steric bulk perpendicular to the plane of the ring to show a significant inhibitory effect against GAC.
Others have utilized 968 in a variety of studies, demonstrating its potency against GAC and KGA. Simpson et al. conducted studies examining metabolically sensitive epigenetic markers, focusing upon histone H3 and histone H4 acetylation and trimethylation and the effects of these modifications upon a number of cancer-related genes [25,26]. Upon treatment with 968, they found that cells tended to exhibit, for example, enhanced H4 lysine 16 acetylation but that histone deacetylase activity was not significantly impacted overall. These investigators further showed that oncogenes such as Akt and ErbB2 were substantially downregulated, thus suggesting that glutaminase inhibition might be a more effective epigenetic therapy than the use of histone deacetylase inhibitors, which tend to have a broader impact on cells.
More recently, Huang et al. utilized 968 while testing the hypothesis that glutamine metabolism via glutaminase in cancer cells is more responsible for the control of intracellular pH (via ammonia release) than for providing inputs to the citric acid cycle [6]. Although this goes against the established doctrine, the investigators provide intriguing evidence that the modulation of cellular acidity represents at least one important function of glutamine metabolism, showing that glutamine withdrawal was far less lethal to HeLa or MCF-7 cell lines if growth media maintained at pH 7.3 was used rather than growth media maintained at pH 6.3. 968 was then used to show that cell growth was preferentially inhibited at lower pH. These results fail to account for studies showing that cell lines resistant to glutamine withdrawal gain sensitivity if glutamine synthetase inhibitors are added. Glutamine synthetase would, by producing glutamine, presumably cause ammonia to be consumed as an outcome of enzymatic activity, and acidify the cellular environment. Nevertheless, these findings still pose an interesting secondary mechanism regarding glutamine utilization in the tumor environment [27].
968 has resisted enzyme co-crystallization efforts, leaving its exact mechanism of action unknown. Kinetic studies have shown that 968 acts as an allosteric inhibitor, and does not compete with glutamine; moreover, mutagenesis experiments suggest that it binds in a pocket formed between the N and C termini of a GAC dimer (Figure 2). Additionally, we have recently developed fluorescent reporter group assays that enable us to show that 968 binds directly to GAC (unpublished[SB7]). GAC is activated by undergoing a dimer-to-tetramer transition that can be stimulated in vitro by 50–100 mM inorganic phosphate [16,17]. Under conditions where GAC has been activated before 968 addition the ability of the drug to inhibit enzyme activity is severely compromised, whereas if the drug is added to GAC just seconds before activating the enzyme with inorganic phosphate the drug exhibits its full inhibitory effect. This finding probably explains at least part of the difficulty in forming a GAC–968 co-crystal, because crystallization trials are typically performed at protein concentrations that would drive GAC to an active tetramer by mass action (i.e. even without the addition of inorganic phosphate). We hypothesize that GAC undergoes relatively large structural changes upon its activation and inhibitor binding, and that 968 cannot bind the activated enzyme. To the best of our knowledge, 968 has not yet been examined as a potential inhibitor of the LGA isoform of glutaminase.
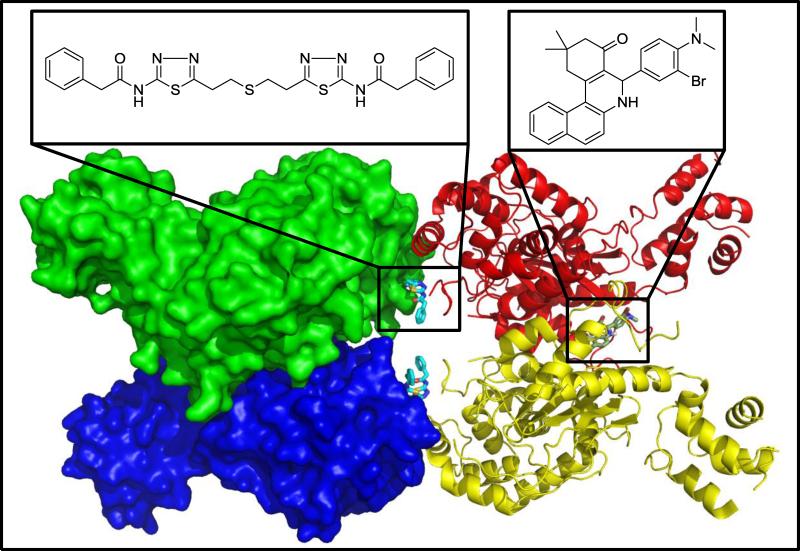
Figure 2.
Structure of the enzyme glutaminase C (GAC) in its tetrameric form (PDB ID 3UO9). GAC monomers are thought to form inactive dimers initially (red–yellow or green–blue pairs) with enzyme activation being accompanied by transition to a tetramer. BPTES (cyan) is shown in this crystal structure to bind between the two dimers, whereas mutagenesis and docking studies currently suggest that 968 (grey) binds between the N and C termini of two GAC monomers.
The second important glutaminase inhibitor to be described during the past few years is BPTES (Figure 1), a symmetrical molecule specific for KGA over the LGA isoform. BPTES has been shown via X-ray crystal structures to bind at the interface where two KGA dimers come together to form a tetramer (Figure 2), and to stabilize a region near the enzyme active site referred to as the ‘gating loop’, which controls access to the catalytic pocket [28–31]. Like 968, BPTES has been shown to exhibit activity in a variety of cancer cells. Unlike 968, BPTES is fully effective against enzyme activity regardless of whether it is added to the enzyme before or after the allosteric activator inorganic phosphate.
BPTES has also been the subject of recent SAR studies. Shukla et al. designed an array of BPTES analogs, focusing in particular upon trying to improve upon the minimal aqueous solubility of BPTES [32]. Many of the discovered analogs were in fact significantly more soluble. Additionally, a number of the analogs lost the symmetrical nature of BPTES (Figure 1). Such molecules maintain much of the BPTES scaffold but lack the benzene rings from one end of the molecule, yielding molecules with one hydrophobic end and one charged end. The authors had not conducted crystallographic studies with any of these asymmetrical derivatives but claim that they bind to the same location as BPTES. It will be interesting to see in future studies if the molecules do in fact bind in a nearly identical orientation to BPTES, or if their asymmetry has enforced a significantly different binding mode.
Other small molecules have been described that inhibit KGA and/or [SB8]GAC, but none of these have attracted the level of attention given to [SB9]968 or BPTES. Gomez et al. described a series of thiourea compounds that exhibited efficacy against glutaminase activity but seem to have been intended as farnesyl diphosphate mimetics (Figure 1) [33,34]. Similarly, Erdmann et al. described a number of glutaminase inhibitors [35]. Unfortunately, the molecular structures were not presented, and even the most potent compound was reported to be less potent than 968, BPTES or DON. These studies show that a number of other scaffolds will be able to inhibit the activity of glutaminase enzymes, and that further investigation in this area is warranted.
Pathways affecting KGA or GAC[SB10]: positive regulation
One important aspect of glutaminase inhibition is that the small molecules 968 and BPTES have been found to be relatively nontoxic. In the case of 968, for example, a drug dose that significantly inhibits the proliferation of cancer cells has been shown to have little effect upon nontransformed cells [2]. This is thought to happen because most cells need little or no glutaminase activity to survive, owing to their use of oxidative phosphorylation as a primary metabolic pathway. However, this effect might be diminished in the brain, where glutamine is a major neurotransmitter and glutaminase activity is required for nonenergetic purposes. In fact, altering glutaminase activity in the brain has been linked to seizures and other disorders [36]. Thus, in some applications, direct inhibition of glutaminase might be undesirable. Similarly, cancer cells are notoriously resistant to drugs and, thus, in a glutamine-addicted glutaminase inhibitor-resistant cell line (e.g. NCI-H157 cells, which are relatively resistant to BPTES despite high glutamine sensativity) it might be desirable to reduce glutaminase activity via some alternate therapy [5]. Therefore, it is worth investigating pathways that intersect with glutaminase.
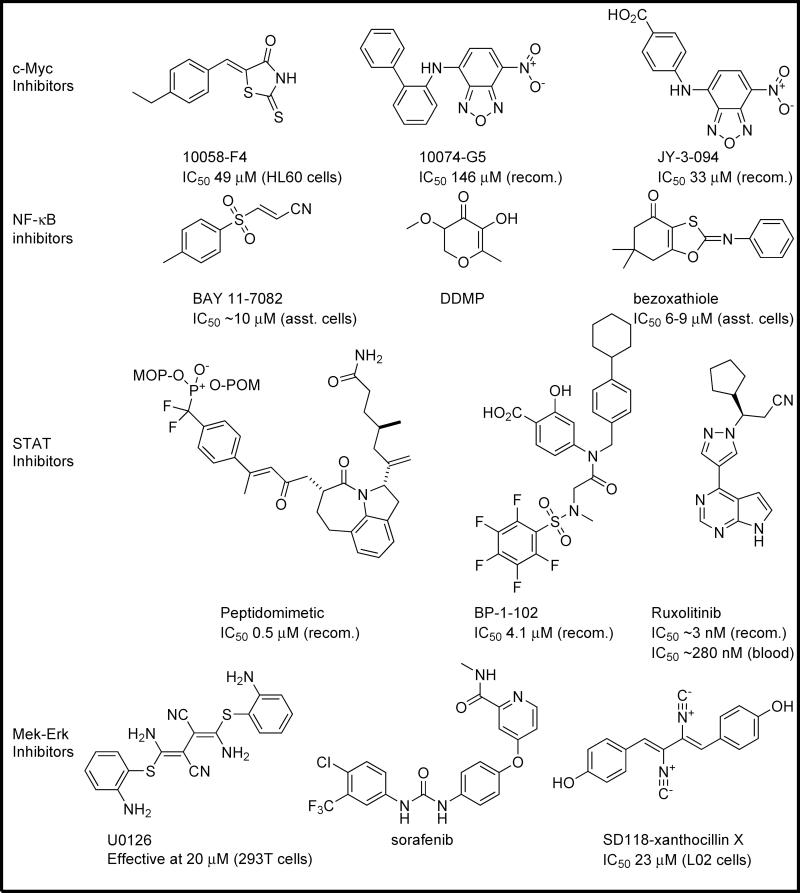
Positive regulators of glutaminase, such as signaling proteins that activate the enzyme, are particularly interesting as targets when designing strategies to modulate glutamine metabolism. For example, by blocking the actions of positive regulators, it would be expected that glutaminase activity, and consequently glutamine metabolism, will be inhibited as well. A number of these positive regulators are druggable, and therefore of interest when selecting inhibitors for glutamine-addicted cancers (Figure 3). Among the best studied of these positive regulators is c-Myc. Although it has long been known to be linked to the regulation of glutaminase activity, Gao et al. showed that c-Myc exerts this influence via the transcriptional suppression of miRNA-23a and miRNA-23b [37], with these microRNA sequences in turn suppressing the transcription of the GLS1 gene. c-Myc is an inherently unstructured protein, which only adopts its functional conformation upon binding a partner such as Max, and thus cannot easily be targeted directly by potential inhibitors. However, the c-Myc–Max dimer can be disrupted via molecular intervention. Owing to the lack of obvious binding pockets, most c-Myc–Max-based drug discovery efforts to date have centered on HTS, with mixed results [38]. These efforts led to studies such as the work of Yin et al., which generated inhibitors including 10058-F4 and 10074-G5 (Figure 4) [39]. Studies detailing the binding mode of these inhibitors have led to a new phase in c-Myc inhibition [40]. An effort by Yap et al. systematically explored the SAR surrounding the c-Myc inhibitor 10074-G5 [41]. This study culminated in the identification of the inhibitor JY-3-094, which exhibited improved potency toward c-Myc and greater solubility than its predecessor (Figure 4).
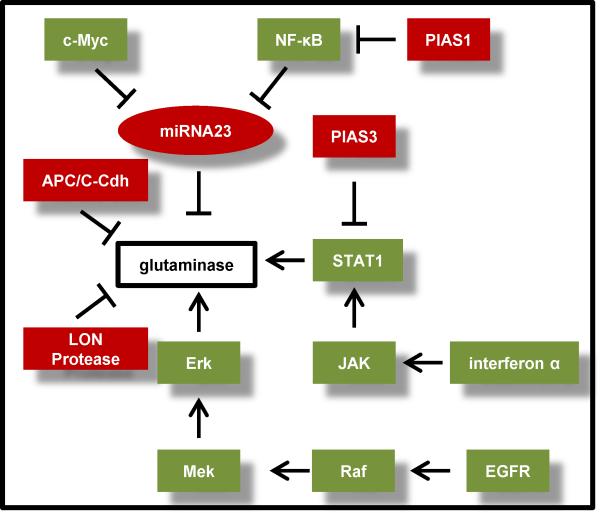
Figure 3.
Pathways that intersect with glutaminase. Glutaminase, a key metabolic enzyme, is regulated by a number of diverse mechanisms. Proteins (boxes) or microRNAs (ovals) that have recently been demonstrated to impact glutaminase expression or activity levels are shown. Signaling proteins (e.g. protein kinases, transcription factors) predicted to lead to increased expression or activity of glutaminase are colored green, and those predicted to lead to decreased activity or expression are colored red.
Abbreviations: PIAS1/3, protein inhibitor of activated STAT1/3; APC/C, anaphase-promoting complex; Cdh, cadherin; JAK, Janus kinase; STAT, signal transducer and activator of transcription; NF-κB, nuclear factor κB; EGFR, epidermal growth factor receptor; Raf, rapidly accelerated fibrosarcoma; MEK, mitogen-activated protein kinase kinase; ERK, extracellular signal-regulated[SB18] kinase.
Figure 4.
An assortment of small molecules that inhibit positive regulators of glutaminase. A number of well-studied cancer-related genes have recently been linked directly to glutaminase activity, and thus to glutamine metabolism. This provides a host of small molecule agents that might be reasonably expected to inhibit glutamine metabolism in cancer cells. IC50 data are presented for each inhibitor versus recombinant (recom.) proteins or cell systems as available: *data presented in the Supplementary Information Figure 1[SB19].
Abbreviations: DDMP[SB20], ; ERK, extracellular signal-regulated[SB21] kinase; MEK, mitogen-activated protein kinase kinase; NF-κB, nuclear factor κB.
Another inhibitor of the miRNA-23a/b sequences, and therefore a potential positive regulator of glutaminase expression, is NF-κB. Moreover, NF-κB was shown to be the common intermediate that modulated glutaminase activation downstream of Rho GTPase signaling [2]. Rathore et al. recently linked NF-κB to glutaminase expression through the inhibition of microRNAs 23a and 23b. Thus, NF-κB can act like c-Myc in transcriptionally suppressing these microRNAs, acting via its subunit p65 [42]. This opens up the heavily investigated field of NF-κB inhibitors as potential modulators of glutamine metabolism. Among the best studied of these inhibitors is BAY 11-7082; however, this molecule, as well as natural products such as curcumin or DDMP[SB11], acts upstream of NF-κB by preventing phosphorylation of the NF-κB activator IκBα (Figure 4). Development of inhibitors is ongoing, with recent efforts from Venkateswararao et al. yielding benzoxathiole derivatives that exhibit low micromolar potency in vitro or in cell culture, similar to that exhibited by the BAY molecules [43]. Although most work has targeted NF-κB activation, a handful of studies have resulted in the direct modulation of NF-κB binding to DNA, primarily through the use of peptidic or heavy metal organometallic agents [44]. Although not ideal, the efficacy of these agents does suggest that direct inhibition of NF-κB should be possible with traditional small molecules.
Another common target for small molecule intervention that has recently been found to be relevant to glutamine metabolism is STAT1. The STAT family is a well-studied set of targets in cancer that are linked to a number of oncogenes [45,46]. GLS1 was implicated in the actions of these transcription factors when Zhao et al. showed that STAT1, in its phosphorylated active form, binds to the GLS1 promoter sequence, and that activation of cellular STAT1 via interferon-α treatment results in the upregulation of GLS1 expression [47]. Because this was shown with interferon-α, it is probable that STAT1 acted in a heterodimer with STAT2, although this was not investigated in the study. A number of discovery efforts have targeted the STAT proteins over recent years. Whereas most have targeted STAT3, peptidomimetic inhibitors recently described by Mandal et al. and nonpeptidic inhibitors such as BP-1-102 described by Zhang et al. (Figure 4) have been shown to have low micromolar affinity for STAT1, suggesting that efforts intended to optimize inhibitors for STAT1 rather than STAT3 should meet with success [48,49]. Interestingly, one drug approved for clinical use by the FDA in 2012[SB12], ruxolitinib (Figure 4), targets Janus kinase (JAK)1 and JAK2, which bind to interferons to activate STAT proteins, with JAK1 being a key intermediate between interferon-α and STAT1 [50]. Thus, the STAT1 activation of glutaminase metabolism can be inhibited, albeit indirectly, by what will probably be a readily commercially available small molecule.
Perhaps most interesting, however, is a recent connection between the Raf-MEK-ERK pathway and glutaminase expression. Thangavelu et al. showed that KGA can bind directly to MEK-ERK via immunoprecipitation assays, and further showed that MEK-ERK pathway activation [via epidermal growth factor (EGF) stimulation] or inactivation [via the inhibitor U0126 – a selective inhibitor of MEK1 and MEK2] could activate or inhibit KGA, respectively [51]. The signaling connection was suggested to be caused by an unknown KGA phosphorylation event via the use of transiently expressed PP2A (a ubiquitous serine–threonine phosphatase). This finding is reminiscent of our own studies, where we found that exposure of isolated GAC from cells to alkaline phosphatase resulted in the loss of all basal enzymatic activity [2]. With the Raf-MEK-ERK pathway linked to glutamine import and metabolism, and the commercial availability of numerous inhibitors for this pathway, this is a particularly promising strategy to inhibit glutamine metabolism indirectly. Several FDA-approved drugs target this pathway (such as sorafenib, a Raf kinase inhibitor), and research into new inhibitors is ongoing, although some recently discovered drugs, such as the marine natural product SD118-xanthocillin X (which has a scaffold strikingly similar to that of U0126; Figure 4), have as yet to be identified protein targets [52].
Pathways affecting KGA and GAC[SB13]: negative regulation
Whereas positive regulators of glutaminase are logical therapeutic targets for blocking the activation of KGA and the metabolic reprogramming of cancer cells, negative regulators of glutaminase could also offer some interesting possibilities for intervention, such as APC/C, an E3 ubiquitin ligase that has been shown by Colombo et al. to form a complex with cadherin[SB14] 1 (Cdh1), which then downregulates glutaminase [53]. APC/C–Cdh1 targets proteins with KEN (Lys-Glu-Asn) box or D ([RH]xxLxx[LIVM]) box sequences for ubiquitination. These proteins, once ubiquitinated, are targeted for degradation by the 26S proteasome. As a negative regulator of glutaminase, direct inhibition of APC/C complexes would be expected to increase glutaminase expression. However, APC/C is downregulated by the early mitotic inhibitor-1 (EMI1), which is itself regulated by the polo-like kinases [54–56]. This suggests a number of juncture points by which APC/C activity could be regulated with the hope of altering glutamine metabolism. Of the two GLS1 splice variants, only KGA has a KEN box or a D box (both located within the final 14 amino acids of its C terminus), which could render the APC/C pathway a particularly attractive target for cancers that rely upon KGA expression and activation rather than the upregulation of GAC.
A second negative regulator of glutaminase to come to light recently is the LON protease. For some years following the pollution of ground water in Kamisu, Japan, the Ochi Laboratory has been investigating the effects of diphenylarsinic acid (DPAA) upon GAC. Recently, Kita et al. showed that DPAA promotes GAC degradation via the action of LON, a serine protease located in the mitochondrial matrix that preferentially targets misfolded or unassembled proteins [57]. DPAA treatment of Hep2A hepatocarcinoma cells caused GAC tetramers to dissociate rapidly, which was followed by GAC degradation. LON was identified as being responsible for GAC degradation; in LON knockdown cells GAC formed insoluble aggregates following DPAA treatment rather than being proteolyzed. Interestingly, DPAA did not effect GAC message levels or its translation. Although DPAA would not be suitable as a drug, this effect provides an interesting mechanism by which GAC might be targeted.
Pathways affecting KGA and GAC[SB15]: brain pathways
Because glutamine is so important for normal brain function, it is not surprising that the brain uses a number of pathways to regulate glutamine levels and glutaminase activity. This offers unique opportunities for glutamine metabolism-based therapies in the brain. One possibility derives from work by Blanco et al., who showed that cocaine-sensitized mice exhibited a decrease in glutaminase activity in the brain, with GLS1 being altered to a much greater extent than GLS2 [58]. Although the exact patterns of changes in expression and activity varied depending upon the region of the brain examined, this still suggests that glutaminase function is affected through the use of cocaine. Although, like DPAA, cocaine is itself unsuitable for use as a drug, therapies that treat cocaine addiction might show similar effects upon glutaminase activity, and thus might be valuable as anticancer agents as well.
Another potentially interesting target is the BCL2/adenovirus E1B 19 kd-interacting protein (BNIP) family. Although the functions of different family members have not been fully elucidated, each shares a small GTPase-interacting domain, and two in particular have been linked to the trafficking of KGA. Buschdorf et al. showed that BNIP-H was able to move KGA to neurite terminals, whereas Boulay et al. reported that Bmcc1s (a short isoform of BNIP-xl) performs the reverse function, bringing KGA to the mitochondria [59,60]. Thus, BNIP family members can directly affect the localization of KGA. Neither member of the BNIP family appears to have been the focus of drug discovery efforts thus far; however, owing to their regulatory functions they could be interesting targets of intervention for glutamine metabolism.
Concluding remarks
Altered metabolism is a key aspect of cancer progression, with most cancers developing an addiction to glutamine. Because of their role in satisfying the metabolic requirements and glutamine addiction of cancer cells, the glutaminase enzymes, and particularly the ubiquitously expressed kidney-type glutaminase isoforms, KGA and GAC, are important targets in cancer chemotherapeutic discovery efforts. We have attempted to highlight the rapidly accelerating research efforts directed at inhibiting glutaminase activity and several of the most recently investigated systems that impact glutaminase function, prioritizing those for which drug development is either ongoing or potentially promising. Positive regulators of glutaminase such as STAT1, c-Myc, NF-κB and ERK allow opportunities to inhibit well-studied signaling proteins while simultaneously negatively impacting glutamine metabolism. Likewise, negative regulators of glutaminase, including APC/C–Cdh1 and LON, represent potential pathways that would allow similar indirect metabolic modulation. Consideration of these approaches to glutaminase inhibition could be valuable when designing drug regimens for patients with highly glutamine-addicted disease, or when interpreting the effects that these inhibitors have in vitro or in animal models. Although current inhibitors of glutaminase are at best lead compounds, ongoing development programs show promise. It is likely that, in the near future, a number of glutaminase inhibitors could enter the toolbox of anticancer therapeutics.
Highlights.
Inhibition of glutamine metabolism is an important mechanism in cancer control
Novel drugs offer a mechanism by which the key metabolic enzyme glutaminase might be inhibited
Regulators of glutaminase offer additional opportunities for modulating cancer metabolism
Acknowledgments
William Katt is a fellow of the American Cancer Society, Illinois Division (grant # PF-10-238-01-CCE). Richard Cerione acknowledges support from the National Institutes of Health. We would like to thank Cindy Westmiller for her excellent secretarial assistance in the preparation of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Teaser: The enzyme glutaminase is a key player in the metabolic changes conferred by the Warburg effect and, together with its regulators, offers opportunities for therapeutic intervention in cancer.
References
- 1.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 2.Wang JB, et al. Targeting mitochondrial glutaminase activity inhibits oncogenic transformation. Cancer Cell. 2010;18:207–219. doi: 10.1016/j.ccr.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Szeliga M, Obara-Michlewska M. Glutamine in neoplastic cells: focus on the expression and roles of glutaminases. Neurochem. Int. 2009;55:71–75. doi: 10.1016/j.neuint.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 4.Aledo JC, et al. Identification of two human glutaminase loci and tissue-specific expression of the two related genes. Mamm. Genome. 2000;11:1107–1110. doi: 10.1007/s003350010190. [DOI] [PubMed] [Google Scholar]
- 5.van den Heuvel AP, et al. Analysis of glutamine dependency in non-small cell lung cancer GLS1 splice variant GAC is essential for cancer cell growth. Cancer Biol. Ther. 2012;13:1185–1194. doi: 10.4161/cbt.21348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang W, et al. A proposed role for glutamine in cancer cell growth through acid resistance. Cell Res. 2013;23:724–727. doi: 10.1038/cr.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le A, et al. Glucose-independent glutamine metabolism via TCA cycling for proliferation and survival in B cells. Cell Metab. 2012;15:110–121. doi: 10.1016/j.cmet.2011.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seltzer MJ, et al. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elgadi KM, et al. Cloning and analysis of unique human glutaminase isoforms generated by tissue-specific alternative splicing. Physiol. Genomics. 1999;1:51–62. doi: 10.1152/physiolgenomics.1999.1.2.51. [DOI] [PubMed] [Google Scholar]
- 10.Szeliga M, et al. Relative expression of mRNAS coding for glutaminase isoforms in CNS tissues and CNS tumors. Neurochem. Res. 2008;33:808–813. doi: 10.1007/s11064-007-9507-6. [DOI] [PubMed] [Google Scholar]
- 11.Kalra J, Brosnan JT. The subcellular localization of glutaminase isoenzymes in rat kidney cortex. J. Biol. Chem. 1974;249:3255–3260. [PubMed] [Google Scholar]
- 12.Aledo JC, et al. Submitochondrial localization and membrane topography of Ehrlich ascitic tumour cell glutaminase. Biochim. Biophys. Acta. 1997;1323:173–184. doi: 10.1016/s0005-2736(96)00189-7. [DOI] [PubMed] [Google Scholar]
- 13.Cardaci S, Ciriolo MR. TCA cycle defects and cancer: when metabolism tunes redox state. Int. J. Cell Biol. 2012;2012:161837. doi: 10.1155/2012/161837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mates JM, et al. Glutaminase isoenzymes as key regulators in metabolic and oxidative stress against cancer. Curr. Mol. Med. 2013;13:514–534. doi: 10.2174/1566524011313040005. [DOI] [PubMed] [Google Scholar]
- 15.Whillier S, et al. Glutamine and alpha-ketoglutarate as glutamate sources for glutathione synthesis in human erythrocytes. FEBS J. 2011;278:3152–3163. doi: 10.1111/j.1742-4658.2011.08241.x. [DOI] [PubMed] [Google Scholar]
- 16.Kvamme E, et al. Kinetics and localization of brain phosphate activated glutaminase. J. Neurosci. Res. 2001;66:951–958. doi: 10.1002/jnr.10041. [DOI] [PubMed] [Google Scholar]
- 17.Curthoys NP, Watford M. Regulation of glutaminase activity and glutamine metabolism. Annu. Rev. Nutr. 1995;15:133–159. doi: 10.1146/annurev.nu.15.070195.001025. [DOI] [PubMed] [Google Scholar]
- 18.Almaden Y, et al. High phosphate level directly stimulates parathyroid hormone secretion and synthesis by human parathyroid tissue in vitro. J. Am. Soc. Nephrol. 1998;9:1845–1852. doi: 10.1681/ASN.V9101845. [DOI] [PubMed] [Google Scholar]
- 19.Turner A, McGivan JD. Glutaminase isoform expression in cell lines derived from human colorectal adenomas and carcinomas. Biochem. J. 2003;370:403–408. doi: 10.1042/BJ20021360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeBerardinis RJ, Cheng T. Q's next: the diverse functions of glutamine in metabolism, cell biology and cancer. Oncogene. 2010;29:313–324. doi: 10.1038/onc.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tennant DA, et al. Targeting metabolic transformation for cancer therapy. Nat. Rev. Cancer. 2010;10:267–277. doi: 10.1038/nrc2817. [DOI] [PubMed] [Google Scholar]
- 22.Cairns RA, et al. Regulation of cancer cell metabolism. Nat. Rev. Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 23.Pinkus LM. Glutamine binding sites. Methods Enzymol. 1977;46:414–427. doi: 10.1016/s0076-6879(77)46049-x. [DOI] [PubMed] [Google Scholar]
- 24.Katt WP, et al. Dibenzophenanthridines as inhibitors of glutaminase C and cancer cell proliferation. Mol. Cancer Ther. 2012;11:1269–1278. doi: 10.1158/1535-7163.MCT-11-0942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simpson NE, et al. An in vitro investigation of metabolically sensitive biomarkers in breast cancer progression. Breast Cancer Res. Treat. 2012;133:959–968. doi: 10.1007/s10549-011-1871-x. [DOI] [PubMed] [Google Scholar]
- 26.Simpson NE, et al. Modifying metabolically sensitive histone marks by inhibiting glutamine metabolism affects gene expression and alters cancer cell phenotype. Epigenetics. 2012;7:1413–1420. doi: 10.4161/epi.22713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tardito S, et al. L-Asparaginase and inhibitors of glutamine synthetase disclose glutamine addiction of beta-catenin-mutated human hepatocellular carcinoma cells. Curr. Cancer Drug Targets. 2011;11:929–943. doi: 10.2174/156800911797264725. [DOI] [PubMed] [Google Scholar]
- 28.Robinson MM, et al. Novel mechanism of inhibition of rat kidney-type glutaminase by bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide (BPTES). Biochem. J. 2007;406:407–414. doi: 10.1042/BJ20070039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeLaBarre B, et al. Full-length human glutaminase in complex with an allosteric inhibitor. Biochemistry. 2011;50:10764–10770. doi: 10.1021/bi201613d. [DOI] [PubMed] [Google Scholar]
- 30.Hartwick EW, Curthoys NP. BPTES inhibition of hGA(124-551), a truncated form of human kidney-type glutaminase. J. Enzyme Inhib. Med. Chem. 2011;27:861–867. doi: 10.3109/14756366.2011.622272. [DOI] [PubMed] [Google Scholar]
- 31.Cassago A, et al. Mitochondrial localization and structure-based phosphate activation mechanism of glutaminase C with implications for cancer metabolism. Proc. Natl. Acad. Sci. U. S. A. 2012;109:1092–1097. doi: 10.1073/pnas.1112495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shukla K, et al. Design, synthesis, and pharmacological evaluation of bis-2-(5-phenylacetamido-1,2,4-thiadiazol-2-yl)ethyl sulfide 3 (BPTES) analogs as glutaminase inhibitors. J. Med. Chem. 2012;55:10551–10563. doi: 10.1021/jm301191p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romero-Gomez M, et al. Use of the compound N-phenyl-N′ (3-methyl-2-butenyl)thiourea for treatment of hepatic encephalopathy. 2011 Vol. WO 2011076967 A1 20110630) (Appl., P.I., ed. [Google Scholar]
- 34.Vega-Perez JM, et al. Isoprenyl-thiourea and urea derivatives as new farnesyl diphosphate analogues: synthesis and in vitro antimicrobial and cytotoxic activities. Eur. J. Med. Chem. 2012;58:591–612. doi: 10.1016/j.ejmech.2012.10.042. [DOI] [PubMed] [Google Scholar]
- 35.Erdmann N, et al. Glutamate production by HIV-1 infected human macrophage is blocked by the inhibition of glutaminase. J. Neurochem. 2007;102:539–549. doi: 10.1111/j.1471-4159.2007.04594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramya LN, et al. L-Asparaginase as potent anti-leukemic agent and its significance of having reduced glutaminase side activity for better treatment of acute lymphoblastic leukaemia. Appl. Biochem. Biotechnol. 2012;167:2144–2159. doi: 10.1007/s12010-012-9755-z. [DOI] [PubMed] [Google Scholar]
- 37.Gao P, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yap JL, et al. Small-molecule inhibitors of dimeric transcription factors: antagonism of protein-protein and protein-DNA interactions. MedChemComm. 2012;3:541–551. [Google Scholar]
- 39.Yin XY, et al. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 40.Hammoudeh DI, et al. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J. Am. Chem. Soc. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 41.Yap JL, et al. Pharmacophore identification of c-Myc inhibitor 10074-G5. Bioorg. Med. Chem. Lett. 2013;23:370–374. doi: 10.1016/j.bmcl.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rathore MG, et al. The NF-kappa B member p65 controls glutamine metabolism through miR-23a. Int. J. Biochem. Cell Biol. 2012;44:1448–1456. doi: 10.1016/j.biocel.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 43.Venkateswararao E, et al. Study on anti-proliferative effect of benzoxathiole derivatives through inactivation of NF-kappa B in human cancer cells. Bioorg. Med. Chem. Lett. 2012;22:4523–4527. doi: 10.1016/j.bmcl.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Garg A, Aggarwal BB. Nuclear transcription factor-kappa B as a target for cancer drug development. Leukemia. 2002;16:1053–1068. doi: 10.1038/sj.leu.2402482. [DOI] [PubMed] [Google Scholar]
- 45.Yu H, et al. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat. Rev. Immunol. 2007;7:41–51. doi: 10.1038/nri1995. [DOI] [PubMed] [Google Scholar]
- 46.Yu H, Jove R. The STATs of cancer—new molecular targets come of age. Nat. Rev. Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 47.Zhao LX, et al. Interferon-alpha regulates glutaminase 1 promoter through STAT1 phosphorylation: relevance to HIV-1 associated neurocognitive disorders. PLoS One. 2012;7:e32995. doi: 10.1371/journal.pone.0032995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mandal PK, et al. Potent and selective phosphopeptide mimetic prodrugs targeted to the Src homology 2 (SH2) domain of signal transducer and activator of transcription 3. J. Med. Chem. 2011;54:3549–3563. doi: 10.1021/jm2000882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang XL, et al. Orally bioavailable small-molecule inhibitor of transcription factor Stat3 regresses human breast and lung cancer xenografts. Proc. Natl. Acad. Sci. U. S. A. 2012;109:9623–9628. doi: 10.1073/pnas.1121606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bronson J, et al. To market, to market-2011. Annu. Rep. Med. Chem. 2012;47:499–569. [Google Scholar]
- 51.Thangavelu K, et al. Structural basis for the allosteric inhibitory mechanism of human kidney-type glutaminase (KGA) and its regulation by Raf-Mek-Erk signaling in cancer cell metabolism. Proc. Natl. Acad. Sci. U. S. A. 2012;109:7705–7710. doi: 10.1073/pnas.1116573109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhao Y, et al. SD118-xanthocillin X (1), a novel marine agent extracted from Penicillium commune, induces autophagy through the inhibition of the MEK/ERK pathway. Mar. Drugs. 2012;10:1345–1359. doi: 10.3390/md10061345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Colombo SL, et al. Molecular basis for the differential use of glucose and glutamine in cell proliferation as revealed by synchronized HeLa cells. Proc. Natl. Acad. Sci. U. S. A. 2011;108:21069–21074. doi: 10.1073/pnas.1117500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Reimann JDR, et al. Emi1 is a mitotic regulator that interacts with Cdc20 and inhibits the anaphase promoting complex. Cell. 2001;105:645–655. doi: 10.1016/s0092-8674(01)00361-0. [DOI] [PubMed] [Google Scholar]
- 55.Zhao YH, et al. Early mitotic inhibitor-1, an anaphase-promoting complex/cyclosome inhibitor, can control tumor cell proliferation in hepatocellular carcinoma: correlation with Skp2 stability and degradation of p27(Kip1). Hum. Pathol. 2013;44:365–373. doi: 10.1016/j.humpath.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 56.Moshe Y, et al. Role of Polo-like kinase in the degradation of early mitotic inhibitor 1, a regulator of the anaphase promoting complex/cyclosome. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7937–7942. doi: 10.1073/pnas.0402442101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kita K, et al. Diphenylarsinic acid promotes degradation of glutaminase C by mitochondrial Lon protease. J. Biol. Chem. 2012;287:18163–18172. doi: 10.1074/jbc.M112.362699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Blanco E, et al. Cocaine modulates both glutaminase gene expression and glutaminase activity in the brain of cocaine-sensitized mice. Psychopharmacology. 2012;219:933–944. doi: 10.1007/s00213-011-2418-x. [DOI] [PubMed] [Google Scholar]
- 59.Boulay AC, et al. Bmcc1s interacts with the phosphate-activated glutaminase in the brain. Biochimie. 2012;95:799–807. doi: 10.1016/j.biochi.2012.11.016. [DOI] [PubMed] [Google Scholar]
- 60.Buschdorf JP, et al. Brain-specific BNIP-2-homology protein Caytaxin relocalises glutaminase to neurite terminals and reduces glutamate levels. J. Cell Sci. 2006;119:3337–3350. doi: 10.1242/jcs.03061. [DOI] [PubMed] [Google Scholar]