Abstract
The Arabidopsis endosperm consists of a single cell layer surrounding the mature embryo and playing an essential role to prevent the germination of dormant seeds or that of nondormant seeds irradiated by a far red (FR) light pulse. In order to further gain insight into the molecular genetic mechanisms underlying the germination repressive activity exerted by the endosperm, a "seed coat bedding" assay (SCBA) was devised. The SCBA is a dissection procedure physically separating seed coats and embryos from seeds, which allows monitoring the growth of embryos on an underlying layer of seed coats. Remarkably, the SCBA reconstitutes the germination repressive activities of the seed coat in the context of seed dormancy and FR-dependent control of seed germination. Since the SCBA allows the combinatorial use of dormant, nondormant and genetically modified seed coat and embryonic materials, the genetic pathways controlling germination and specifically operating in the endosperm and embryo can be dissected. Here we detail the procedure to assemble a SCBA.
Keywords: Plant Biology, Issue 81, Technology, Industry, and Agriculture, Life Sciences (General), Control of Seed germination, Seed Coat, Endosperm, Dormancy, Far red light, Abscisic acid, gibberellins, DELLA factors
Introduction
In Arabidopsis mature seeds, the seed coat is composed of the testa, an external layer of dead tissue of maternal origin, and the endosperm, a single cell layer of live tissue directly surrounding the embryo1. The endosperm and the embryo are derived from separate fertilization events: the endosperm is a triploid tissue with two maternal and one paternal genome whereas the embryo is a diploid tissue with one maternal and one paternal genome2.
The main function traditionally assigned to the endosperm is that of a nutritive tissue. However, it is becoming increasingly evident that the endosperm also plays a central role to control seed germination. This notion became first apparent in the case of dormancy, a trait exhibited by newly produced seeds. Dormant seeds fail to germinate despite the presence of favorable germination conditions. Seeds lose their dormancy after a ripening period and become nondormant, i.e. they will germinate when exposed to favorable germination conditions. In many plant species, including the model plant Arabidopsis, the seed coat is absolutely required to prevent the germination of dormant seeds since seed coat removal triggers embryonic growth and greening3,4. In Arabidopsis, Bethke et al. observed that germination remained repressed after removing the testa while maintaining the endosperm surrounding the endosperm5. These observations strongly indicated that the endosperm is the tissue within the seed coat exerting a repressive activity on the embryo. However, seed coat removal experiments do not necessarily help clarifying the nature of the germination repressive activity provided by the seed coat nor identifying the genes that implement it.
We recently introduced a seed coat bedding assay (SCBA) where seed coats and embryos are physically separated but kept in close proximity so that the germination repressive activity provided by the endosperm is maintained6. The SCBA allows the combinatorial use of dormant, nondormant, and genetically modified seed coat and embryonic materials. As a result, the genetic pathways controlling germination and specifically operating in the endosperm and embryo can be dissected. The SCBA was used in the context of dormancy to show that the endosperm releases the phytohormone abscisic acid (ABA) towards the embryo to repress its growth6. Furthermore we could use the SCBA to identify the signaling pathways operating in endosperm and embryonic tissues to promote dormancy.
The role of the endosperm to control germination was further strengthened by considering the case of nondormant seeds exposed to a pulse of far red (FR) light. Early upon seed imbibition a FR light pulse is known to inhibit germination7,8. When seed coats were removed from seeds a pulse of FR light was unable to inhibit germination, strongly suggesting that the endosperm can also repress the germination of nondormant seeds 9. Remarkably, the SCBA could also be used to recapitulate FR-dependent inhibition of germination. This allowed to show that that FR-dependent inhibition of seed germination is also a process involving ABA release from the endosperm9. Furthermore, the SCBA allowed identifying the different light-signaling pathways operating in the endosperm and the embryo to control nondormant seed germination in response to light cues9,10.
The SCBA appears therefore to be a reliable technique to explore the function of the endosperm in the context of the control of seed germination. It is also a powerful tool to assess in vitro whether genes suspected to control germination operate in the endosperm, the embryo or both tissues. Here we detail the various steps required to assemble a SCBA.
Protocol
Once the SCBA is assembled, the growth of embryos is monitored over several days. Therefore, before the seed dissection procedure and assembly of the SCBA, one needs to sterilize seeds to avoid future contaminations that could prevent proper assessment of the effect of seed coat material on embryonic growth.
1. Seed Sterilization
Pour 50-60 μl of mature and dry Arabidopsis seeds in a 1.5 ml microcentrifuge tube and prepare a 70% ethanol solution.
Add 1 ml of 70% ethanol solution to the microcentrifuge tube containing the seeds and shake at room temperature for 10 min at 1,200 rpm in a vortexer.
Centrifuge microcentrifuge tube for 3 sec at 4,000 x g to concentrate seeds at the bottom of the tube.
Aspirate the 70% ethanol solution using a vacuum suction tip carefully leaving the seeds at the bottom of the microcentrifuge tube.
Add 1 ml of sterile distilled water to the microcentrifuge tube still containing the seeds.
Shake at room temperature for 10 min at 1,200 rpm.
Centrifuge microcentrifuge tube for 3 sec at 4,000 x g to concentrate seeds at the bottom of the tube. Aspirate water supernatant using a vacuum suction tip carefully leaving the seeds at the bottom of the tube.
Add 1 ml of sterile water distilled water to the microcentrifuge tube still containing the seeds. Ensure that the flow of water homogenously suspends the seeds inside the tube. The goal here is to avoid further shaking so as to preserve the integrity of the seeds.
Let the seeds settle down by gravity at the bottom of the tube by leaving the tube still for 30 sec. Aspirate water supernatant using a vacuum suction tip carefully leaving the seeds at the bottom of the tube.
Repeat four times steps 1.8 - 1.9. Overall the sterilization procedure takes about 30 min.
2. Seed Plating
All subsequent steps are performed inside a laminar flow cabinet to preserve sterile conditions.
Prepare a Petri dish plate (100 mm diameter, 20 mm height) containing 30 ml of germination medium prepared by autoclaving a solution containing 4.3 g/L Murashige and Skoog medium in 2.5 mM 2-(N-morpholino)ethanesulfonic acid (MES-KOH; pH 5.7) and 0.8 g/L agar. Let the solution cool down in a 50 °C water bath before pouring it on the Petri dish plate.
Add 200 μl of sterile distilled water to the tube containing the seeds. Resuspend the seeds and transfer them on the surface of the germination medium using a standard pipette with a 1 ml tip.
Remove the water surrounding the seeds on the surface of the germination medium using a vacuum suction tip.
Leave the Petri dish containing the seeds without its lid inside the laminar flow cabinet for 2 hr for further drying. Close the Petri dish with the lid and let it stand under the laminar flow cabinet for about 90 min so that about 4 hr have passed since the initiation of the seed sterilization procedure (step 1.2).
3. Seed Dissection
The following steps necessitate working with a stereomicroscope placed inside the laminar flow hood cabinet. A Dumont forceps #5 with truncated (i.e. blunt) tips greatly facilitates the handling of the seeds (Figure 1A).
Prepare a new Petri dish plate containing 30 ml of germination medium. Place on the surface of the medium two juxtaposed rectangular sterile Whatman 3MM papers (1.5 cm x 1.0 cm; Figure 1B, step 1). Whatman 3MM paper is sterilized in an autoclave.
Transfer seeds on the Whatman 3MM papers using sterile forceps (Figure 1B, step 2).
Hold an individual seed against the Whatman 3MM paper by gently pressing the seed using the juxtaposed blunt tips of the truncated forceps (Figure 1B, step 3).
Cut the seed coat (testa and endosperm) of the seed using a syringe needle (e.g. U100 insulin syringe + 1 ml syringe, 0.33 mm (29 G) x 12.7 mm) (Figure 1B, step 4). The shape of Arabidopsis seed is ellipsoidal and it is recommended to cut the seed coat along the longest semi-principal axis as close as possible to the place where the cotyledons are joined to the radicle (i.e. away from the radical tip). This helps the safe release of the embryo in the next step of the dissection procedure.
Push the seed against the Whatman 3MM paper using the juxtaposed blunt tips of the truncated forceps (Figure 1B, step 5). This step releases the embryo out the seed coat through the opening created in step 3.4 (Figure 1B, step 6). It is recommended to apply the push on the seed where the tip of the cotyledons are in closest to the tip of the radicle (i.e. away from the opening).
4. Assembly of the Seed Coat Bedding Assay (SCBA)
See discussion for the proper choice of the number of seed coats and embryos.
Prepare a new Petri dish plate containing 30 ml of germination medium. Place on the surface of the medium a sterile rectangular piece (3.5 cm x 5.0 cm) of Nylon mesh (Figure 2, step 1).
To avoid damage, embryos and seed coats can be moved to the surface of the metallic edges of the forceps by gently pushing them using the syringe needle (Figure 2, step 2). Transfer embryos and seed coats on the Nylon mesh (Figure 2, step 3).
Assemble a circular and single layer of seed coats using the blunted forceps and the needle making sure that seeds coats are in as much closer proximity as possible (Figure 2, step 4). Try to expose as much as possible the seed coat opening generated by the needle upwards. This step was not systematically tested. However, it could increase the efficiency of the SCBA since diffusible substances inhibiting the growth of the embryo are expected to directly diffuse out towards the embryo rather than away from it.
Place the embryos as a circular single layer on the center of the seed coat bed assembled in step 4.3 (Figure 2, step 5). Ensure that the embryos are in as much closer proximity as possible.
Incubate the SCBAs under continuous light (40 μmol/m2sec) at 20-21 °C. In the case where a FR pulse is used to arrest germination, assemble the SCBA under normal light, apply the FR pulse as described9 and leave Petri dish in darkness.
Representative Results
Previous work showed that mutant seeds unable to synthesize GA were unable to germinate as a result of high ABA accumulation in seeds 11,12. However, inability to germinate requires the seed coat since its removal triggers embryonic growth13. This strongly indicated that the endosperm of seeds unable to synthesize GA is releasing ABA to block embryonic growth. We therefore expect seeds coats unable to synthesize GA to block the growth of embryos in a SCBA unlike seed coats unable to synthesize GA and ABA. Here we use ga1-21 (Salk insertion line Salk_109115) mutant seeds carrying a T-DNA insertion disrupting the GA1 gene (accession AT4G02780) encoding ent-copalyl diphosphate synthase, which is essential for GA biosynthesis14 (SALK_109115 insertion line in GA1 (Col ecotype) is referenced in the SALK SIGnAL database15 (http://signal.salk.edu/).
Consistent with previous results13, ga1-21 mutant seeds could not germinate 80 hr after seed imbibition (Figure 3, Panel 1). In contrast and further consistent with previous results13, seed coat removal triggered ga1-21 embryo growth (Figure 3, panel 2) unless ABA was present in the medium (Figure 3, panel 3).
Panel 4 in Figure 3 was taken 80 hr after seed imbibition and shows a SCBA with ga1-21 embryos on a bed of 80 ga1-21 seed coats. Consistent with our hypothesis, ga1-21 mutant seed coats blocked the growth of ga1-21 embryos.
Previous work showed that in dormant seeds the seed coat synthesizes and releases ABA towards the embryo as a result of constitutive expression of the GA-response DELLA factor RGL2, which promotes ABA accumulation in imbibed seeds6,11. Under conditions where GA synthesis is blocked the DELLA factor RGL2 constitutively accumulates and promotes ABA accumulation11. Absence of RGL2 and ABA synthesis in ga1-21 seed coats is therefore expected to permit the growth of ga1-21 embryos in a SCBA. To test this hypothesis we used the aba2-1 mutant allele (Col ecotype), deficient in ABA synthesis as a result of a point mutation in the gene encoding Xanthosin dehydrogenase (AT1G52340) and the rgl2-14 T-DNA insertion line (Salk_027654, Col ecotype)16.
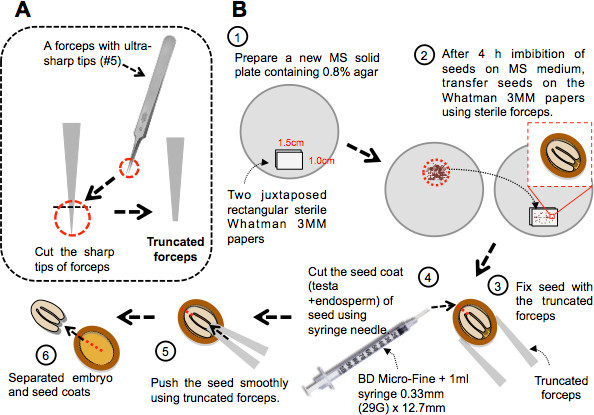 Figure 1. Procedure for seed dissection. A) Diagram describing how to make truncated forceps for seed dissection. Forceps can be truncated using scissors or pincers. When the forceps is closed the juxtaposed truncated tips should cover a surface about the size of a seed. B) Step 1: two side by side rectangular sterile Whatman 3MM papers are laid on a Petri dish plate containing MS solid medium. Step 2: seeds are transferred on the surface of the Whatman papers 4 hr after imbibition. Step 3: the dissection procedure requires holding the seed against the Whatman paper using the truncated forceps. Step 4: A syringe needle is used to cut the seed coat as shown. Steps 5 and 6: The juxtaposed tips of the closed forceps are used to gently push the embryo out of the seed coat through the site of the cut.
Figure 1. Procedure for seed dissection. A) Diagram describing how to make truncated forceps for seed dissection. Forceps can be truncated using scissors or pincers. When the forceps is closed the juxtaposed truncated tips should cover a surface about the size of a seed. B) Step 1: two side by side rectangular sterile Whatman 3MM papers are laid on a Petri dish plate containing MS solid medium. Step 2: seeds are transferred on the surface of the Whatman papers 4 hr after imbibition. Step 3: the dissection procedure requires holding the seed against the Whatman paper using the truncated forceps. Step 4: A syringe needle is used to cut the seed coat as shown. Steps 5 and 6: The juxtaposed tips of the closed forceps are used to gently push the embryo out of the seed coat through the site of the cut.
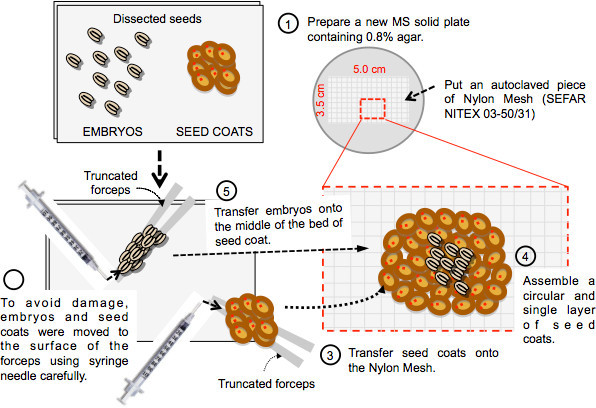 Figure 2. Procedure for assembly of the seed coat bedding assay. Step 1: A rectangular piece of Nylon mesh is laid on a Petri dish plate containing MS solid medium. Step 2 and 3: Dissected embryos and seed coats are gently moved to the edges of the forceps using the syringe needle prior to transfer them onto the Nylon mesh. Step 4 and 5: Assemble a circular and single layer of seed coats and dispose the embryos as a circular single layer on the center of the assembled seed coat bed.
Figure 2. Procedure for assembly of the seed coat bedding assay. Step 1: A rectangular piece of Nylon mesh is laid on a Petri dish plate containing MS solid medium. Step 2 and 3: Dissected embryos and seed coats are gently moved to the edges of the forceps using the syringe needle prior to transfer them onto the Nylon mesh. Step 4 and 5: Assemble a circular and single layer of seed coats and dispose the embryos as a circular single layer on the center of the assembled seed coat bed.
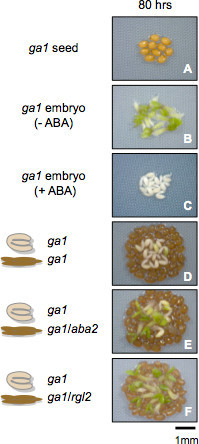 Figure 3. Representative results with seed material unable to synthesize GA. Seed coat bedding assays using seed coats and embryos dissected from ga1 (ga1-21) mutant seeds, unable to synthesize GA. Panels show plant material as described in the figure 80 hr after seed imbibition.
Figure 3. Representative results with seed material unable to synthesize GA. Seed coat bedding assays using seed coats and embryos dissected from ga1 (ga1-21) mutant seeds, unable to synthesize GA. Panels show plant material as described in the figure 80 hr after seed imbibition.
Discussion
The seed coat bedding assay (SCBA) procedure described here is in principle applicable to any circumstance where Arabidopsis seed germination is blocked (or delayed) and where the endosperm is suspected to implement this arrest. The latter can be evidenced by removing the seed coat (testa and endosperm) and observing that embryonic growth proceeds faster relative to that observed when embryos are surrounded by the seed coat. Germination may be blocked in response to particular environmental physical parameters (e.g. water potential or light quality) or in specific genetic backgrounds (e.g. absence of GA synthesis as in ga1 mutants, see Figure 3).
There are no general rules concerning the number of seed coats and embryos to be used for a SCBA. This depends on the seed batch (and in particular its freshness) and the type of experiment to be performed. It is therefore critical to adjust the amount of dissected material for each particular case by monitoring the growth on the embryos on the bed of seed coats on a daily basis. However, usually few seed coats are necessary when highly dormant Arabidopsis ecotypes are used. Shown below are typical amounts of seed coats and embryos that were used successfully when assembling SCBAs:
SCBA with dormant seed material6: * Freshly harvested (i.e. dormant) Cape Verde Island ecotype (Cvi): Use 10 embryos and 20 seed coats.
* Freshly harvested Landsberg (Ler) seeds: Use 10 embryos and 80 seed coats.
SCBA in absence of gibberellin (GA) biosynthesis (Figure 3): * ga1 mutant seeds (Columbia - Col - ecotype): Use 10 embryos and 80 seed coats.
* Wild type Col seeds with 10 μM paclobutrazol (an inhibitor of GA biosynthesis) in the germination medium: Use 10 embryos and 100 seed coats. Add Paclobutrazol when the medium is cooled down to 50 °C before pouring into the Petri dish.
SCBA after a pulse of far red (FR) light9: * Use 12 embryos and 100 seed coats.
The only limitation of the SCBA technique is that it requires patience and dexterity so as to rapidly assemble the dissected seed materials while not killing the embryos. An experienced researcher can dissect 100 seeds within 20-30 min. However, less experienced users may need 1 hr and expect embryonic damage. To our knowledge there are so far no alternative techniques to explore the role of the endosperm for the control of seed germination.
Disclosures
The authors declare that they have no competing financial interests.
Acknowledgments
This work was supported by grants from the Swiss National Science Foundation and by the State of Geneva.
References
- Debeaujon I, Leon-Kloosterziel KM, Koornneef M. Influence of the testa on seed dormancy, germination, and longevity in Arabidopsis. Plant Physiol. 2000;122:403–414. doi: 10.1104/pp.122.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Spillane C, Grossniklaus U. Evolutionary origins of the endosperm in flowering plants. Genome Biol. 2002;3:reviews1026. doi: 10.1186/gb-2002-3-9-reviews1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Lepiniec L, Pourcel L, Routaboul J-M. Seed development, dormancy and germination. Blackwell Publishing; 2007. pp. 25–43. [Google Scholar]
- Finch-Savage WE, Leubner-Metzger G. Seed dormancy and the control of germination. New Phytol. 2006;171:501–523. doi: 10.1111/j.1469-8137.2006.01787.x. [DOI] [PubMed] [Google Scholar]
- Bethke PC, et al. The Arabidopsis aleurone layer responds to nitric oxide, gibberellin, and abscisic acid and is sufficient and necessary for seed dormancy. Plant Physiol. 2007;143:1173–1188. doi: 10.1104/pp.106.093435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Tureckova V, Strnad M, Lopez-Molina L. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proc. Natl. Acad. Sci. U.S.A. 2010;107:19108–19113. doi: 10.1073/pnas.1012896107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JW, Nagatani A, Elich TD, Fagan M, Chory J. Phytochrome A and Phytochrome B Have Overlapping but Distinct Functions in Arabidopsis Development. Plant Physiol. 1994;104:1139–1149. doi: 10.1104/pp.104.4.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinomura T, et al. Action spectra for phytochrome A- and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 1996;93:8129–8133. doi: 10.1073/pnas.93.15.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, et al. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012;26:1984–1996. doi: 10.1101/gad.194266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KP, Lopez-Molina L. Control of seed germination in the shade. Cell Cycle. 2012;11:4489–4490. doi: 10.4161/cc.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, et al. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell. 2008;20:2729–2745. doi: 10.1105/tpc.108.061515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Tureckova V, Lacombe E, Lopez-Molina L. Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 2009;28:2259–2271. doi: 10.1038/emboj.2009.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debeaujon I, Koornneef M. Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 2000;122:415–424. doi: 10.1104/pp.122.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Rook F, et al. Impaired sucrose-induction mutants reveal the modulation of sugar-induced starch biosynthetic gene expression by abscisic acid signalling. Plant J. 2001;26:421–433. doi: 10.1046/j.1365-313x.2001.2641043.x. [DOI] [PubMed] [Google Scholar]


