Abstract
Background
Chronic methamphetamine (METH) exposure causes neuroadaptations at glutamatergic synapses.
Methods
To identify the METH-induced epigenetic underpinnings of these adaptations in the brain, we injected increasing METH doses to rats for two weeks and measured striatal glutamate receptor expression. We then quantified the effects of METH exposure on histone acetylation using chromatin immunoprecipitation (ChIP) and qPCR. We also measured METH-induced changes in DNA methylation and hydroxylation by using methylated (Me) and hydroxymethylated (hMe) DNA precipitation (DIP) and qPCR.
Results
Chronic METH decreased transcript and protein expression of GluA1 and GluA2 AMPAR and GluN1 NMDAR subunits. These changes were associated with decreased electrophysiological glutamatergic responses in striatal neurons. ChIP-PCR revealed that METH decreased enrichment of acetylated histone H4 on GluA1, GluA2, and GluN1 promoters. METH also increased protein levels of histone deacetylases (HDAC1, HDAC2 and SIRT2), protein repressors (REST and CoREST), and of the methylated DNA binding protein, MeCP2. Moreover, METH exposure increased CoREST, MeCP2, and HDAC2, but not SIRT1 or SIRT2, enrichment onto GluA1 and GluA2 gene sequences. Furthermore, METH caused interactions of CoREST and MeCP2 with HDAC2 and of REST with HDAC1. Surprisingly, MeDIP and hMeDIP-PCR revealed METH-induced decreased enrichment of 5-methylcytosine and 5-hydroxymethylcytosine at GluA1 and GluA2 promoter sequences. Furthermore, the HDAC inhibitor, valproic acid, blocked METH-induced decreased expression of AMPAR and NMDAR subunits. Finally, valproic acid also attenuated METH-induced decreased H4K16Ac recruitment on AMPAR gene sequences.
Conclusions
These observations suggest that histone H4 hypoacetylation might be the main determinant of METH-induced decreased striatal glutamate receptor expression.
Keywords: Addiction, AMPAR, CoREST, HDAC2, MeCP2, NMDAR, REST, valproic acid
Addictions are neuropsychiatric disorders that are secondary, in part, to altered synaptic plasticity in mesostriatal and corticostriatal projection areas (1–3). The dorsal striatum is important in the neural circuitry of addiction because the nidus of control for drug taking appears to shift from the ventral to the dorsal striatum as drug taking becomes habitual (4–7). Repeated psychostimulant injections can produce biochemical, molecular, and physiological alterations at striatal glutamatergic synapses (1, 8). Specifically, cocaine administration is accompanied by changes in the expression or trafficking of AMPA receptors (AMPAR) in the mesolimbic system (1). Both contingent and non-contingent administration of cocaine is associated with increased expression of AMPAR on neuronal membranes (9), increased expression of GluA2-lacking AMPARs (10, 11) and physiological evidence of differential AMPAR expression (12) in the ventral striatum. Therefore, dynamic alterations in AMPAR subunit composition might be involved in the maintenance of drug seeking and/or in the occurrence of relapses (10, 11). Parenthetically, very little is known about the effects of methamphetamine (METH) on the expression of these receptors. In rodents, injections of increasing doses of METH (10–30 mg/kg) for 7 consecutive days produced increased AMPA GluA2 protein expression in the dorsal striatum (13). Nevertheless, the transcriptional effects of METH on GluA1 or GluA2 or the epigenetic bases for any potential METH-induced changes in striatal AMPAR expression are unknown.
Gene transcription is regulated by complex epigenetic changes including post-translational histone modifications and DNA methylation that regulate diverse genomic functions (14, 15). Eukaryotic DNA is packaged into chromatin whose basic unit, the nucleosome, contains 4 core histones that form an octamer surrounded by 147 bp of DNA. The N-tails of histones possess lysine residues that can be reversibly acetylated or deacetylated by histone acetyltransferases (HATs) or histone deacetylases (HDACs), respectively (16). Because epigenetic phenomena are involved in the clinical manifestations of neuropsychiatric diseases including addiction (17), we thought it is likely that METH could engender transcriptional and epigenetic changes that are unique to this clinically devastating drug (18). Studies of the transcriptional effects of METH on AMPAR expression are important because its biochemical effects are different from those of cocaine. Specifically, METH interacts with vesicular monoamine transporter and causes release of dopamine (DA) by reverse transport (18, 19) whereas cocaine inhibits monoamine reuptake (20, 21). The two main purposes of this study were to characterize the effects of METH exposure on striatal AMPAR expression and to identify potential epigenetic bases for any changes in receptor expression.
Methods and Materials
Animals and Drug Treatment
All animal treatments and procedures were approved by the National Institute of Drug Abuse Animal Care and Use Committee and followed the Guide for the Care and Use of Laboratory Animals (ISBN 0-309-05377-3). Male Sprague-Dawley rats (Charles River Labs), weighing 250–300g were housed in a humidity and temperature-controlled (22.2 + 0.2°C) room with free access to food and water. Following habituation, rats were assigned to two groups (8 rats each) and were injected daily for 2 weeks with either saline or METH, as shown in Table S1. The animals were euthanized 16 hours after the last saline or METH injection. This METH regimen was meant to mimic the patterns of METH abuse by human abusers who start at low to moderate doses (10–50 mg) and progressed to higher doses (22, 23). This pattern of METH administration to rats does not cause any striatal toxicity (24, see Figure S1).
For co-treatment with HDAC inhibitor, rats received intraperitoneal sodium valproate (VPA) (300 mg/kg, dissolved in water, Sigma) injections twice a day 30 min prior to either saline or METH injections. We chose VPA, a well-tolerated agent with extensive clinical use, recognizing its varied effects on the brain (25). The VPA dose was based on the published literature (26). There were four groups for the co-treatment experiments: vehicle/saline (control); vehicle/METH (METH); VPA/saline (VPA); VPA/METH (VPA + METH).
Quantitative PCR analysis of mRNA levels
Total RNA was isolated from one striatal hemisphere using RNeasy Mini kit (Qiagen) from 8 rats per group. Quantitative PCR was carried out essentially as described by us (27).
SubCellular Fractionation
Separation of nuclear, cell membrane and cytoplasmic fractions from striatal tissues was performed by differential centrifugation at 4°C. Details are provided in the SI Text.
Immunoblot Analysis
Striatal protein lysates (n = 6) were separated by SDS-PAGE and electrophoretically transferred on PVDF membranes, essentially as described by us (see SI text for details). The membranes were incubated overnight at 4°C with specific antibodies against GluA1, GluA2, GluN1/NR1, HDAC1, HDAC2 (Santa Cruz); H4K5ac; H4K12ac; H4K16ac (Millipore) and SIRT1, SIRT2, MeCP2 (Cell Signaling).
Co-immunoprecipitation
Nuclear extracts were prepared from the striatum of saline- and METH-treated rats according to Barrett et al. (28) with minor modifications. Details are included in the SI Text.
Chromatin Immunoprecipitation (ChIP) Assays
Striatal tissue was processed for acetyl H4, REST, CoREST, HDAC1, HDAC2 and MeCP2 ChIP (29) or MeDIP and hMeDIP (30, 31) according to published protocols. Details are provided in the SI Text.
Enrichment of various proteins at GluA1, GluA2 and GluN1 promoters were determined by quantitative real-time PCR using specific ChIP primers designed to amplify proximal or distal sequences from the transcription start site (TSS). Each PCR reaction was repeated at least twice. The specific primers used are listed under Table S2.
Electrophysiology
Perfused rat dorsal striatum was used in the electrophysiology experiments. Details of the electrophysiological experiments were essentially as described by Britt et al. (32) and are provided in the SI Text.
Statistical Analysis
All the quantitative data are presented as mean + SEM. For data comparing control and METH-treated groups, un-paired Student’s t-test was used (StatView version 4.02). For the experiments involving VPA co-treatment, two-way ANOVA followed by Bonferonni post-hoc. For electrophysiology, data were assessed using one-way ANOVA for multiple group comparisons, with a Bonferroni post hoc. For all experiments, the null hypothesis was rejected at p < 0.05.
Results
Chronic METH administration causes decreased striatal AMPAR expression and function
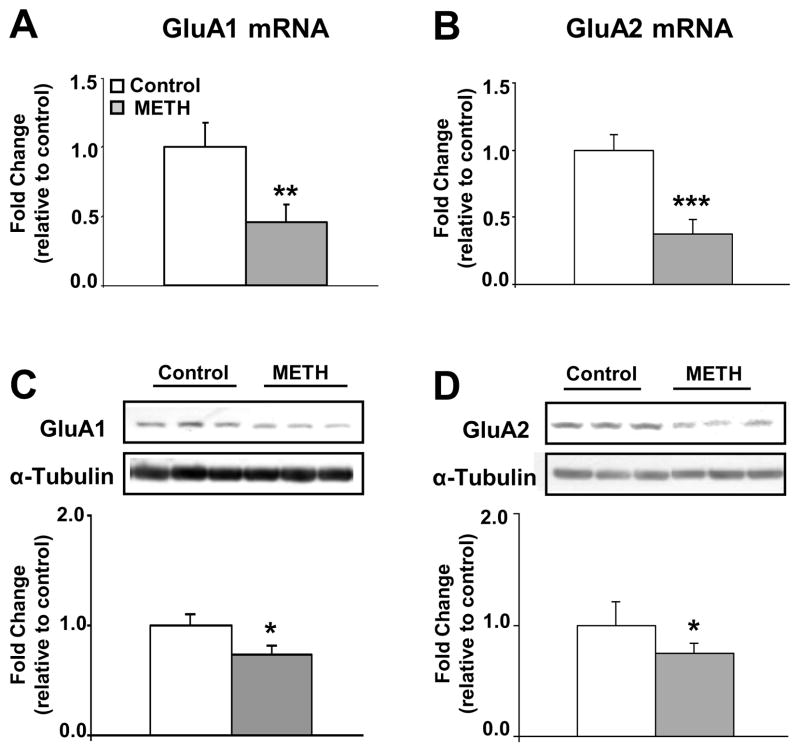
To identify the effects of METH on AMPAR expression, we treated rats with either saline or increasing METH doses as described above (Table S1). Chronic METH decreased striatal mRNA expression of GluA1 (Figure 1A) and GluA2 (Figure 1B). METH also caused decreased GluA1 (Figure 1C) and GluA2 (Figure 1D) protein levels.
Figure 1.
Effects of chronic METH treatment on the mRNA and protein expression of AMPAR (GluA1 and GluA2) subtype of glutamate receptors. The rats were treated with saline or METH for two weeks as described in table S1. Total RNA was extracted from the striatum (n=8 rats per group) and quantitative PCR for (A) GluA1 and (B) GluA2 were carried out as described in the text. The relative amounts of mRNA were normalized to OAZ1 (ornithine decarboxylase antizyme 1) and quantified. Western blot analyses (n=6 rats per group) showed significant decreases in the membrane protein levels of (C) GluA1 and (D) GluA2. Representative photomicrographs show results of three samples per group. For quantification, the signal intensity was normalized to 3-tubulin. Values represent means ± SEM of fold changes relative to the controls. Statistical significance was determined by un-paired Student’s t-test. Key to statistics: * p< 0.05; ** p< 0.01; *** p< 0.001 vs. control group.
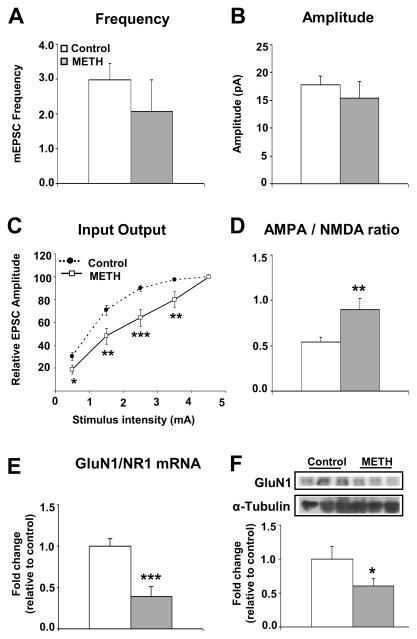
To determine whether these changes in AMPAR subtypes alter excitatory synaptic transmission, ex vivo whole-cell patch clamp recordings were performed on striatal medium spiny neurons. Sixteen hours after the last METH or saline injection, rats were sacrificed and coronal slices containing the striatum were obtained. Miniature excitatory postsynaptic currents (mEPSCs) on medium spiny neurons (Figure 2) were measured blindly according to previous descriptions (32). Unexpectedly, chronic METH did not cause significant changes in mEPSC amplitude or frequency (Figs. 2A and 2B), in contrast to published observations with cocaine (12, 33). We also increased stimulus intensities and measured evoked EPSCs. We found that the input–output relationship between evoked EPSCs and increasing stimulus intensities was significantly decreased in the METH group in comparison to controls (Figure 2C). Surprisingly, we found that the ratio of peak AMPAR- to peak NMDAR-mediated evoked currents, a measure of glutamate synaptic plasticity (34), was significantly increased in the chronic METH-treated group (Figure 2D). The METH-induced increases in AMPAR/NMDAR ratios appear to be related, in part, to METH-induced decreased mRNA (Figure 2E) and protein (Figure 2F) levels of the obligatory NMDA receptor, GluN1/NR1 because the percentage decrease in AMPA protein expression (−22 to 26 %) was less than that of GluN1 (−45%) (compare Figs. 1C and 1D to Figure 2F). Interestingly, the psychostimulant, cocaine, also enhances AMPAR/NMDAR ratios, presumably via other mechanisms (12, 35, 36).
Figure 2.
Glutamate receptor function is decreased following chronic METH administration. Chronic METH administration had no effect on the (A) frequency or (B) amplitude of mEPSCs in medium spiny neurons in the dorsal lateral striatum. (C) A significant decrease in the input-output ratio was observed in the METH group. (D) AMPAR/NMDAR ratio in MSN was significantly increased by chronic METH administration. The increase in the AMPAR/NMDAR ratio is consistent with the observed decreases in (E) GluN1/NR1 mRNA and (F) protein levels.
Chronic METH treatment causes decreased enrichment of H4K5ac, H4K12ac and H4K16ac on GluA1 and GluA2 promoters
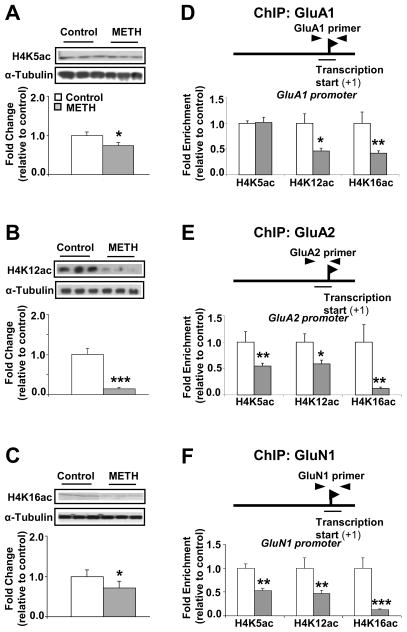
Changes in gene expression are mediated, in part, by epigenetic modifications (15). Studies of the epigenetic effects of cocaine have measured mostly changes in histone H3 modifications (37–39). However, because a strong link exists between gene activation and acetylation of lysine residues (K5, K8, K12, and K16) of histone H4 (40–43), we decided to measure the effects of chronic METH administration on the abundance of these histone marks. Western blot analyses revealed that METH caused significant decreased abundance of striatal H4K5ac (Figure 3A), H4K12ac (Figure 3B) and H4K16ac (Figure 3C). These results are different from the effects of a single injection of METH (20 mg/kg) that caused time-dependent increased abundance of H4K5ac and H4K8ac in the nucleus accumbens (27), suggesting that the epigenetic effects of METH in the brain might depend on the pattern of METH injections.
Figure 3.
Chronic exposure to METH promotes hypoacetylation of H4K5, H4K12 and H4K16 on the promoters of AMPA GluA1, GluA2 and NMDA GluN1 subunits. The rats were treated as mentioned in Figure 1. Chronic METH treatment decreases the levels of nuclear (A) H4K5ac; (B) H4K12ac and (C) H4K16ac proteins in the dorsal striatum (n=6 rats per group). Representative photomicrographs show results of three samples per group. For quantification, the signal intensity was normalized to 3-tubulin. ChIP assays (n=6 - 8 rats per group) were carried out using antibodies against histone H4 acetylated at lysine 5 (H4K5ac), at lysine 12 (H4K12ac) and at lysine 16 (H4K16ac) on (D) GluA1, (E) GluA2 and (F) GluN1 DNA sequences. Quantitative PCR was conducted as described in the text using specific ChIP primers directed at GluA1, GluA2 or GluN1 promoters (see Table S2). Values represent means ± SEM of fold changes relative to the controls. Statistical significance was determined by un-paired Student’s t-test. Key to statistics: * p< 0.05; ** p< 0.01; *** p< 0.001 vs. control group.
We then hypothesized that the drug might also cause changes in modified histone H4 binding to GluA1, GluA2, and GluN1 promoters. Using ChIP-PCR assays, we found significant METH-induced decreased enrichment on GluA1 (Figure 3D), GluA2 (Figure 3E), and GluN1 (Figure 3F) promoter sequences located near their TSSs. METH also caused decreased acetylated H4 enrichment on sequences distal from the GluA1 TSS (see Figure S2A).
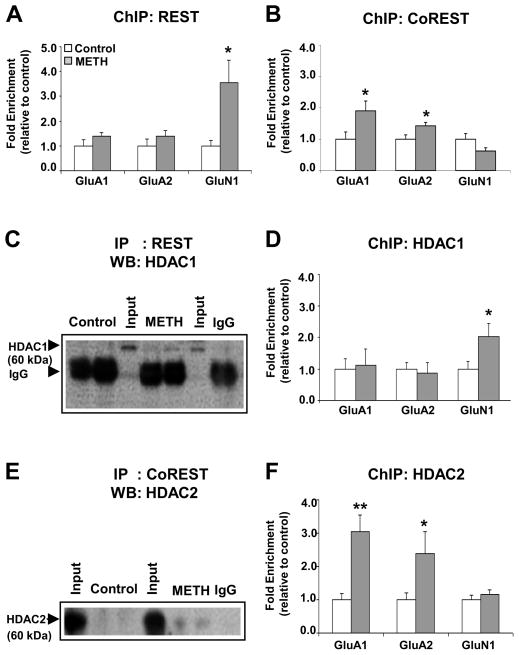
Chronic METH administration enhances the recruitment of CoREST onto GluA1 and GluA2 DNA sequences but that of REST onto GluN1 promoter region
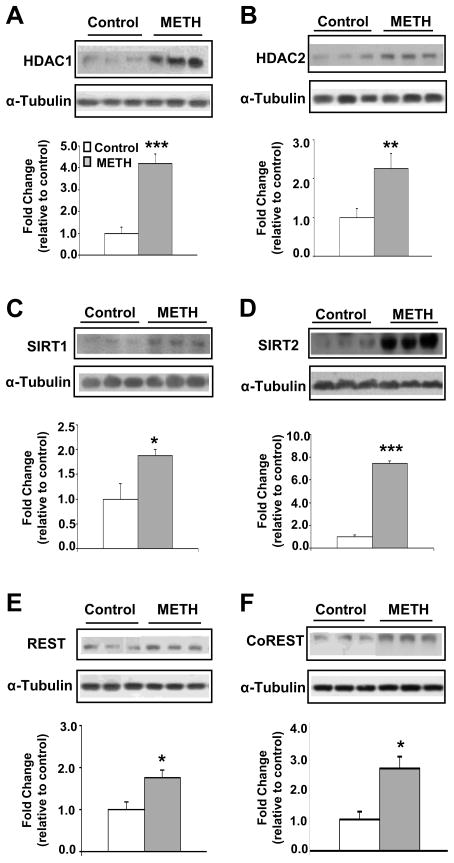
Histone deacetylation is mediated by members of 4 different classes of HDACs (16). Among these, class I HDAC (HDAC1, 2, 3) and class III HDAC (SIRTs) are known to participate in histone H4 deacetylation (16, 44). We thus measured HDAC protein expression after chronic METH and found significant increases in HDAC1 (Figure 4A), HDAC2 (Figure 4B), SIRT1 (Figure 4C), SIRT2 (Figure 4D) but not in SIRT3. A single METH (20 mg/kg) injection also increased HDAC2 but decreased HDAC1 protein levels (27). The METH-induced increased SIRT2 expression is consistent with the observation that cocaine administration also caused SIRT2 upregulation in the rat brain (45).
Figure 4.
Chronic METH treatment induced the expression of HDACs (HDAC1, HDAC2, SIRT1 and SIRT2), REST, and CoREST proteins in the dorsal striatum. Chronic METH administration increased the protein levels of (A) HDAC1, (B) HDAC2, (C) SIRT1, (D) SIRT2, (E) REST, and (F) CoREST. Representative photomicrographs show results of three samples per group. For quantification, the signal intensity was normalized to 3-tubulin. Values represent means ± SEM of fold changes relative to the controls. Statistical significance was determined by un-paired Student’s t-test. Key to statistics: * p< 0.05; ** p< 0.01; *** p< 0.001 vs. control group.
Because transcriptional repression occurs in response to recruitment of HDACs by other repressor proteins (46), we measured the protein level of RE-1 silencing transcription factor (REST) that is implicated in the control of GluA2 (47) and GluN1/NR1 expression (48). We also quantified the expression of CoREST that interacts with REST in a repressor complex (49, 50). Both REST (Figure 4E) and CoREST (Figure 4F) protein levels were increased after repeated METH injections. To test if METH increased the recruitment of REST and CoREST onto GluA1, GluA2, and GluN1 DNA sequences, we carried out ChIP-PCR. There was increased REST enrichment on the GluN1 but not on either GluA1 or GluA2 DNA promoter sequences (Figure 5A). CoREST binding increased significantly at a CpG-rich region located −23Kb upstream of the GluA1 TSS and one near the GluA2 TSS (Figure 5B). There were no changes in CoREST binding on GluN1 promoter sequences. Importantly, METH caused REST co-immunoprecipitation with HDAC1 (Figure 5C) but not with HDAC2 (Figure S3A) whereas METH produced CoREST co-immunoprecipitation with HDAC2 (Figure 5E) but not with HDAC1. Preimmune sera (rabbit IgG) did not precipitate either HDAC1 or HDAC2. These observations are consistent with the observed METH-induced recruitment of HDAC1 on the GluN1 promoter sequence (Figure 5D) and increased HDAC2 recruitment on GluA1 and GluA2 DNA sequences (Figure 5F). In contrast, we found no significant changes in SIRT1 and SIRT2 recruitment onto GluA1 and GluA2 promoter sequences (Figure S4). Together, these results implicate mainly HDAC2 in the regulation of AMPAR expression although the potential roles of sirtuins (51) in the metabolic effects of METH need to be considered (18).
Figure 5.
Chronic METH increases enrichment of CoREST on GluA1 and GluA2 gene promoters whereas enrichment of REST was observed on GluN1 promoter. ChIP assays (n=8 rats per group) were performed on striata of control and METH-treated rats with (A) anti-REST and (B) anti-CoREST antibodies. Quantitative PCR was conducted as described in the text using specific ChIP primers directed at GluA1 or GluA2 or GluN1 promoter (see Table S2). Values represent means ± SEM of fold changes relative to the controls. Statistical significance was determined by un-paired Student’s t-test. Key to statistics: * p< 0.05; ** p< 0.01; *** p< 0.001 vs. control group. Co-immunoprecipitation assays of (C) REST and HDAC1, and (E) CoREST and HDAC2. Immunoprecipitates were prepared from striatal nuclear extracts of control and METH-treated rats using antibody against anti-REST, anti-CoREST, and recovery of HDAC1 and HDAC2 was determined by western blot assay. The levels of HDAC1 and HDAC2 from non-specific IgG are indicated. Input levels (5 %) of HDAC1 and HDAC2 are shown for comparison. ChIP assays (n=6 - 8 rats per group) were carried out using antibodies against HDAC1 (D), and HDAC2 (F) on GluA1, GluA2 and GluN1 DNA sequences. Quantitative PCR was conducted as described in the text using specific ChIP primers directed at GluA1, GluA2 or GluN1 promoters (see Table S2). Values represent means ± SEM of fold changes relative to the controls. Statistical significance was determined by un-paired Student’s t-test. Key to statistics: * p< 0.05; ** p< 0.01; *** p< 0.001 vs. control group.
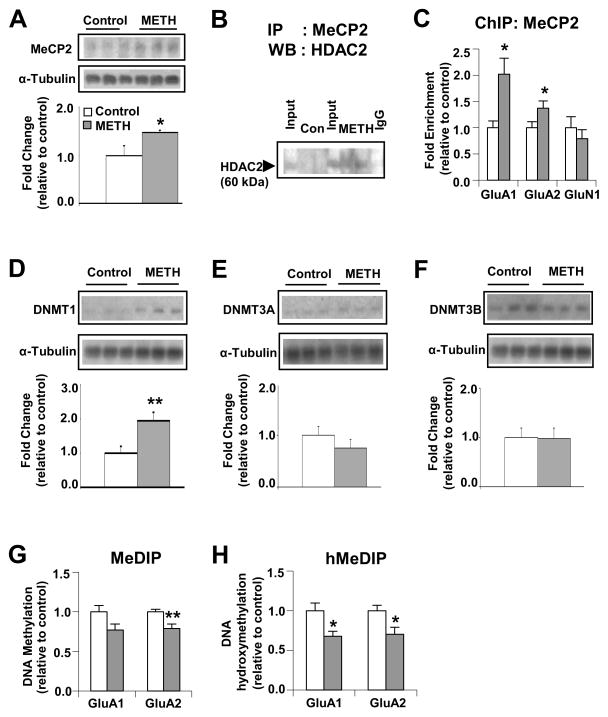
Chronic METH-induced repression of AMPAR also involves MeCP2 recruitment
In addition to histone acetylation, decrease in gene expression can be regulated by DNA methylation (52). DNA methylation is mediated by three DNA methyl transferases (DNMTs), namely the maintenance enzyme, DNMT1 (53, 54), and the de novo DNA methylation enzymes, DNMT3a and DNMT3b (55). In association with DNA methylation, the multifunctional, methyl DNA-binding transcriptional regulator, MeCP2, can cause decreased gene expression by recruiting HDACs (56). MeCP2 exists as a large complex (57), binds to DNMT1 (58), and interacts with both methylated and unmethylated DNA (59–61). To test the involvement of DNA methylation in METH-induced decreased gene expression, we first measured MeCP2 protein levels because it participates in DNA methylation-related epigenetic phenomena (62). Chronic METH increased MeCP2 protein levels (Figure 6A). Because MeCP2 exerts its transcriptional repressing activities, in part, by recruiting HDAC complexes (56, 63), we carried out co-immunoprecipitation studies and found that METH caused MeCP2 co-precipitation with HDAC2 (Figure 6B) but not HDAC1 (Figure S3B). ChIP-PCR experiments also showed that METH increased MeCP2 enrichment at sequences located near the GluA1 and GluA2 TSSs (Figure 6C). There were no changes in MeCP2 binding on the GluN1 promoter (Figure 6C). Similar to the cases of histone H4 marks (Figure S2A), METH also altered MeCP2 binding at the CpG-rich region located 23Kb upstream of the GluA1 TSS (Figure S2B).
Figure 6.
Chronic METH exposure causes down-regulation of GluA1 and GluA2 transcription by formation of a MeCP2-HDAC2 complex. Western blot analysis of (A) MeCP2, (D) DNMT1, (E) DNMT3A and (F) DNMT3B Representative photomicrographs show results of 3 samples per group. For quantification, the signal intensity was normalized to 3-tubulin. Co-immunoprecipitation assays of (B) MeCP2 and HDAC2 show METH-induced interactions of MeCP2 with HDAC2. The level of HDAC2 from non-specific IgG is indicated. Input levels (5 %) of HDAC2 are shown for comparison. ChIP assays (n=6 – 8 rats per group) were carried out using antibodies against MeCP2 (C). Quantitative PCR was conducted using specific ChIP primers (see Table S2). Denatured genomic DNA of ~200 – 600 bp (generated by sonication) was incubated with an antibody directed against 5mC (G) or 5hmC (H), in order to isolate methylated or hydroxymethylated DNA by immunoprecipitation. Relative enrichment of 5mC and 5hmC in the bound over input fractions was calculated by real-time PCR. Values represent means ± SEM of fold enrichment relative to the controls. Statistical significance was determined by un-paired Student’s t-test. Key to statistics: * p< 0.05; ** p< 0.01; *** p< 0.001 vs. control group.
To further test for the involvement of DNA methylation, we also measured DNMT protein levels after METH injections. Chronic METH increased the expression of DNMT1 (Figure 6D) but not of DNMT3A (Figure 6E) or DNMT3B (Figure 6F). This was of interest because DNMT1 is the maintenance DNMT (53) and was not expected to change after METH. However, our observations are consistent with the report that nicotine (64) can decrease DNMT1 expression.
To further test if METH treatment did cause changes in DNA methylation, we used MeDIP-qPCR to measure DNA methylation at CpG sites around the TSSs of GluA1 and GluA2. MeDIP values are reported to be proportional to the number of methylated CpG dinucleotides quantified by sodium bisulfite (65). Unexpectedly, METH significantly decreased cytosine methylation at the CpG-rich site located at – 23K from (Figure S2B) but caused non-significant changes (− 23%, p = 0.051) near the GluA1 TSS (Figure 6G). There was also decreased (− 21%, p = 0.0081) cytosine methylation at CpG sites located near the GluA2 TSS (Figure 6G). In addition to 5-methylcytosine methylation, recent reports have identified novel DNA modifications (66, 67) including DNA hydroxymethylation which is relatively abundant in the brain (68–71). Therefore, we used hMeDIP to measure DNA hydroxymethylation at the same AMPAR sequences described above. We observed significantly decreased DNA hydroxymethylation at the CpG-rich sequences located at – 23Kb (− 29%, p = 0.046, Figure S2B) and at sequences near the GluA1 TSS (− 33%, p = 0.012; Figure 6H). We also detected significant decrease in DNA hydroxymethylation at the CpG-rich sequences located near the GluA2 TSS (− 30%, p = 0.017; Figure 6H).
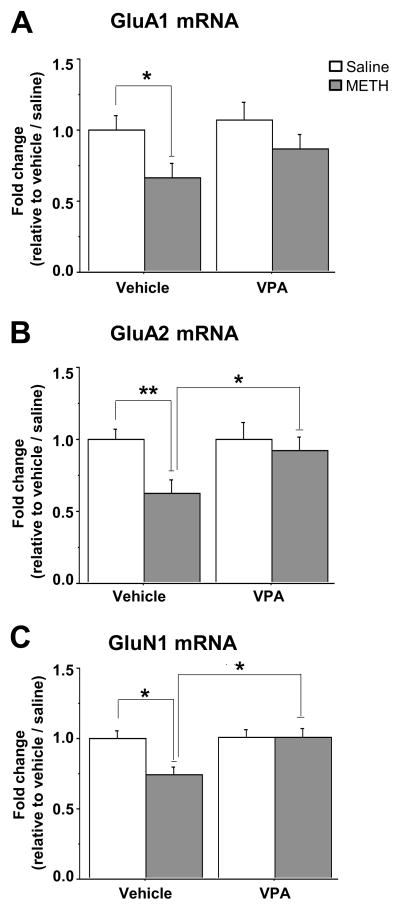
The HDAC inhibitor, valproic acid, prevents METH-induced down-regulation of GluA1, GluA2 mRNA levels by preventing METH-induced increased HDAC2 recruitment
Together, the collected results had suggested that chronic METH might have caused decreased AMPAR expression by inducing the formation of HDAC-containing repressor complexes onto AMPAR DNA sequences. To test this idea, we used the HDAC inhibitor, valproic acid (VPA) (72), to attempt to antagonize the effects of METH. VPA is a well-known FDA-approved drug used in the treatment of neuropsychiatric disorders with varied biochemical and molecular effects (25, 73). Two-way ANOVA identified significant METH-induced changes in GluA1 (F(1,28)=5.88, p=0.023) and GluA2 (F(1,28)=5.251, p=0.031) expression, no significant effects of VPA alone, and no significant interactions (Figure 7A and 7B). METH also caused significant changes in GluN1 expression (F(1,27)=5.267, p=0.031). There were also significant effects of VPA (F(1,27)=6.136, p=0.021) and significant METH x VPA interactions (F(1,27)=5.3, p=0.03) (Figure 7C). Bonferroni post-hoc revealed that METH caused significant decreases in GluA1 (p=0.038, Figure 7A) and GluA2 (p=0.009, Figure 7B) mRNA expression in comparison to the saline/vehicle group. In addition, VPA/METH co-treatment caused increased GluA2 mRNA levels compared to METH/vehicle (p=0.031, Figure 7B). METH caused significant decreased GluN1 mRNA compared to saline/vehicle (p=0.048) and the VPA/METH group was significantly higher than the METH/vehicle group (p=0.012, Figure. 7C).
Figure 7.
Co-treatment with valproic acid (VPA) blocked the METH-induced decreases in (A) GluA1 mRNA, (B) GluA2 mRNA, and (C) GluN1 mRNA. Sodium valproate (300 mg/kg) was injected intraperitoneally twice a day 30 min prior to either saline or to METH injections. Drug administration, RNA extraction and RT-PCR of GluA1, GluA2 and GluN1 are as described in the text. The relative amounts of mRNA were normalized to OAZ1 and quantified. Values represent means ± SEM of fold changes relative to the controls. Statistical significance for the four groups (n=7–8 rats per group) were compared by two-way analysis of variance and Bonferroni correction. Key to statistics: * p< 0.05; ** p< 0.01 (Bonferroni).
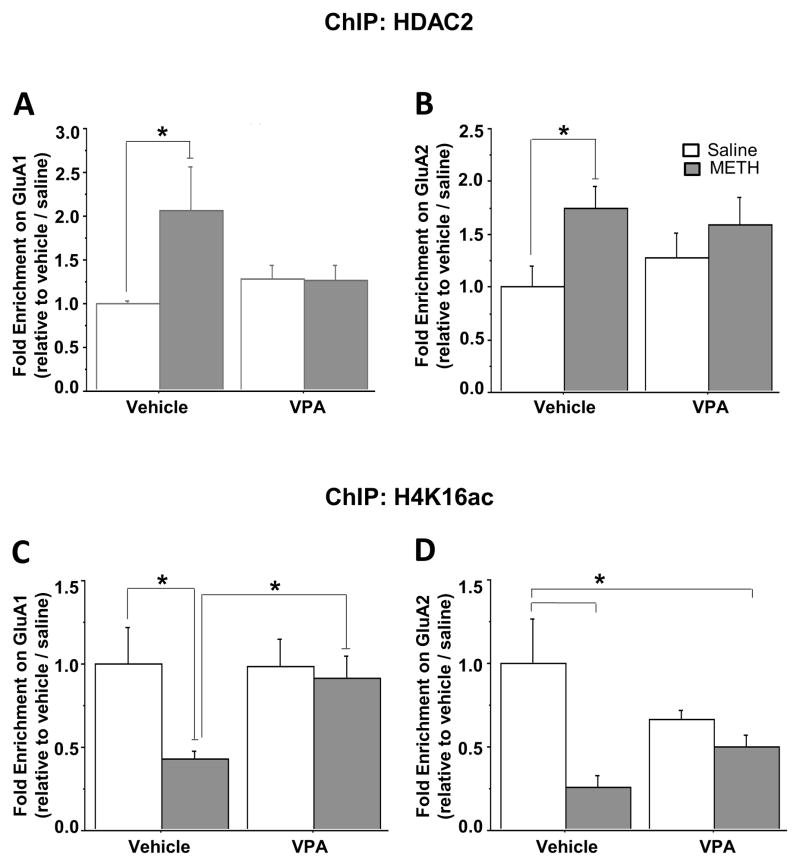
We tested the possibility that VPA may also block METH-induced epigenetic changes by carrying out ChIP-PCR with antibodies against HDAC2 and H4K16ac, respectively. Two-way ANOVA revealed main effects of METH (F(1,16)=4.42, p=0.05) on HDAC2 enrichment at sequences located near the GluA1 TSS. There were no significant effects of VPA (p=0.892) but significant METH x VPA interactions (F(1,16)=4.91, p=0.044) (Figure 8A). Similarly, there were significant effects of METH (F(1,21)=4.56, p=0.046) at the GluA2 TSS but no effects of VPA alone. METH X VPA interactions were not significant (Figure 8B). Bonferroni corrections showed METH-induced increases in HDAC2 recruitment around GluA2 TSS compared to saline/vehicle group (p=0.032, Figure 8B). METH also caused changes in H4K16ac recruitment on GluA1 (F(1,15)=4.75, p=0.048). Bonferroni post-hoc revealed METH-induced decreased H4K16ac recruitment around the GluA1 TSS compared to the saline/vehicle group (p=0.022, Figure 8C). The VPA-treated METH group showed significant increased H4K16ac recruitment compared to METH alone (p=0.033, Figure 8C). Abundance of H4K16ac around the GluA2 TSS was also significantly affected by METH (F(1,14)=9.93, p=0.008) but not by VPA alone (p=0.747). Both vehicle- and VPA-treated METH groups showed decreased abundance of H4K16ac in comparison to the saline/vehicle group.
Figure 8.
Co-treatment with valproic acid (VPA) blocked METH-induced increased enrichment of HDAC2 on (A) GluA1 and (B) GluA2 gene promoters. VPA also blocked decreased enrichment of H4K16ac on (C) GluA1 but not on (D) GluA2 gene promoters. Sodium valproate (300 mg/kg) was injected intraperitoneally twice a day 30 min prior to either saline or to METH injections. Drug administration and ChIP-PCR for GluA1 and GluA2 are as described in the text. ChIP assays (n=4 – 6 rats per group) were carried out using an antibody against HDAC2 and H4K16ac. Values for all experiments represent means ± SEM of fold changes relative to the controls. Statistics are as described in Figure 7. Key to statistics: * p< 0.05 (Bonferroni).
Discussion
METH is a psychostimulant that causes DA release in the dorsal striatum (74) and altered striatal expression of immediate early genes [see (75), for review]. Nevertheless, because addiction occurs after chronic drug exposure, it is unlikely that these acute METH effects are directly responsible for the development of chronic METH addiction. Molecular changes that occur during repeated exposure to the drug might offer a better window to the molecular substrate(s) of METH use disorder. In the present study, repeated METH exposure decreased striatal GluA1 and GluA2 mRNA and protein levels and electrophysiological studies, done by investigators who were blind to the treatment conditions, revealed decreased slope of stimulus-to-response curve after METH. Surprisingly, we also found that METH increased AMPAR/NMDAR ratios, a marker of glutamate synaptic plasticity (12, 35, 36). These changes, although unexpected, appear to be secondary to the relatively greater decreases in the expression of the NMDAR subunit, GluN1/NR1, which is the subunit essential for ion selectivity (76, 77). Our observations of METH-induced decreased NR1 expression are consistent with those of a previous report of decreased striatal GluN1/NR1 mRNA expression in mice that received daily METH (2 mg/kg) injections for 12 days (78). Administration of METH (4 mg/kg daily for 3 weeks) also decreased striatal NMDA GluN1 protein levels in animals sacrificed several days after stopping the injections (79).
Gene transcription is regulated, in part, by epigenetic modifications that include acetylation of histones (80). Histone hypoacetylation can cause conformational changes that make it difficult for transcription factors and other proteins important for active gene expression to access chromatin (81). We thus tested the idea that modifications at histone H4 tails might regulate METH-induced effects on AMPA and NMDA subunit expression because of their importance in gene activation (40–42). Chronic METH administration did cause decreased H4K5ac, H4K12ac and H4K16ac abundance on GluA1, GluA2, and GluN1 promoters. METH-induced histone H4 hypoacetylation is probably the result of the observed METH-induced increased expression of HDACs that remove acetyl groups from histone tails (82). In fact, HDAC2, but not HDAC1, was enriched on GluA1 and GluA2 sequences, whereas HDAC1, but not HDAC2, was enriched on the GluN1 promoter sequence. These observations are consistent with the report that reduced GluA1 and GluA2 expression is correlated with increased HDAC2 binding and concomitant decreases in H4K5ac and H4K12ac binding on GluA1 and GluA2 promoter regions in mice models (83). Conversely, HDAC2 removal is linked to global increases in H4K5ac and H4K12ac (84). Unlike H4K5ac and H4K12ac, acetylated H4K16 can be deacetylated by both class I (HDAC1 and HDAC2) (85) and class III HDAC, SIRT2 (44, 86). Initially, we thought that increased protein expression of HDAC1, HDAC2, and SIRT2 along with increased HDAC2 binding on GluA1 and GluA2 DNA sequences after METH might have provided a partial explanation for the decreased H4K16ac abundance on GluA1, GluA2, and GluN1 promoters. However, the lack of changes in SIRT2 binding on GluA1 and GluA2 promoter sequences suggests that HDAC2 might be the main effector of METH-induced changes in the expression of these AMPAR subunits. A role for HDAC2 in mediating the effects of the drug on AMPAR subtype expression is further supported by the fact that administration of the inhibitor of Classes I and II HDACs, VPA (87), attenuated METH-induced repression of their AMPAR expression and associated epigenetic alterations. The fact that VPA was able to attenuate METH-induced decreased H4K16ac abundance on GluA1 and GluA2 sequences is consistent with recent results documenting VPA-induced increased H4K16 acetylation in human vascular endothelial and cancer cell lines (88–90). Other inhibitors of Class I and II HDACs, including sodium butyrate and suberol-anilide hydroxamic acid (SAHA) (88, 89), also caused dose-dependent increased H4K16 acetylation (89), but with variable time courses (88). Therefore, while acknowledging the multifaceted biochemical and molecular effects of VPA (25, 91), it is fair to suggest that VPA might have prevented the effects of METH on glutamate receptors by blocking the effects of HDAC1 and HDAC2 on histone H4 acetylation (see schema in Figure 9).
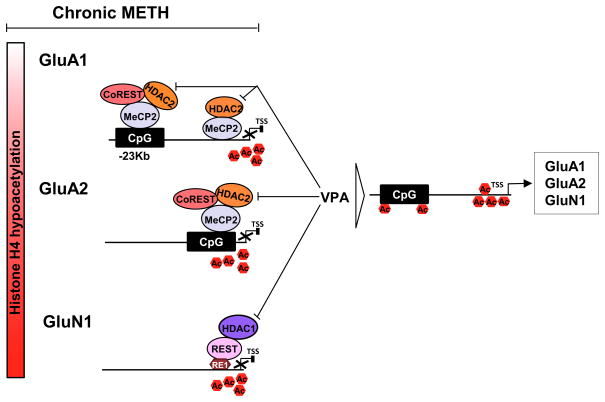
Figure 9.
Schematic models showing chronic METH-induced epigenetic modifications in the dorsal striatum. Under control condition, there exists a balance between histone acetylases (HATs) and histone deacetylases (HDACs) that regulate the histone acetylation/deacetylation status that maintain the baseline transcription levels. However, chronic METH exposure leads to formation of protein repressor complexes MeCP2-CoREST-HDAC2 that cause H4K5, H4K12 and H4K16 hypoacetylation at enhancer or promoter sequences of GluA1 and GuA2 genes. This then leads to decreased expression of these receptors. Rats chronically exposed to METH also show formation of a protein repressor complex that contains REST-HDAC1 that produces hypoacetylation of H4K5, H4K12 and H4K16 at the promoter region of GluN1 and subsequent decreased GluN1 (NR1) expression in the dorsal striatum. Co-treatment of METH-treated animals with valproic acid that inhibits HDAC1 and HDAC2 blocked METH-mediated hypoacetylation and METH-induced decreased expression of these glutamate receptors.
Of further interest is the fact that HDAC actions occur via their recruitment into repressor complexes that contain RE1 silencing transcription factor (REST) (49), among others. Specifically, the negative cis-element, repressor element 1 (RE1) is located on the promoter region of rat GluA2 (92) and GluN1/NR1 (93) and interacts with the trans-acting factor, REST (49). After binding to DNA, REST recruits co-repressor CoREST and negatively regulates transcription (49). These two proteins work together to repress the expression of many neuronal genes (94) including GluA2 (95) and GluN1/NR1 (48). In our study, however, we observed increased enrichment of CoREST at sequences located upstream to GluA1 and GluA2 TSSs while there was increased enrichment of REST, but not of CoREST, on the GluN1 promoter. These observations are consistent with recent reports that CoREST and REST can function independently (50). For example, specific CoREST target genes that do not contain the RE1 motif to which REST binds are still targeted by CoREST for transcriptional repression (96). The observed repression was shown to result from interactions of CoREST with other repressor proteins (94, 97) including HDACs (see above discussion). In addition to a role for CoREST, our findings indicate that MeCP2, an important protein that is involved in RETT syndrome (98), also participates in METH-induced repression of AMPAR expression. The METH-induced increased MeCP2 recruitment at distal sites to the GluA1TSS is consistent with a potential effect on enhancer sequences that are known to interact with promoter sequences to regulate transcription (99, 100). The observation of METH-induced increased MeCP2 binding had suggested that METH might have also caused increased DNA methylation, a suggestion that we thought was supported by our demonstration of increased striatal expression of the maintenance DNMT, DNMT1 (101). However, our failure to observe any METH-induced increased DNA methylation in GluA promoter or enhancer regions argues against the involvement of DNA modifications in the regulation of AMPAR expression in the present model. The present observations also suggest that DNA methylation might not be obligatory for the gene suppressive effects of MeCP2 that binds to both methylated and unmethylated DNA (60, 61, 102, 103). Finally, the observations that the HDAC inhibitor, VPA, also attenuated the METH-induced decreased glutamate receptor expression, HDAC2 binding to GluA1 and GluA2 promoters, and decreased H4K16Ac recruitment to these receptor promoter sequences also argue for a preferential role for histone hypoacetylation for METH-induced decreased AMPAR expression.
In summary, our study provides direct evidence for epigenetic regulation of transcriptional effects of chronic METH exposure on glutamate receptors. Figure 9 provides a schematic representation that describes potential roles of REST, CoREST, MeCP2, HDAC1, and HDAC2 in mediating METH-induced downregulation of GluA1, GluA2 and GluN1 mRNA levels. The scheme suggests two distinct regulatory mechanisms: (1) one for GluA expression that involves MeCP2 and CoREST recruitment of HDAC2 onto the chromatin, with resulting H4K5, K12 and K16 deacetylation and decreased H4K5ac, K12ac and K16ac binding onto GluA1 and GluA2 DNA sequences and another one for the regulation of NR1 expression that is mediated by REST and HDAC1 with decreased H4K5ac, K12ac and K16ac binding onto GluN1 promoter region. Because the clinically effective FDA-approved neuropsychiatric medication, VPA, was able to block METH-induced decreases in AMPAR and NMDAR expression, it is tempting to speculate that this medication or similar epigenetic agents could be used to mitigate some neuropsychiatric manifestations of chronic METH exposure in humans.
Supplementary Material
Acknowledgments
This work was supported by funds of the Intramural Research Program of the DHHS/NIH/NIDA. The authors thank Dr. Genevieve Beauvais, Dr. Ingrid Tulloch, and Tracey Martin for their help with animal injections and dissection and Dr. Benita Gonzalez with help with statistical analyses. The authors also thank the reviewers whose constructive criticisms helped to improve the content, presentation, and discussion of our results.
Footnotes
FINANCIAL DISCLOSURES
The authors report no biomedical financial interests or potential conflicts of interest
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bowers MS, Chen BT, Bonci A. AMPA receptor synaptic plasticity induced by psychostimulants: the past, present, and therapeutic future. Neuron. 2010;67:11–24. doi: 10.1016/j.neuron.2010.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen BT, Hopf FW, Bonci A. Synaptic plasticity in the mesolimbic system: therapeutic implications for substance abuse. Ann N Y Acad Sci. 2010;1187:129–139. doi: 10.1111/j.1749-6632.2009.05154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Luscher C, Malenka RC. Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron. 2011;69:650–663. doi: 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belin D, Everitt BJ. Cocaine seeking habits depend upon dopamine-dependent serial connectivity linking the ventral with the dorsal striatum. Neuron. 2008;57:432–441. doi: 10.1016/j.neuron.2007.12.019. [DOI] [PubMed] [Google Scholar]
- 5.Doyon J, Bellec P, Amsel R, Penhune V, Monchi O, Carrier J, et al. Contributions of the basal ganglia and functionally related brain structures to motor learning. Behav Brain Res. 2009;199:61–75. doi: 10.1016/j.bbr.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 6.Everitt BJ, Robbins TW. From the ventral to the dorsal striatum: Devolving views of their roles in drug addiction. Neurosci Biobehav Rev. 2013 doi: 10.1016/j.neubiorev.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Wise RA. Roles for nigrostriatal--not just mesocorticolimbic--dopamine in reward and addiction. Trends Neurosci. 2009;32:517–524. doi: 10.1016/j.tins.2009.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalivas PW, Volkow ND. New medications for drug addiction hiding in glutamatergic neuroplasticity. Mol Psychiatry. 2011;16:974–986. doi: 10.1038/mp.2011.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu L, Dempsey J, Shaham Y, Hope BT. Differential long-term neuroadaptations of glutamate receptors in the basolateral and central amygdala after withdrawal from cocaine self-administration in rats. J Neurochem. 2005;94:161–168. doi: 10.1111/j.1471-4159.2005.03178.x. [DOI] [PubMed] [Google Scholar]
- 10.Conrad KL, Tseng KY, Uejima JL, Reimers JM, Heng LJ, Shaham Y, et al. Formation of accumbens GluR2-lacking AMPA receptors mediates incubation of cocaine craving. Nature. 2008;454:118–121. doi: 10.1038/nature06995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wolf ME, Ferrario CR. AMPA receptor plasticity in the nucleus accumbens after repeated exposure to cocaine. Neurosci Biobehav Rev. 2010;35:185–211. doi: 10.1016/j.neubiorev.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kourrich S, Rothwell PE, Klug JR, Thomas MJ. Cocaine experience controls bidirectional synaptic plasticity in the nucleus accumbens. J Neurosci. 2007;27:7921–7928. doi: 10.1523/JNEUROSCI.1859-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simoes PF, Silva AP, Pereira FC, Marques E, Milhazes N, Borges F, et al. Methamphetamine changes NMDA and AMPA glutamate receptor subunit levels in the rat striatum and frontal cortex. Ann N Y Acad Sci. 2008;1139:232–241. doi: 10.1196/annals.1432.028. [DOI] [PubMed] [Google Scholar]
- 14.Martin C, Zhang Y. Mechanisms of epigenetic inheritance. Curr Opin Cell Biol. 2007;19:266–272. doi: 10.1016/j.ceb.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 15.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–141. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- 16.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasnova IN, Cadet JL. Methamphetamine toxicity and messengers of death. Brain Res Rev. 2009;60:379–407. doi: 10.1016/j.brainresrev.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulzer D. How addictive drugs disrupt presynaptic dopamine neurotransmission. Neuron. 2011;69:628–649. doi: 10.1016/j.neuron.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuhar MJ, Ritz MC, Boja JW. The dopamine hypothesis of the reinforcing properties of cocaine. Trends Neurosci. 1991;14:299–302. doi: 10.1016/0166-2236(91)90141-g. [DOI] [PubMed] [Google Scholar]
- 21.Ritz MC, Lamb RJ, Goldberg SR, Kuhar MJ. Cocaine receptors on dopamine transporters are related to self-administration of cocaine. Science. 1987;237:1219–1223. doi: 10.1126/science.2820058. [DOI] [PubMed] [Google Scholar]
- 22.Cho AK, Melega WP. Patterns of methamphetamine abuse and their consequences. J Addict Dis. 2002;21:21–34. doi: 10.1300/j069v21n01_03. [DOI] [PubMed] [Google Scholar]
- 23.Kramer JC, Fischman VS, Littlefield DC. Amphetamine abuse. Pattern and effects of high doses taken intravenously. JAMA. 1967;201:305–309. doi: 10.1001/jama.201.5.305. [DOI] [PubMed] [Google Scholar]
- 24.Cadet JL, McCoy MT, Cai NS, Krasnova IN, Ladenheim B, Beauvais G, et al. Methamphetamine preconditioning alters midbrain transcriptional responses to methamphetamine-induced injury in the rat striatum. PLoS One. 2009;4:e7812. doi: 10.1371/journal.pone.0007812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosenberg G. The mechanisms of action of valproate in neuropsychiatric disorders: can we see the forest for the trees? Cell Mol Life Sci. 2007;64:2090–2103. doi: 10.1007/s00018-007-7079-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rowley HL, Marsden CA, Martin KF. Differential effects of phenytoin and sodium valproate on seizure-induced changes in gamma-aminobutyric acid and glutamate release in vivo. Eur J Pharmacol. 1995;294:541–546. doi: 10.1016/0014-2999(95)00589-7. [DOI] [PubMed] [Google Scholar]
- 27.Martin TA, Jayanthi S, McCoy MT, Brannock C, Ladenheim B, Garrett T, et al. Methamphetamine causes differential alterations in gene expression and patterns of histone acetylation/hypoacetylation in the rat nucleus accumbens. PLoS One. 2012;7:e34236. doi: 10.1371/journal.pone.0034236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barrett LE, Van Bockstaele EJ, Sul JY, Takano H, Haydon PG, Eberwine JH. Elk-1 associates with the mitochondrial permeability transition pore complex in neurons. Proc Natl Acad Sci U S A. 2006;103:5155–5160. doi: 10.1073/pnas.0510477103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsankova NM, Kumar A, Nestler EJ. Histone modifications at gene promoter regions in rat hippocampus after acute and chronic electroconvulsive seizures. J Neurosci. 2004;24:5603–5610. doi: 10.1523/JNEUROSCI.0589-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, et al. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genetics. 2005;37:853–862. doi: 10.1038/ng1598. [DOI] [PubMed] [Google Scholar]
- 31.Vucetic Z, Kimmel J, Totoki K, Hollenbeck E, Reyes TM. Maternal high-fat diet alters methylation and gene expression of dopamine and opioid-related genes. Endocrinology. 2010;151:4756–4764. doi: 10.1210/en.2010-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Britt JP, Benaliouad F, McDevitt RA, Stuber GD, Wise RA, Bonci A. Synaptic and behavioral profile of multiple glutamatergic inputs to the nucleus accumbens. Neuron. 2012;76:790–803. doi: 10.1016/j.neuron.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas MJ, Beurrier C, Bonci A, Malenka RC. Long-term depression in the nucleus accumbens: a neural correlate of behavioral sensitization to cocaine. Nat Neurosci. 2001;4:1217–1223. doi: 10.1038/nn757. [DOI] [PubMed] [Google Scholar]
- 34.Hsia AY, Malenka RC, Nicoll RA. Development of excitatory circuitry in the hippocampus. J Neurophys. 1998;79:2013–2024. doi: 10.1152/jn.1998.79.4.2013. [DOI] [PubMed] [Google Scholar]
- 35.Chen BT, Bowers MS, Martin M, Hopf FW, Guillory AM, Carelli RM, et al. Cocaine but not natural reward self-administration nor passive cocaine infusion produces persistent LTP in the VTA. Neuron. 2008;59:288–297. doi: 10.1016/j.neuron.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ungless MA, Whistler JL, Malenka RC, Bonci A. Single cocaine exposure in vivo induces long-term potentiation in dopamine neurons. Nature. 2001;411:583–587. doi: 10.1038/35079077. [DOI] [PubMed] [Google Scholar]
- 37.Covington HE, 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kennedy PJ, Feng J, Robison AJ, Maze I, Badimon A, Mouzon E, et al. Class I HDAC inhibition blocks cocaine-induced plasticity by targeted changes in histone methylation. Nat Neurosci. 2013;16:434–440. doi: 10.1038/nn.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rogge GA, Wood MA. The role of histone acetylation in cocaine-induced neural plasticity and behavior. Neuropsychopharmacology. 2013;38:94–110. doi: 10.1038/npp.2012.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dion MF, Altschuler SJ, Wu LF, Rando OJ. Genomic characterization reveals a simple histone H4 acetylation code. Proc Natl Acad Sci U S A. 2005;102:5501–5506. doi: 10.1073/pnas.0500136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Henriksen P, Wagner SA, Weinert BT, Sharma S, Bacinskaja G, Rehman M, et al. Proteome-wide analysis of lysine acetylation suggests its broad regulatory scope in Saccharomyces cerevisiae. Mol Cell Proteomics. 2012;11:1510–1522. doi: 10.1074/mcp.M112.017251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein M. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- 43.Vettese-Dadey M, Grant PA, Hebbes TR, Crane-Robinson C, Allis CD, Workman JL. Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro. EMBO J. 1996;15:2508–2518. [PMC free article] [PubMed] [Google Scholar]
- 44.Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- 45.Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, 3rd, Maze I, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bithell A. REST: transcriptional and epigenetic regulator. Epigenomics. 2011;3:47–58. doi: 10.2217/epi.10.76. [DOI] [PubMed] [Google Scholar]
- 47.Calderone A, Jover T, Noh KM, Tanaka H, Yokota H, Lin Y, et al. Ischemic insults derepress the gene silencer REST in neurons destined to die. J Neurosci. 2003;23:2112–2121. doi: 10.1523/JNEUROSCI.23-06-02112.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bai G, Zhuang Z, Liu A, Chai Y, Hoffman PW. The role of the RE1 element in activation of the NR1 promoter during neuronal differentiation. J Neurochem. 2003;86:992–1005. doi: 10.1046/j.1471-4159.2003.01922.x. [DOI] [PubMed] [Google Scholar]
- 49.Andres ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, et al. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci U S A. 1999;96:9873–9878. doi: 10.1073/pnas.96.17.9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abrajano JJ, Qureshi IA, Gokhan S, Zheng D, Bergman A, Mehler MF. REST and CoREST modulate neuronal subtype specification, maturation and maintenance. PLoS One. 2009;4:e7936. doi: 10.1371/journal.pone.0007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang F, Wang S, Gan L, Vosler PS, Gao Y, Zigmond MJ, et al. Protective effects and mechanisms of sirtuins in the nervous system. Prog Neurobiol. 2011;95:373–395. doi: 10.1016/j.pneurobio.2011.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 53.Denis H, Ndlovu MN, Fuks F. Regulation of mammalian DNA methyltransferases: a route to new mechanisms. EMBO Reports. 2011;12:647–656. doi: 10.1038/embor.2011.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kar S, Deb M, Sengupta D, Shilpi A, Parbin S, Torrisani J, et al. An insight into the various regulatory mechanisms modulating human DNA methyltransferase 1 stability and function. Epigenetics. 2012;7:994–1007. doi: 10.4161/epi.21568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chedin F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog Mol Biol Transl Sci. 2011;101:255–285. doi: 10.1016/B978-0-12-387685-0.00007-X. [DOI] [PubMed] [Google Scholar]
- 56.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 57.Young JI, Hong EP, Castle JC, Crespo-Barreto J, Bowman AB, Rose MF, et al. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc Natl Acad Sci U S A. 2005;102:17551–17558. doi: 10.1073/pnas.0507856102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura H, Shiota K. Methyl-CpG-binding protein, MeCP2, is a target molecule for maintenance DNA methyltransferase, Dnmt1. J Biol Chem. 2003;278:4806–4812. doi: 10.1074/jbc.M209923200. [DOI] [PubMed] [Google Scholar]
- 59.Adams VH, McBryant SJ, Wade PA, Woodcock CL, Hansen JC. Intrinsic disorder and autonomous domain function in the multifunctional nuclear protein, MeCP2. J Biol Chem. 2007;282:15057–15064. doi: 10.1074/jbc.M700855200. [DOI] [PubMed] [Google Scholar]
- 60.Hansen JC, Ghosh RP, Woodcock CL. Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin. IUBMB Life. 2010;62:732–738. doi: 10.1002/iub.386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meehan RR, Lewis JD, Bird AP. Characterization of MeCP2, a vertebrate DNA binding protein with affinity for methylated DNA. Nucleic Acids Res. 1992;20:5085–5092. doi: 10.1093/nar/20.19.5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Feng J, Fan G. The role of DNA methylation in the central nervous system and neuropsychiatric disorders. Int Rev Neurobiol. 2009;89:67–84. doi: 10.1016/S0074-7742(09)89004-1. [DOI] [PubMed] [Google Scholar]
- 63.Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 64.Satta R, Maloku E, Zhubi A, Pibiri F, Hajos M, Costa E, et al. Nicotine decreases DNA methyltransferase 1 expression and glutamic acid decarboxylase 67 promoter methylation in GABAergic interneurons. Proc Natl Acad Sci U S A. 2008;105:16356–16361. doi: 10.1073/pnas.0808699105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dong E, Nelson M, Grayson DR, Costa E, Guidotti A. Clozapine and sulpiride but not haloperidol or olanzapine activate brain DNA demethylation. Proc Natl Acad Sci U S A. 2008;105:13614–13619. doi: 10.1073/pnas.0805493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333:1300–1303. doi: 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nabel CS, Manning SA, Kohli RM. The curious chemical biology of cytosine: deamination, methylation, and oxidation as modulators of genomic potential. ACS Chem Biol. 2012;7:20–30. doi: 10.1021/cb2002895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Szulwach KE, Li X, Li Y, Song CX, Wu H, Dai Q, et al. 5-hmC-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci. 2011;14:1607–1616. doi: 10.1038/nn.2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Phiel CJ, Zhang F, Huang EY, Guenther MG, Lazar MA, Klein PS. Histone deacetylase is a direct target of valproic acid, a potent anticonvulsant, mood stabilizer, and teratogen. J Biol Chem. 2001;276:36734–36741. doi: 10.1074/jbc.M101287200. [DOI] [PubMed] [Google Scholar]
- 73.Nalivaeva NN, Belyaev ND, Turner AJ. Sodium valproate: an old drug with new roles. Trends Pharmacol Sci. 2009;30:509–514. doi: 10.1016/j.tips.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 74.Kazahaya Y, Akimoto K, Otsuki S. Subchronic methamphetamine treatment enhances methamphetamine- or cocaine-induced dopamine efflux in vivo. Biol Psychiatry. 1989;25:903–912. doi: 10.1016/0006-3223(89)90270-9. [DOI] [PubMed] [Google Scholar]
- 75.Cadet JL, Jayanthi S, McCoy MT, Beauvais G, Cai NS. Dopamine D1 receptors, regulation of gene expression in the brain, and neurodegeneration. CNS Neurol Disord Drug Targets. 2010;9:526–538. doi: 10.2174/187152710793361496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Cull-Candy SG, Leszkiewicz DN. Role of distinct NMDA receptor subtypes at central synapses. Sci STKE. 2004:re16. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 77.Madden K. NMDA receptor antagonists and glycine site NMDA antagonists. Curr Med Res Opin. 2002;18(Suppl 2):s27–31. doi: 10.1185/030079902125000705. [DOI] [PubMed] [Google Scholar]
- 78.Motawaj M, Arrang JM. Ciproxifan, a histamine H-receptor antagonist/inverse agonist, modulates methamphetamine-induced sensitization in mice. Eur J Neurosci. 2011;33:1197–1204. doi: 10.1111/j.1460-9568.2011.07618.x. [DOI] [PubMed] [Google Scholar]
- 79.Yamamoto H, Kitamura N, Lin XH, Ikeuchi Y, Hashimoto T, Shirakawa O, et al. Differential changes in glutamatergic transmission via N-methyl-D-aspartate receptors in the hippocampus and striatum of rats behaviourally sensitized to methamphetamine. Int J Neuropsychopharmacol. 1999;2:155–163. doi: 10.1017/S1461145799001480. [DOI] [PubMed] [Google Scholar]
- 80.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 81.Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–4356. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–372. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- 83.Graff J, Rei D, Guan JS, Wang WY, Seo J, Hennig KM, et al. An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature. 2012;483:222–226. doi: 10.1038/nature10849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, et al. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, et al. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17:1144–1151. doi: 10.1038/nsmb.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Gene Dev. 2006;20:1256–1261. doi: 10.1101/gad.1412706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kramer OH, Zhu P, Ostendorff HP, Golebiewski M, Tiefenbach J, Peters MA, et al. The histone deacetylase inhibitor valproic acid selectively induces proteasomal degradation of HDAC2. EMBO J. 2003;22:3411–3420. doi: 10.1093/emboj/cdg315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Barbetti V, Gozzini A, Cheloni G, Marzi I, Fabiani E, Santini V, et al. Time- and residue-specific differences in histone acetylation induced by VPA and SAHA in AML1/ETO-positive leukemia cells. Epigenetics. 2013;8:210–219. doi: 10.4161/epi.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bartels M, Geest CR, Bierings M, Buitenhuis M, Coffer PJ. Histone deacetylase inhibition modulates cell fate decisions during myeloid differentiation. Haematologica. 2010;95:1052–1060. doi: 10.3324/haematol.2009.008870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Larsson P, Ulfhammer E, Magnusson M, Bergh N, Lunke S, El-Osta A, et al. Role of histone acetylation in the stimulatory effect of valproic acid on vascular endothelial tissue-type plasminogen activator expression. PLoS One. 2012;7:e31573. doi: 10.1371/journal.pone.0031573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Monti B, Polazzi E, Contestabile A. Biochemical, molecular and epigenetic mechanisms of valproic acid neuroprotection. Curr Mol Pharmacol. 2009;2:95–109. doi: 10.2174/1874467210902010095. [DOI] [PubMed] [Google Scholar]
- 92.Myers SJ, Peters J, Huang Y, Comer MB, Barthel F, Dingledine R. Transcriptional regulation of the GluR2 gene: neural-specific expression, multiple promoters, and regulatory elements. J Neurosci. 1998;18:6723–6739. doi: 10.1523/JNEUROSCI.18-17-06723.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bai G, Norton DD, Prenger MS, Kusiak JW. Single-stranded DNA-binding proteins and neuron-restrictive silencer factor participate in cell-specific transcriptional control of the NMDAR1 gene. J Biol Chem. 1998;273:1086–1091. doi: 10.1074/jbc.273.2.1086. [DOI] [PubMed] [Google Scholar]
- 94.Lunyak VV, Burgess R, Prefontaine GG, Nelson C, Sze SH, Chenoweth J, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science. 2002;298:1747–1752. doi: 10.1126/science.1076469. [DOI] [PubMed] [Google Scholar]
- 95.Huang Y, Doherty JJ, Dingledine R. Altered histone acetylation at glutamate receptor 2 and brain-derived neurotrophic factor genes is an early event triggered by status epilepticus. J Neurosci. 2002;22:8422–8428. doi: 10.1523/JNEUROSCI.22-19-08422.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abrajano JJ, Qureshi IA, Gokhan S, Molero AE, Zheng D, Bergman A, et al. Corepressor for element-1-silencing transcription factor preferentially mediates gene networks underlying neural stem cell fate decisions. Proc Natl Acad Sci U S A. 2010;107:16685–16690. doi: 10.1073/pnas.0906917107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ballas N, Grunseich C, Lu DD, Speh JC, Mandel G. REST and its corepressors mediate plasticity of neuronal gene chromatin throughout neurogenesis. Cell. 2005;121:645–657. doi: 10.1016/j.cell.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 98.Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23:185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- 99.Marsman J, Horsfield JA. Long distance relationships: enhancer-promoter communication and dynamic gene transcription. Biochim Biophys Acta. 2012;1819:1217–1227. doi: 10.1016/j.bbagrm.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 100.Ong CT, Corces VG. Enhancer function: new insights into the regulation of tissue-specific gene expression. Nat Rev Genet. 2011;12:283–293. doi: 10.1038/nrg2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 102.Georgel PT, Horowitz-Scherer RA, Adkins N, Woodcock CL, Wade PA, Hansen JC. Chromatin compaction by human MeCP2. Assembly of novel secondary chromatin structures in the absence of DNA methylation. J Biol Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 103.Yasui DH, Peddada S, Bieda MC, Vallero RO, Hogart A, Nagarajan RP, et al. Integrated epigenomic analyses of neuronal MeCP2 reveal a role for long-range interaction with active genes. Proc Natl Acad Sci U S A. 2007;104:19416–19421. doi: 10.1073/pnas.0707442104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.