Abstract
Highly parallel sequencing of cDNA derived from endogenous small RNAs (small RNA-seq) is a key method that has accelerated understanding of regulatory small RNAs in eukaryotes. Eukaryotic regulatory small RNAs, which include microRNAs (miRNAs), short interfering RNAs (siRNAs), and Piwi-associated RNAs (piRNAs), typically derive from the processing of longer precursor RNAs. Alignment of small RNA-seq data to a reference genome allows the inference of the longer precursor and thus the annotation of small RNA producing genes. ShortStack is a program that was developed to comprehensively analyze reference-aligned small RNA-seq data, and output detailed and useful annotations of the causal small RNA-producing genes. Here, we provide a step- by-step tutorial of ShortStack usage with the goal of introducing new users to the software and pointing out some common pitfalls.
Keywords: Small RNA, High-throughput sequencing, Bioinformatics, microRNA, siRNA, Genome Annotation
1. Introduction
Rapid progress in high throughout sequencing methodologies has played a key role in unveiling the scope of regulatory small RNAs in eukaryotes. Highly parallel sequencing of cDNAs derived from small RNAs (small RNA-seq) allows comprehensive assessment of regulatory small RNA accumulation, especially when combined with alignments to a reference genome. MIRNA loci, which produce the primary stem-loop transcripts that are processed to yield mature microRNAs (miRNAs), represent the best characterized type of small RNA gene; they are readily discerned from genome-aligned small RNA-seq data [1], and are extensively annotated in multiple species by the miRBase database [2]. However, miRNAs frequently comprise only a small percentage of the entire small RNA repertoire, especially in plants [3] and in animal germ-line associated tissues [4]. Unlike MIRNA loci, the loci producing other types of endogenous regulatory small RNAs have had little systematic curation. Thus, there is an emerging need for computational tools that can provide global annotations of both MIRNA and non-MIRNA loci from small RNA-seq datasets.
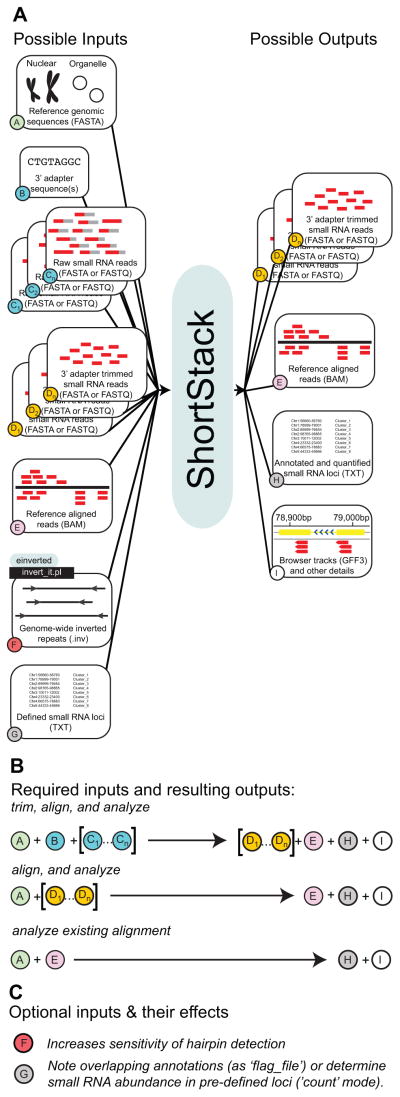
With this need in view, ShortStack was developed for comprehensive analysis of small RNA-seq data [5]. Since the publication of the first ShortStack paper, based on version 0.4.0 of the software [5], the capabilities and performance of ShortStack have been substantially improved. The current version (1.1.0) now is able to handle adapter-trimming and alignment of small RNA-seq data prior to annotation and quantification (Fig. 1). Additionally, ShortStack also can now accept multiple small RNA-seq libraries in a single run. These improvements are layered on top of the core annotation and quantification methods that were previously described [5]. Based on the small RNA alignment patterns, ShortStack identifies and annotates both MIRNA and non-MIRNA loci, and provides detailed descriptions of the small RNA populations emanating from each locus. In this article, we provide a step-by-step tutorial of ShortStack usage for the current version 1.1.0 (Fig. 1). Factors affecting analysis and detection sensitivity in different stages of the computational pipeline are addressed.
Figure 1.
Inputs and outputs of ShortStack.
A. Schematic showing possible inputs and outputs of a ShortStack analysis. Letters in colored circles indicate different types of files or data.
B. Required inputs and their outputs for analysis. Three possible input combinations are possible, depending on the requirements for adapter trimming and alignment. Letters in colored circles refer to file/data types in panel A.
C. Optional inputs. Letters in colored circles refer to file/data types in panel A.
2. Materials
2.1. Basic system requirements
ShortStack is a command-line program for Linux or Mac OSX operating systems. In this article, we assume that the reader has a basic working knowledge of the Bourne Again Shell (BASH) or similar shell applications on their system. ShortStack is implemented in Perl; hence Perl should be installed in the system. By default ShortStack searches for the Perl libraries in /usr/bin/perl, which is the usual location for pre-installed Perl. If the installation directory is different than the default one, then the first line of ShortStack.pl (e.g. the ‘hashbang’) should be modified to reflect that. To compile, ShortStack requires the Perl module Getopt::Long. This module is already installed in nearly all Perl installations, but if not, it can be obtained from the Comprehensive Perl Archive Network (CPAN) [6]. ShortStack was developed using Perl version 5.10.0; there are no known compatibility issues with other versions of Perl but Perl 5.x is recommended.
ShortStack has been tested on Mac OS X (10.6, 10.7, 10.8) and Ubuntu (12.04). At least 4GB of system memory is recommended for running ShortStack. Larger analyses (large small RNA-seq datasets and/or large repetitive genomes) might require even more memory. Precise peak memory requirements are not easily predictable, but are positively correlated with genome size, depth of small RNA-seq data, and the number of hairpins/MIRNA loci identified in a given run.
2.2. Software dependencies
ShortStack depends on several freely available, commonly used third-party software packages (Table 1). Prior installation of RNALfold and RNAeval from the Vienna RNA package [7], and samtools [8] is mandatory for running ShortStack.pl program. The alignment software bowtie (version 0.12.x or 1.x) and its helper program bowtie-build [9] must be installed if ShortStack is asked to perform alignment of small RNA-seq data. All of the required programs (RNALfold, RNAeval, samtools, bowtie, and bowtie-build) must be system-executable, which typically means installing them into a common directory for system executables, such as /usr/bin/ or /usr/local/bin/. Additionally, installation of the EMBOSS package [10] is not needed to execute ShortStack, but the EMBOSS einverted application can be used to enhance identification of large inverted repeats that spawn small RNAs. To complete this tutorial, each of the above-mentioned programs (RNALfold, RNAeval, samtools, bowtie, bowtie-build, and einverted) should be installed according to the instructions provided with the source packages, made system executable, and tested by calling individual program in the terminal to ensure that the program is working properly.
Table 1.
ShortStack software dependencies
| Software | Version tested with ShortStack | Description | URL | Reference |
|---|---|---|---|---|
| perl | 5.10.0 | General purpose high-level interpreted programming language | http://www.perl.org | N/A |
| samtools | 1.1.19 | Utilities for manipulation of alignments in SAM (Sequence Alignment/Map) format | http://samtools.sourceforge.net/ | [8] |
| RNALfold & RNAeval (ViennaRNA package) | 1.8.x, 2.x.x | Predicts locally stable RNA secondary structures | http://www.tbi.univie.ac.at/RNA/ | [7] |
| bowtie & bowtie-build | 0.12.x-1.0.0 | Fast, gap-free short read (e.g. <50 bp) aligner based on the Burrows-Wheeler transform | http://sourceforge.net/projects/bowtie-bio/files/bowtie/ | [9] |
| einverted (EMBOSS package) | 6.5.7 | Finds inverted repeats in DNA sequences | http://emboss.sourceforge.net/ | [10] |
2.2.1 Justifications for selections of dependencies
RNALfold is optimized for fast RNA secondary structure predictions in local regions of very long queries, making it ideal for use in hairpin identification in the context of entire genomes [7]. RNAeval rapidly estimates free energies of given secondary structures [7], which is required during hairpin and MIRNA annotation by ShortStack. ShortStack relies upon of alignments in the widely used BAM format [8] due to the indexing and fast-retrieval capabilities, complete information storage, and highly extensible nature of such alignments when manipulated with the various samtools applications. Finally, ShortStack uses bowtie [9] as an aligner due to bowtie’s very fast performance and optimization for the short, ungapped alignments required for small RNA-seq. The bowtie-build application [9] is required to build the genomic indices required for bowtie to function.
2.3 ShortStack installation
Installation of ShortStack simply involves extracting the contents of the source .tgz file (available from [11]). For convenience, a copy of the script ShortStack.pl can be moved to /usr/bin, /usr/local/bin/, or elsewhere in the user’s PATH to make it system executable. Once installed, calling ShortStack.pl with no arguments will give a brief usage statement:
$ ShortStack.pl ShortStack.pl version 1.1.0 USAGE: ShortStack.pl [options] genome.fasta MODES: 1. Trim, align, and analyze: Requires --untrimmedFA OR -- untrimmedFQ. Also requires --adapter. To stop after alignments, specify --align_only 2. Align, and analyze: Requires --trimmedFA OR --trimmedFQ. To stop after alignments, specify --align_only 3. Analyze: Requires --bamfile Type ‘ShortStack.pl --help’ for full list of options DOCUMENTATION: type ‘perldoc ShortStack.pl’
The full list of options, as of version 1.1.0, is given in section 6 below.
2.4 Tutorial data
A small RNA-seq dataset from Arabidopsis thaliana, along with the reference genome and other key files, is available from [12]. Table 2 shows the contents of the tutorial. To follow along with the example analyses below, download the tutorial and unpack it.
Table 2.
Example datasets and accessory scripts for ShortStack analysis. These resources along with a complete tutorial are available at [12].
| Datasets/scripts | Description |
|---|---|
| Athaliana_167.fa | FASTA file containing unmasked reference nuclear and organelle genomes for Arabidopsis thaliana (TAIR 10). The genomes were downloaded from www.phytozome.net and sequence headers were shortened. |
| Athaliana_167.inv | einverted generated file containing inverted repeats identified in Athaliana_167.fa. |
| SRR051927_even.fastq | Untrimmed small RNA-seq dataset from above ground tissues of Arabidopsis, in FASTQ format. From NCBI SRA SRR051927. Only the even- numbered reads. |
| SRR051927_odd.fastq | Untrimmed small RNA-seq dataset from above ground tissues of Arabidopsis, in FASTQ format. From NCBI SRA SRR051927. Only the odd-numbered reads. |
| ath_hp_mb19_SStack_Athal_167.txt | Coordinates of miRBase 19 Arabidopsis thaliana MIRNA hairpins relative to the TAIR10 genome. |
| invert_it.pl | Perl script for faster processing of einverted predicted inverted repeats |
| ShortStack_TUTORIAL.pdf | Detailed tutorial instructions |
3. Formatting of input data
The required combinations of ShortStack input files vary depending upon the type of analysis being performed (Fig. 1B). Below, the formatting requirements for the various eligible input files and data are discussed.
3.1 Reference Genome
All ShortStack analyses require a reference genome (‘A’ in Fig. 1), supplied as the last argument in the command for running ShortStack. The reference genome should be in multi-FASTA format, with each sequence representing an individual chromosome/scaffold. Generally the unmasked version of the genome should be used, unless you truly want to exclude the discovery of repeat-associated small RNAs, which are numerous in many species. We recommend compiling both organelle and nuclear genomes into one multi-FASTA file to allow comprehensive annotation/discovery of all small RNA-producing loci.
ShortStack will abort or behave unexpectedly if any of the chromosome names have whitespace and/or meta-characters (such as pipe symbols) in the headers. If the original headers in the genome file are long and complicated, these should be modified to a simplified, short version. For instance, if we examine the first two lines of Arabidopsis chromosome 1 sequence, as found directly in a download from Phytozome [13], we find a long header with lots of whitespaces:
>Chr1 CHROMOSOME dumped from ADB: Feb/3/09 16:9; last updated: 2007-12-20 CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCCCTAAATC
In contrast, in the example file from our tutorial (“Athaliana_167.fa”), the chromosome names have been shortened to a simple string:
>Chr1 CCCTAAACCCTAAACCCTAAACCCTAAACCTCTGAATCCTTAATCCCTAAATC
3.2. Adapter sequence(s)
If the input data are untrimmed FASTA or FASTQ small RNA-seq reads, ShortStack requires the appropriate 3′ adapter sequence(s) to search for when trimming (‘B’ in Fig. 1). A valid adapter sequence consists of at least 8 characters, all of which must be either A, T, G, or C. The adapter sequence is passed with the option --adapter. If more than one untrimmed small RNA-seq file is being input, trimming with multiple adapters is possible by passing in a comma-delimited list of corresponding adapters. The number of adapters provided must be either one (in which case it is applied to all untrimmed files) or equal to the number of input untrimmed files.
3.3 Raw (untrimmed) small RNA-seq data
Untrimmed small RNA-seq data must be in either the FASTA or FASTQ format (‘C’ in Fig. 1). Path(s) to the file(s) are provided with the option --untrimmedFA or --untrimmedFQ, for FASTA and FASTQ data respectively. The data should not be condensed or manipulated in any way (for instance, users should NOT collapse reads with identical sequences into a single entry). There is no support for paired-end libraries. Multiple files can be input as a comma-delimited list of files. No comment lines are allowed in the data, and valid adapter(s) must also be provided. The data are assumed to represent the sense strand of small RNAs, with the 5′-most base of the small RNA occurring at the first position of the read. 3′-adapter trimming by ShortStack is very simplistic: It identifies the 3′-most occurrence of the adapter specified in the read, allowing for no substitutions and irrespective of quality values. More sophisticated methods of adapter trimming are available as an alternative to relying upon ShortStack for read-trimming [14,15]. Currently colorspace data are not supported but we plan to implement this in future updates.
3.4 Trimmed small RNA-seq data
Adapter-trimmed small RNA-seq data must be in either the FASTA or FASTQ format (‘D’ in Fig. 1). Path(s) to the file(s) are provided with the option --trimmedFA or --trimmedFQ, for FASTA and FASTQ data respectively. The data should not be condensed or manipulated in any way except for adapter trimming or removal of reads via upstream quality control processes (for instance, users should NOT collapse reads with identical sequences into a single entry). There is no support for paired-end libraries. Multiple files can be input as a comma-delimited list of files. No comment lines are allowed in the data. The data are assumed to represent the sense strand of small RNAs with no residual adapter bases or quality values.
3.5 Alignments
Reference-aligned small RNA-seq data must be provided in the BAM format (‘E’ in Fig. 1). When provided by the user, the file is passed by the option --bamfile. BAM is the compressed, binary representation of the Sequence Alignment/Map (SAM) format [8]. Users unfamiliar with the SAM/BAM format should consult the format specification [16] to understand the details presented below. ShortStack has very particular requirements for a BAM alignment, and checks these requirements during every run during a validation step. Because of the very specific formatting requirements, it is highly recommended that users simply align their data using ShortStack instead of creating BAM alignments by other means. Note that BAM alignments created for versions of ShortStack prior to 1.0.0 will NOT be valid when used with ShortStack version 1.0.0 and higher. The specific requirements of a valid BAM alignment for ShortStack are as follows:
It must have a header
It must be sorted by genomic coordinate (as indicated by the SO tag under the @HD record type in the header)
It must match the genome provided to ShortStack (this is checked by cross-referencing the chromosome names given in the SN header tags under @SQ record types in the header with those present in the input genome FASTA file)
The alignment lines must possess custom ‘XX’ tags. This ShortStack-derived custom tag gives the number of possible valid alignments for a read.
It must have an index (created by the samtools index command), or ShortStack will attempt to build the index, aborting if the index build fails.
The alignment lines for mapped reads must possess valid Compact Idiosyncratic Gapped Alignment Report (CIGAR) strings per the SAM format specification.
Support for read groups is provided only if they are properly specified in the BAM header with ID tags under @RG record types.
It is assumed that each read has only one reported alignment, which in the case of potentially multi-mapping reads was selected at random. ShortStack has no way of checking this (except by looking for the XX tag; see above), but the results will not be reliable if this assumption is not met.
Unmapped reads are permitted (and present when alignments are created by ShortStack) but are ignored during analysis.
3.6 Inverted repeats
The program einverted from the EMBOSS package (Table 1) is used to generate a file of qualifying inverted repeats from the reference genome (‘F’ in Fig. 1). When provided, the file is passed in with the option --inv_file. The required format is the text-based alignments given by einverted. The file ‘Athaliana_167.inv’ provided in the tutorial provides an example of this format.
einverted output is not a requirement for ShortStack, but it does increase the sensitivity of ShortStack to detect hairpin-associated small RNA loci, especially for very large hairpins. einverted usually consumes a very large amount of memory when applied to whole genomes. Using a wrapper script to call one chromosome at a time for einverted analysis, and then merging the outputs, considerably reduces the memory footprint of this process. In addition, many of the einverted-produced structures will not meet the default hairpin criteria of ShortStack, so it is useful to filter those out too. The wrapper script “invert_it.pl” (Table 2), provided with the ShortStack tutorial [12], can accomplish all of this. Below is an example calling the invert_it.pl wrapper script on the Arabidopsis genome provided in the tutorial data. Note that the completed set of inverted repeats is also already provided in the tutorial data download (Table 2).
$ perl invert_it.pl -g Athaliana_167.fa -o Athaliana_167_user.inv
3.6.1 Parameters for invert_it.pl
The invert_it.pl script (section 3.6) accepts following parameters for executing einverted-based identification of repeats:
-g : Path to FASTA-formatted reference genome
-o : Name of output .inv file (will be created in working directory; existing file of same name will be overwritten without warning).
-p : Minimum number of potential base pairs required to keep an inverted repeat. Defaults to 15 if not specified by the user.
-f : Minimum fraction of stem bases that must be paired to keep an inverted repeat. Defaults to 0.67 if not specified by the user.
3.6.2 Notes
Since einverted is very memory intensive, it is prudent to perform this step on a machine that has a lot of memory. Even for the very small Arabidopsis genome analyzed in the above example, peak memory usage of about 2.6G was observed (Mac OS 10.6.8, Dual quad-core Xeon processors).
3.7 Defined small RNA loci
Files containing lists of defined genomic loci (‘G’ and ‘H’ in Fig. 1) are tab-delimited text files. Comment lines (beginning with “#”) are ignored. The first column gives a genomic location in the format Chromosome:Start-Stop, where ‘Chromosome’ is the name of a reference sequence from the genome, and start and stop are one-base inclusive coordinates. The second column gives a name for the feature. Any additional columns, if present, are ignored.
These files can be used in two different ways. When passed in under the --flag_file option, ShortStack will report on any positional overlap between the loci in the defined file, and the small RNA loci it discovers/analyzes during the run. When passed in under the --count option, ShortStack is instructed to avoid de novo annotation of small RNA genes, and instead simply quantify and describe small RNA expression from the pre-defined loci.
For convenience, the output of a previous ShortStack run (‘H’ in Fig. 1) is compatible for use as a file of defined small RNA loci in subsequent runs.
4. ShortStack analysis
Small RNA gene discovery by ShortStack is highly flexible since it allows for optimization of various parameters for increasing detection sensitivity and specificity. Input data can be untrimmed raw small RNA-seq reads, trimmed reads, or a pre-existing alignment (Fig. 1). Multiple small RNA-seq libraries can be automatically merged and analyzed. ShortStack analysis can be run in three different modes: de novo (full de novo annotation and subsequent analysis of small RNA loci including secondary structure evaluation, default mode), nohp (full de novo annotation of loci, but no hairpin or MIRNA evaluation) and count (quantitation of small RNA expression from pre-defined loci, forces nohp mode). Additionally, ShortStack has multiple user-adjustable parameters related to small RNA cluster discovery, acceptable small RNA size range, secondary structure criteria, and phasing size (for phased siRNA loci detection). Furthermore, MIRNA discovery in ShortStack can be optimized for plants (default option) or animals by providing the appropriate specimen type in the miRType parameter. This optimization involves fine tuning of several MIRNA secondary structure related parameters based on comprehensive analysis of miRBase annotations [5]. The full list of ShortStack options, their defaults, and their meanings is given in section 6 below.
4.1 Full de novo mode with multiple, untrimmed FASTQ libraries
A common small RNA experiment generates multiple libraries from technical or biological replicates, distinct tissues, individuals, or genotypes. In such experiments, it is often desirable to derive a set of de novo small RNA gene annotations based on the union of all of the data, and then to subsequently quantify small RNA expression at those loci in each of the input datasets separately. As of version 1.1.0, ShortStack automatically handles all of these tasks in a single run. To illustrate this, install ShortStack.pl and its required dependencies (section 2), download the tutorial data [12], unpack it, and cd into the unpacked TUTORIAL directory. Once there, type the following command:
$ ShortStack.pl --outdir tutorial1 --untrimmedFQ SRR051927_odd.fastq,SRR051927_even.fastq --adapter CTGTAGGC --inv_file Athaliana_167.inv --flag_file ath_hp_mb19_SStack_Athal_167.txt Athaliana_167.fa
4.1.1 Explanation of the command
--outdir : This specifies the name of the directory that will house the results. The directory will be created in the working directory. If it already exists, ShortStack will complain and abort.
--untrimmedFQ : This specifies the paths to the files of FASTQ small RNA reads that have not been trimmed to remove the 3′ adapter sequence (‘C’ in Fig. 1). Multiple paths are separated by commas. In this case there are two input files.
--adapter : This is a required option if --untrimmedFQ or --untrimmedFA is provided. It gives the sequence to search for to identify the start of the 3′ adapter sequence (‘B’ in Fig. 1). This option requires at least 8 nts, and all characters must be A, T, G, or C. If multiple untrimmedFQ/FA files are provided, the adapter must also be a comma-delimited list of multiple adapters (corresponding in order to the untrimmed data files), or, in cases where the same adapter applies to all libraries, just one sequence (as above).
--inv_file : This is a file of inverted repeats in the genome created by the EMBOSS program einverted via the wrapper script ‘invert_it.pl’. ‘F’ in Fig. 1. See section 3.6.
--flag_file : This is a file containing a list of defined loci to ‘flag’ if any of the ShortStack-discovered loci overlap them (‘G’ in Fig. 1). The example file, ‘ath_hp_mb19_SStack_Athal_167.txt’, is a list of Arabidopsis thaliana MIRNA hairpin locations.
Athaliana_167.fa : The final argument in in ShortStack run is the path to the reference genome of interest. ‘A’ in Fig. 1.
4.1.2 Progress of the analysis
The first action you will see during the run after a verbose printout of the parameters is the creation of the .fai fasta index file for the reference genome, as noted by the message:
Expected genome index Athaliana_167.fa.fai for genome file Athaliana_167.fa not found. Creating it using samtools faidx done
This index file is placed in the same location as the reference genome file.
Next, the adapter trimming will occur. This should complete relatively fast and when done give this output:
Adapter trimming file SRR051927_odd.fastq with adapter CTGTAGGC … Done No insert (includes adapter-only and no-adapter cases combined): 355867 Too short (less than 15nts): 20706 Ambiguous bases after trimming: 71 OK - output: 2475354 Results in file SRR051927_odd_trimmed.fastq Adapter trimming file SRR051927_even.fastq with adapter CTGTAGGC … Done No insert (includes adapter-only and no-adapter cases combined): 356565 Too short (less than 15nts): 20866 Ambiguous bases after trimming: 77 OK - output: 2474489 Results in file SRR051927_even_trimmed.fastq
The two trimmed FASTQ files, with names of “SRR051927_odd _trimmed.fastq” and “ SRR051927_even_trimmed.fastq” will be written to the path where the untrimmed FASTQ files were found.
Next, alignment will begin. ShortStack first searches for the bowtie indices for the reference genome. They will not be found (unless you already built them by a previous run, or by calling bowtie-build), and so ShortStack.pl will run bowtie-build:
Beginning alignment of reads from SRR051927_odd_trimmed.fastq to genome Athaliana_167.fa … Check for ebwt bowtie indices for the genome: ABSENT. Attempting to build with bowtie-build… Done. See Athaliana_167.fa_bowtie_build_log.txt for output from bowtie-build
The 6 .ebwt bowtie indices will be placed in the same path as the genome.fasta file.
After that, read alignment begins. The two files are aligned separately to the genome, temporarily creating two BAM alignments in the working directory. After both alignment jobs have completed, the alignments are merged to create a single BAM alignment file. After the merge, the original two separate alignments are deleted. Importantly, the origin of each read is tracked with the read group (RG) tag, allowing future analysis of the merged alignment with respect to individual libraries. The header of the merged final bam alignment file also contains information on the read groups in the @RG tags. The name of the final alignment file is determined by the value of option --outdir ; in this example, the final alignment is “tutorial1.bam”. It is written to the working directory.
Next, the full de novo analysis pipeline will begin using all alignments in the merged file. Progress is reported to STDERR, and to the logfile. RNA secondary structural analysis is by far the most time-consuming portion of the process.
When the de novo analysis completes, ShortStack automatically performs a “count” mode analysis of each read group separately, effectively quantitating the small RNA expression at each discovered locus in each of the input libraries separetly. This is triggered when the user does NOT specify option --read_group AND the analyzed bam alignment file has > 1 read group specified, as was this case in this example.
When complete, the results will be in the directory “tutorial1”, as specified by the --outdir option. When performed on a Mac Pro tower with 2 x 2.8GHz Quad-Core Xeon processors this analysis takes about 80 minutes to complete and has an approximate peak memory usage of about 1.2Gb. Small RNA alignment is the most memory-intensive phase, while RNA folding is the most time-consuming phase.
4.1.3 Selecting parameters for de novo analysis
In most analyses that we can foresee, it will be best to use the default values for all parameters for standard de novo ShortStack analyses. However, some parameters may need to be more frequently adjusted. For accurate annotation of MIRNA loci from animals, the miRType parameter should be set to “animal” instead of its default setting of “plant”. In cases where the small RNAs of regulatory significance differ in size from the default 20–24 nt range, the dicermin and dicermax parameters should be adjusted accordingly. Finally, the sensitivity and specificity of locus discovery can be adjusted using the mindepth and pad paramters. Lower values of mindepth will trigger annotation of loci with lower small RNA mapping densities, and thus increase the sensitivity; higher values of mindepth will have the opposite effect, and decrease sensitivity. The default value of the mindepth parameter is 20, which means that a minimum coverage 20 reads deep is required to trigger locus annotation. The pad parameter dictates the extent of merging of adjacent “islands” of coverage that exceed mindepth occurs. Higher values of pad cause more merging, and decrease specificity; lower values have the opposite effect. The default value for pad is 100, meaning that adjacent islands of coverage are merged if they are within 200 nts (100 + 100) from each other. Full descriptions of all parameters are found in the README file of ShortStack package, in [5], or by typing “perldoc ShortStack.pl”.
4.2 Count mode
In this example, the use of ShortStack in ‘count’ mode is demonstrated. This mode takes in a list of coordinates from a file of defined small RNA loci (‘G’ in Fig. 1) via the option --count, and quantifies their small RNA expression values. In addition, this example will demonstrate the use of the --read_group option, which is used to limit analysis to only the specified read group. The example below assumes you have completed step 4.1 above, which will create the merged BAM alignment of the example data, ‘tutorial1.bam’. Move into the TUTORIAL directory and type the command:
$ ShortStack.pl --outdir tutorial2 --count ath_hp_mb19_SStack_Athal_167.txt --bamfile tutorial1.bam -- read_group SRR051927_odd_trimmed Athaliana_167.fa
4.2.1 Explanation of the command
--outdir : This specifies the name of the directory that will house the results. The directory will be created in the working directory. If it already exists, ShortStack will complain and abort.
--count : Instructs ShortStack to quantify small RNA expression within the loci provided in the file (‘G’ in Fig. 1), which in this case are Arabidopsis thaliana MIRNA hairpins.
--bamfile : Path to a properly formatted BAM alignment file (‘E’ in Fig. 1). In this case, the tutorial1.bam file was created during the previous example (section 4.1).
--read_group : Restricts analysis only to small RNAs derived from the indicated read group.
Athaliana_167.fa : The final argument in in ShortStack run is the path to the reference genome of interest. ‘A’ in Fig. 1.
4.2.2 Progress of the analysis
This analysis will proceed very rapidly, as the time consuming alignment and hairpin analyses are not being performed.
5. Results and Discussion
A brief summary of results is provided (as terminal output and also in a Log.txt file) after completion of ShortStack run. Detailed annotation of all loci, including locus types, strandedness and repetitiveness are provided in a separate Results file (‘H’ in Fig. 1). This text file can be imported as data table in R [17] or general spreadsheet application for further downstream analyses. The abundances reported for each locus can be also processed for differential expression analysis. ShortStack also generates GFF3-formatted browser track files (‘I’ in Fig. 1), which facilitate visualization of the loci in conjunction with a genome browser. For de novo discovery, Hairpin (HP) and MIRNA loci details are provided in separate folders, which is useful for gaining further biological insight about biogenesis, precision and function of such loci. Finally, a short summary file lists coordinates and mature sequences of all ShortStack annotated MIRNA loci. The mature sequences can then be used for analyzing sequence conservation, annotating novel miRNA families, target prediction and/or identification of sliced targets.
In a typical de novo ShortStack analysis in plants, the abundance of different classes of small RNA loci follows a pattern with non-hairpin/siRNA loci being the most abundant (typically thousands of loci), hairpin loci as relatively less abundant (typically hundreds) and MIRNA loci the least abundant. In case of any deviation from the usual loci abundance patterns, it may be useful to analyze the genomic distribution of mapped reads to discern any bias in the small RNA-seq data. For example, if a significant portion of reads is derived from ribosomal RNA fragments, the discovery of small RNA loci will be limited due to insufficient coverage of the regulatory small RNAs of interests. Additionally, systemic biases due to specimen quality, variation in library preparation techniques and sequencing platforms can affect the small RNA gene identification process [18,19]. For this reason, in case of analyzing condition- or tissue-specific differential expression of small RNA loci, combining libraries made with different technologies should be avoided. Also, replicates for each condition or tissue type should be prepared with identical methodologies.
Even though the ShortStack-based protocol described above provides comprehensive annotations, identification of certain classes of small RNA-producing loci may not be possible due to limitations in current methodologies. Further improvement of detection sensitivity depends on development of faster RNA secondary structure prediction algorithms, advancement of sequencing technology, and more knowledge about the processing and biogenesis of different categories of non-coding RNA genes.
Acknowledgments
We thank Zhaorong Ma for productive discussions during ShortStack development and testing. Research in the Axtell Lab is currently supported by grants from the NIH (R01 GM084051) and NSF (1121438).
6. Appendix - Full list of ShortStack version 1.1.0 options, their default values, and their meanings
USAGE: ShortStack.pl [options] genome.fasta
OPTIONS:
--help : Print a help message and then quit.
--version : Print the version number and then quit.
--outdir [string] : Name of directory to be created to receive results of the run. Defaults to “ShortStack_[time]”, where time is “UNIX time” (the number of non-leap seconds since January 1, 1970 UCT), if not provided
--untrimmedFA [string] : Path to untrimmed small RNA-seq data in FASTA format. Multiple datasets can be provided as a comma-delimited list.
--untrimmedFQ [string] : Path to untrimmed small RNA-seq data in FASTQ format. Multiple datasets can be provided as a comma-delimited list.
--adapter [string] : Sequence of 3′ adapter to search for during adapter trimming. Must be at least 8 nts in length, and all ATGC characters. Required if either --untrimmedFA or --untrimmedFQ are specified. Multiple adapters (for when multiple input untrimmedFA/FQ files are specified) can be provided as a comma-delimited list.
--trimmedFA [string] : Path to trimmed and ready to map small RNA-seq data in FASTA format. Multiple datasets can be provided as a comma-delimited list.
--trimmedFQ [string] : Path to trimmed and ready to map small RNA-seq data in FASTQ format. Multiple datasets can be provided as a comma-delimited list.
--align_only : Exits program after completion of small RNA-seq data alignment, creating BAM file.
--bamfile [string] : Path to properly formatted and sorted BAM alignment file of small RNA-seq data.
--read_group [string] : Analyze only the indicated read-group. Read-group must be specified in the bam alignment file header. Default = [not active -- all reads analyzed]
--inv_file [string] : PATH to an einverted-produced .inv file of inverted repeats within the genome of interest. Not required but strongly suggested for more complete annotations of hairpin-derived small RNA genes. Default = {blank}. Not needed for runs in “nohp” mode or runs in “count” mode (because “count” mode forces “nohp” mode as well). A typical eniverted run uses default parameters except “-maxrepeat 10000”, in order to capture long IRs.
--flag_file [string] : PATH to a simple file of genomic loci of interest. The ShortStack-analyzed small RNA clusters will be analyzed for overlap with the loci in the flag_file .. if there is any overlap (as little as one nt), it will be reported.
--mindepth [integer] : Minimum depth of mapping coverage to define an ‘island’. Default = 20. Must be at least 2, more than 5 preferred.
--pad [integer] : Number of nucleotides upstream and downstream to extend initial islands during cluster definition. Default = 100
--dicermin [integer] : Smallest size in the Dicer size range (or size range of interest). Deafult = 20. Must be between 15 and 35, and less than or equal to --dicermax
--dicermax [integer] : Largest size in the Dicer size range (or size range of interest). Deafult = 24. Must be between 15 and 35, and more than or equal to --dicermin
--minUI [float] : Minimum uniqueness index required to attempt RNA folding. Must be a value between 0 and 1. Zero forces all clusters to be folded; default: 0.1
--maxhpsep [integer] : Maximum allowed span for a base-pair during hairpin search with RNALfold; Also serves as the maximum size of genomic query to fold with RNALfold .. loci whose unpadded size is more than --maxhpsep will not be analyzed at all with RNALfold. Default = 300. Must be between 50 and 2000.
--minfracpaired [float] : Minimum fraction of paired nucleotides required within a valid hairpin structure. Default = 0.67. Allowed values are greater than 0 and less than or equal to 1.
--minntspaired [integer] : Minimum absolute number of paired nucleotides required within a valid hairpin structure. Default = 15. Allowed values are greater than zero and less than or equal to --maxhpsep
--maxdGperStem [float] : Maximum deltaG / stem length allowed in a valid hairpin structure. Stem length is 0.5 * (left_stem_length + right_stem_length). Default = -0.5
--minfrachpdepth [float] : Minimum fraction of corrected coverage within hairpin arms to keep hairpin for further analysis. Default = 0.67. Allowed values between 0 and 1.
--miRType [string] : Either “plant” or “animal”. Defaults to “plant”. This option sets --maxmiRHPPairs, --maxmiRUnpaired, and --maxLoopLength to 150, 5, and 100,000 respectively for type “plant”. For type “animal”, the three are instead set to 45, 6, and 15, respectively.
--maxmiRHPPairs [integer] : Maximum number of base pairs in a valid MIRNA hairpin. default: set by --miRType “plant” to 150. --miRType “animal” sets to 45 instead. When provided, user settings will override miRType settings.
--maxmiRUnpaired [integer] : Maximum number of unpaired miRNA nts in a miRNA/miRNA* duplex. default: set by --miRType “plant” to 5. --miRType “animal” instead sets it to 6. When provided, user settings will override miRType settings.
--maxLoopLength [integer] : maximum allowed loop length for a valid hairpin. default: set by --miRType “plant” be essentially unlimited (100,000). --miRType “plant” sets it to 15. When provided, user settings will override miRType settings.
--minstrandfrac [float] : Minimum fraction of mappings to one or the other strand call a polarity for non-hairpin clusters. Also the minimum fraction of “non-dyad” mappings to the sense strand within potential hairpins/miRNAs to keep the locus annotated as a hp or miRNA. See below for details. Default = 0.8. Allowed values between 0.5 and 1.
--mindicerfrac [float] : Minimum fraction of mappings within Dicer size range to annotate a locus as Dicer-derived. Default = 0.85. Allowed values between 0 and 1.
--phasesize [integer] : Examine phasing only for clusters dominated by the indicated size range. Size must be within the bounds described by --dicermin and --dicermax. Set to ‘all’ to examine p-values of each locus within the Dicer range, in its dominant size. Set to ‘none’ to suppress all phasing analysis. Default = 21. Allowed values between --dicermin and --dicermax.
--count [string] : Invokes count mode, in which user-provided clusters are annotated and quantified instead of being defined de novo. When invoked, the file provided with --count is assumed to contain a simple list of clusters. Count mode also forces nohp mode. Default : Not invoked.
--nohp : If “--nohp” appears on the command line, it invokes running in “no hairpin” mode. RNA folding, hairpin annotation, and MIRNA annotation will be skipped (likely saving significant time). Note that --count mode forces --nohp mode as well. Default: Not invoked.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Li Y, Zhang Z, Liu F, Vongsangnak W, Jing Q, Shen B. Performance comparison and evaluation of software tools for microRNA deep-sequencing data analysis. Nucleic Acids Res. 2012;40:4298–4305. doi: 10.1093/nar/gks043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axtell MJ. Classification and Comparison of Small RNAs from Plants. Annu Rev Plant Biol. 2013;64:137–159. doi: 10.1146/annurev-arplant-050312-120043. [DOI] [PubMed] [Google Scholar]
- 4.Juliano C, Wang J, Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu Rev Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Axtell MJ. ShortStack: Comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. [Accessed October 2, 2013]; http://www.cpan.org/
- 7.Lorenz R, Bernhart SH, zu Siederdissen CH, Tafer H, Flamm C, Stadler PF, Hofacker IL. ViennaRNA Package 20. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice P, Longden I, Bleasby A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 11. [Accessed October 2, 2013]; http://axtell-lab-psu-weebly.com/shortstack.html.
- 12. [Accessed October 2, 2013]; http://axtelldata.bio.psu.edu/data/ShortStack_TestData/TUTORIAL.tgz.
- 13.Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokhsar DS. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2011;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17.1:10–12. [Google Scholar]
- 15.Patel RK, Jain M. NGS QC Toolkit: A Toolkit for Quality Control of Next Generation Sequencing Data. Plos One. 2012;7:e30619. doi: 10.1371/journal.pone.0030619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. [Accessed October 2, 2013]; http://samtools.sourceforge.net/SAMv1.pdf.
- 17.R Core Development Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2013. [Google Scholar]
- 18.Toedling J, Servant N, Ciaudo C, Farinelli L, Voinnet O, Heard E, Barillot E. Deep-Sequencing Protocols Influence the Results Obtained in Small-RNA Sequencing. Plos One. 2012;7:e32724. doi: 10.1371/journal.pone.0032724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormick KP, Willmann MR, Meyers BC. Experimental design, preprocessing, normalization and differential expression analysis of small RNA sequencing experiments. Silence. 2011;2:2. doi: 10.1186/1758-907X-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]