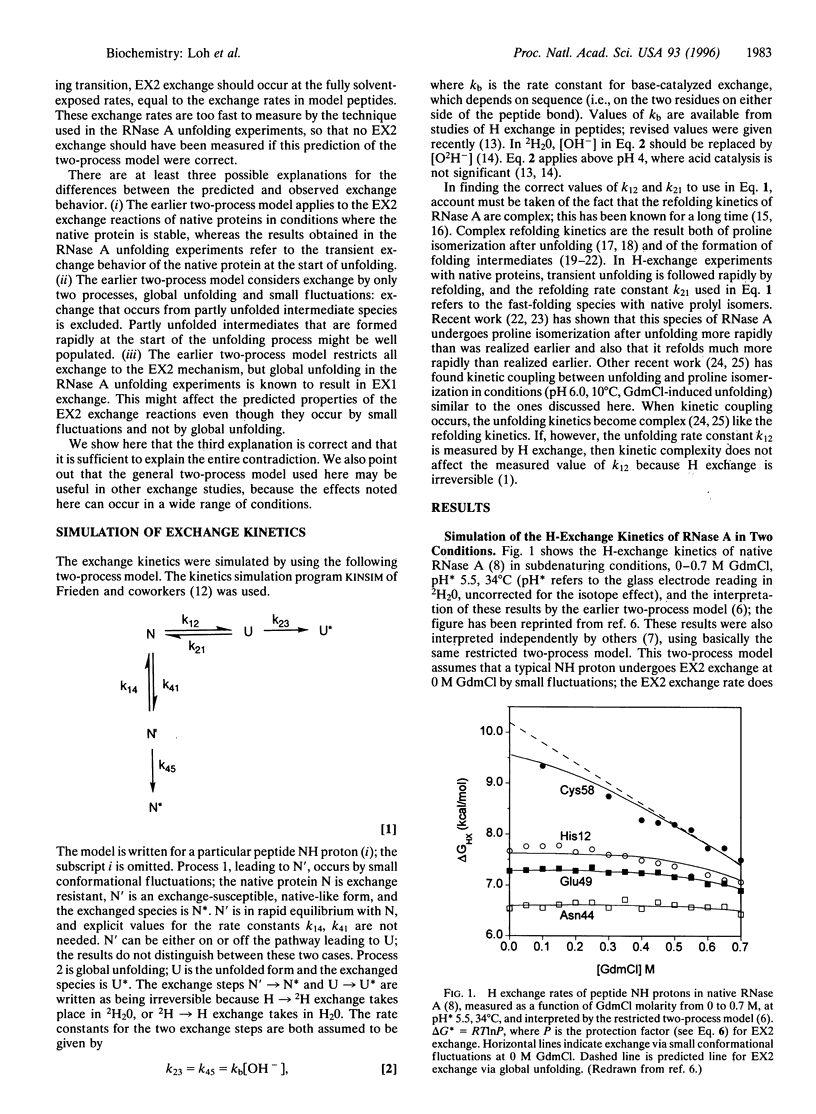
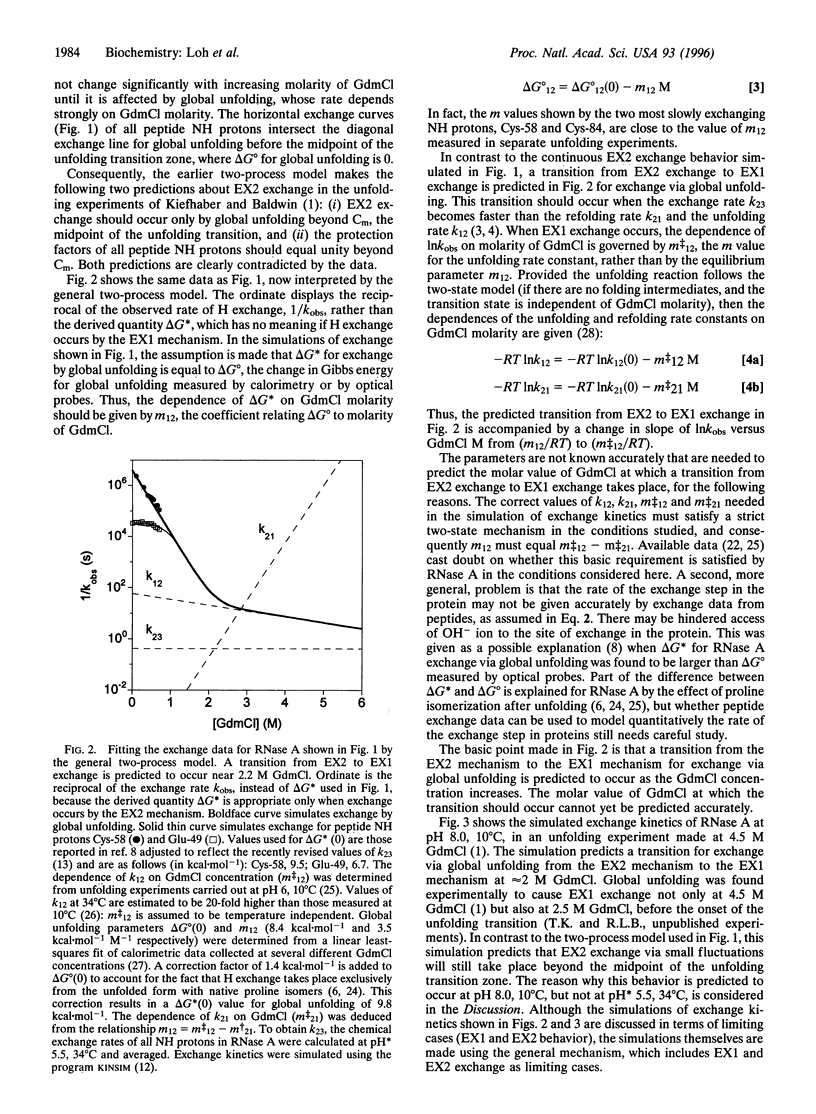
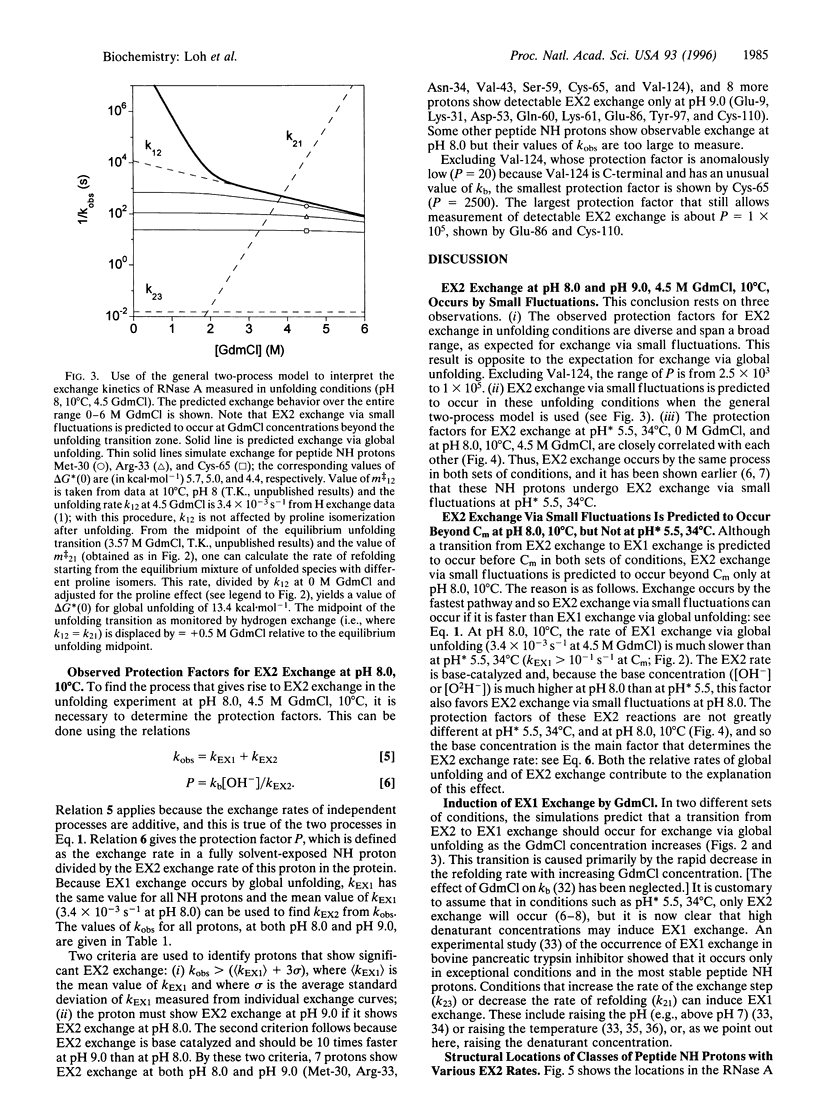
Abstract
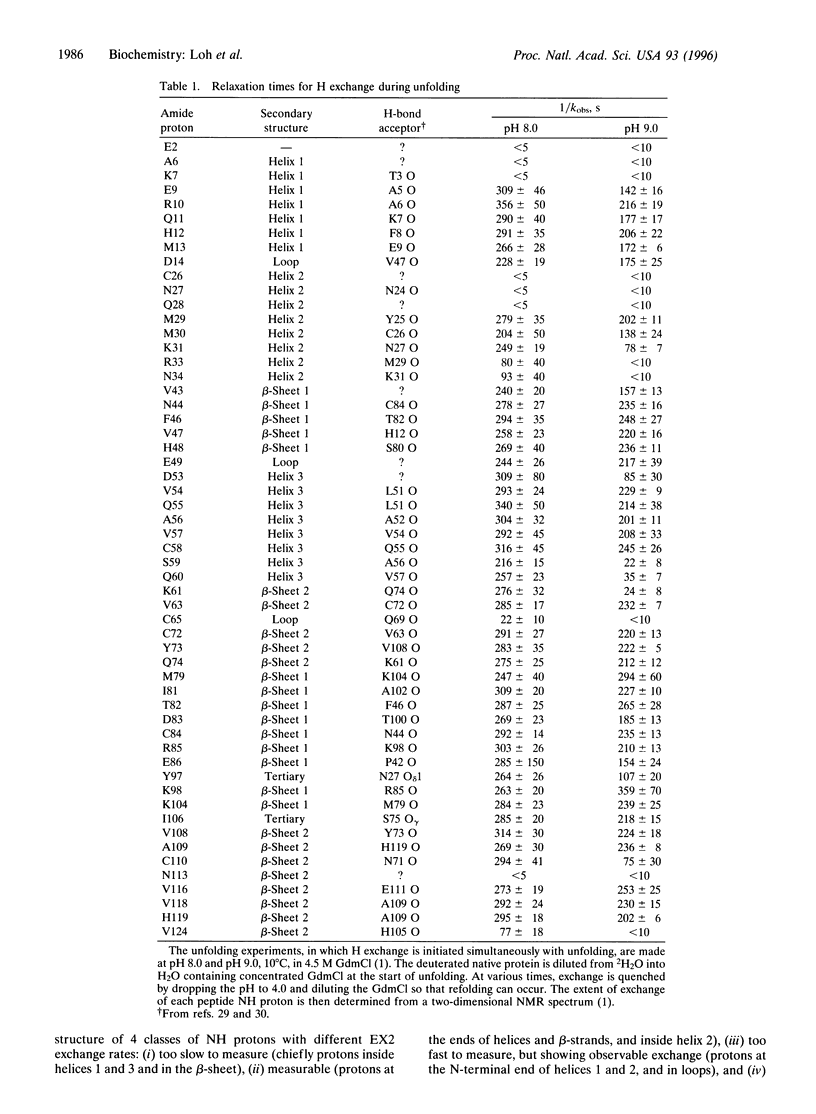
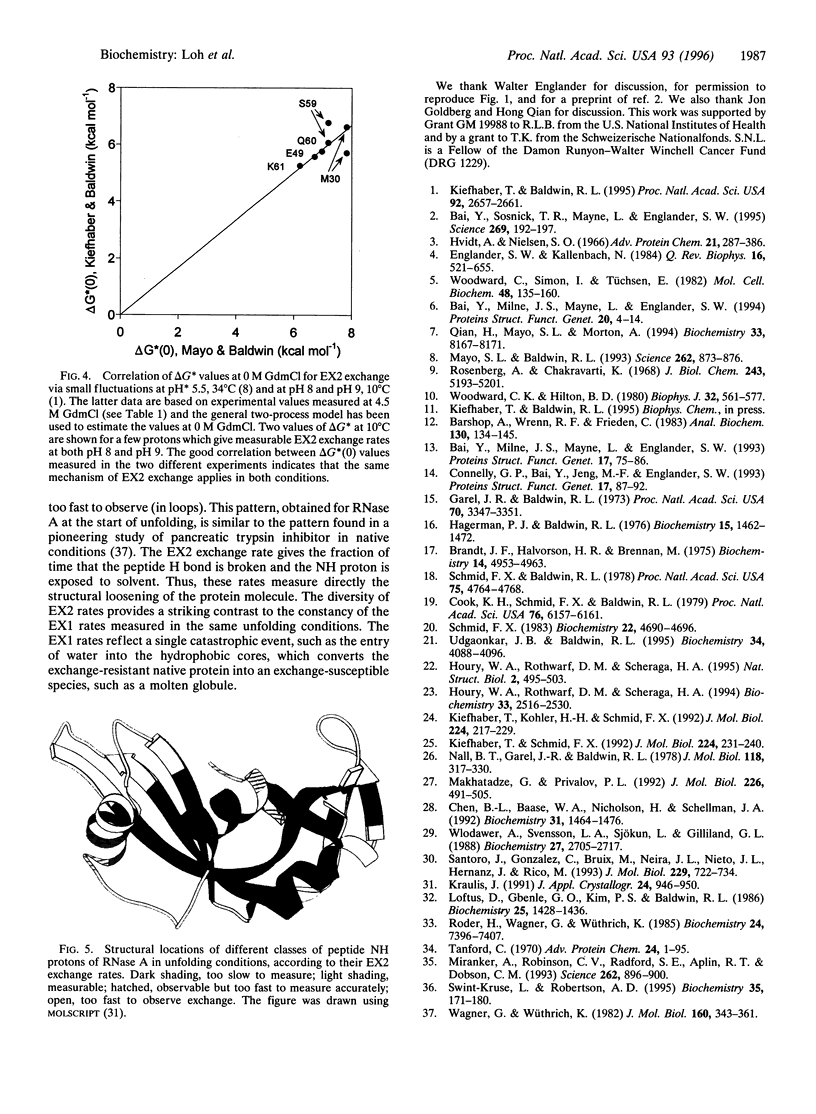
When NMR hydrogen exchange was used previously to monitor the kinetics of RNase A unfolding, some peptide NH protons were found to show EX2 exchange (detected by base catalysis) in addition to the expected EX1 exchange, whose rate is limited by the kinetic unfolding process. In earlier work, two groups showed independently that a restricted two-process model successfully fits published hydrogen exchange rates of native RNase A in the range 0-0.7 M guanidinium chloride. We find that this model predicts properties that are very different from the observed properties of the EX2 exchange reactions of RNase A in conditions where guanidine-induced unfolding takes place. The model predicts that EX2 exchange should be too fast to measure by the technique used, whereas it is readily measurable. Possible explanations for the contradiction are considered here, and we show that removing the restriction from the earlier two-process model is sufficient to resolve the contradiction; instead of specifying that exchange caused by global unfolding occurs by the EX2 mechanism, we allow it to occur by the general mechanism, which includes both the EX1 and EX2 cases. It is logical to remove this restriction because global unfolding of RNase A is known to give rise to EX1 exchange in these unfolding conditions. Resolving the contradiction makes it possible to determine whether populated unfolding intermediates contribute to the EX2 exchange, and this question is considered elsewhere. The results and simulations indicate that moderate or high denaturant concentrations readily give rise to EX1 exchange in native proteins. Earlier studies showed that hydrogen exchange in native proteins typically occurs by the EX2 mechanism but that high temperatures or pH values above 7 may give rise to EX1 exchange. High denaturant concentrations should be added to the list of variables likely to cause EX1 exchange.
Full text
PDF
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bai Y., Milne J. S., Mayne L., Englander S. W. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993 Sep;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Milne J. S., Mayne L., Englander S. W. Protein stability parameters measured by hydrogen exchange. Proteins. 1994 Sep;20(1):4–14. doi: 10.1002/prot.340200103. [DOI] [PubMed] [Google Scholar]
- Bai Y., Sosnick T. R., Mayne L., Englander S. W. Protein folding intermediates: native-state hydrogen exchange. Science. 1995 Jul 14;269(5221):192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barshop B. A., Wrenn R. F., Frieden C. Analysis of numerical methods for computer simulation of kinetic processes: development of KINSIM--a flexible, portable system. Anal Biochem. 1983 Apr 1;130(1):134–145. doi: 10.1016/0003-2697(83)90660-7. [DOI] [PubMed] [Google Scholar]
- Brandts J. F., Halvorson H. R., Brennan M. Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry. 1975 Nov 4;14(22):4953–4963. doi: 10.1021/bi00693a026. [DOI] [PubMed] [Google Scholar]
- Chen B. L., Baase W. A., Nicholson H., Schellman J. A. Folding kinetics of T4 lysozyme and nine mutants at 12 degrees C. Biochemistry. 1992 Feb 11;31(5):1464–1476. doi: 10.1021/bi00120a025. [DOI] [PubMed] [Google Scholar]
- Connelly G. P., Bai Y., Jeng M. F., Englander S. W. Isotope effects in peptide group hydrogen exchange. Proteins. 1993 Sep;17(1):87–92. doi: 10.1002/prot.340170111. [DOI] [PubMed] [Google Scholar]
- Cook K. H., Schmid F. X., Baldwin R. L. Role of proline isomerization in folding of ribonuclease A at low temperatures. Proc Natl Acad Sci U S A. 1979 Dec;76(12):6157–6161. doi: 10.1073/pnas.76.12.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englander S. W., Kallenbach N. R. Hydrogen exchange and structural dynamics of proteins and nucleic acids. Q Rev Biophys. 1983 Nov;16(4):521–655. doi: 10.1017/s0033583500005217. [DOI] [PubMed] [Google Scholar]
- Garel J. R., Baldwin R. L. Both the fast and slow refolding reactions of ribonuclease A yield native enzyme. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3347–3351. doi: 10.1073/pnas.70.12.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J., Baldwin R. L. A quantitative treatment of the kinetics of the folding transition of ribonuclease A. Biochemistry. 1976 Apr 6;15(7):1462–1473. doi: 10.1021/bi00652a017. [DOI] [PubMed] [Google Scholar]
- Houry W. A., Rothwarf D. M., Scheraga H. A. A very fast phase in the refolding of disulfide-intact ribonuclease A: implications for the refolding and unfolding pathways. Biochemistry. 1994 Mar 8;33(9):2516–2530. doi: 10.1021/bi00175a022. [DOI] [PubMed] [Google Scholar]
- Houry W. A., Rothwarf D. M., Scheraga H. A. The nature of the initial step in the conformational folding of disulphide-intact ribonuclease A. Nat Struct Biol. 1995 Jun;2(6):495–503. doi: 10.1038/nsb0695-495. [DOI] [PubMed] [Google Scholar]
- Hvidt A., Nielsen S. O. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Baldwin R. L. Kinetics of hydrogen bond breakage in the process of unfolding of ribonuclease A measured by pulsed hydrogen exchange. Proc Natl Acad Sci U S A. 1995 Mar 28;92(7):2657–2661. doi: 10.1073/pnas.92.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefhaber T., Kohler H. H., Schmid F. X. Kinetic coupling between protein folding and prolyl isomerization. I. Theoretical models. J Mol Biol. 1992 Mar 5;224(1):217–229. doi: 10.1016/0022-2836(92)90585-8. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Schmid F. X. Kinetic coupling between protein folding and prolyl isomerization. II. Folding of ribonuclease A and ribonuclease T1. J Mol Biol. 1992 Mar 5;224(1):231–240. doi: 10.1016/0022-2836(92)90586-9. [DOI] [PubMed] [Google Scholar]
- Loftus D., Gbenle G. O., Kim P. S., Baldwin R. L. Effects of denaturants on amide proton exchange rates: a test for structure in protein fragments and folding intermediates. Biochemistry. 1986 Mar 25;25(6):1428–1436. doi: 10.1021/bi00354a036. [DOI] [PubMed] [Google Scholar]
- Makhatadze G. I., Privalov P. L. Protein interactions with urea and guanidinium chloride. A calorimetric study. J Mol Biol. 1992 Jul 20;226(2):491–505. doi: 10.1016/0022-2836(92)90963-k. [DOI] [PubMed] [Google Scholar]
- Mayo S. L., Baldwin R. L. Guanidinium chloride induction of partial unfolding in amide proton exchange in RNase A. Science. 1993 Nov 5;262(5135):873–876. doi: 10.1126/science.8235609. [DOI] [PubMed] [Google Scholar]
- Miranker A., Robinson C. V., Radford S. E., Aplin R. T., Dobson C. M. Detection of transient protein folding populations by mass spectrometry. Science. 1993 Nov 5;262(5135):896–900. doi: 10.1126/science.8235611. [DOI] [PubMed] [Google Scholar]
- Nall B. T., Garel J. R., Baldwin R. L. Test of the extended two-state model for the kinetic intermediates observed in the folding transition of ribonuclease A. J Mol Biol. 1978 Jan 25;118(3):317–330. doi: 10.1016/0022-2836(78)90231-0. [DOI] [PubMed] [Google Scholar]
- Qian H., Mayo S. L., Morton A. Protein hydrogen exchange in denaturant: quantitative analysis by a two-process model. Biochemistry. 1994 Jul 12;33(27):8167–8171. doi: 10.1021/bi00193a001. [DOI] [PubMed] [Google Scholar]
- Roder H., Wagner G., Wüthrich K. Amide proton exchange in proteins by EX1 kinetics: studies of the basic pancreatic trypsin inhibitor at variable p2H and temperature. Biochemistry. 1985 Dec 3;24(25):7396–7407. doi: 10.1021/bi00346a055. [DOI] [PubMed] [Google Scholar]
- Rosenberg A., Chakravarti K. Studies of hydrogen exchange in proteins. I. The exchange kinetics of bovine carbonic anhydrase. J Biol Chem. 1968 Oct 10;243(19):5193–5201. [PubMed] [Google Scholar]
- Santoro J., González C., Bruix M., Neira J. L., Nieto J. L., Herranz J., Rico M. High-resolution three-dimensional structure of ribonuclease A in solution by nuclear magnetic resonance spectroscopy. J Mol Biol. 1993 Feb 5;229(3):722–734. doi: 10.1006/jmbi.1993.1075. [DOI] [PubMed] [Google Scholar]
- Schmid F. X., Baldwin R. L. Acid catalysis of the formation of the slow-folding species of RNase A: evidence that the reaction is proline isomerization. Proc Natl Acad Sci U S A. 1978 Oct;75(10):4764–4768. doi: 10.1073/pnas.75.10.4764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid F. X. Mechanism of folding of ribonuclease A. Slow refolding is a sequential reaction via structural intermediates. Biochemistry. 1983 Sep 27;22(20):4690–4696. doi: 10.1021/bi00289a013. [DOI] [PubMed] [Google Scholar]
- Swint-Kruse L., Robertson A. D. Temperature and pH dependences of hydrogen exchange and global stability for ovomucoid third domain. Biochemistry. 1996 Jan 9;35(1):171–180. doi: 10.1021/bi9517603. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. C. Theoretical models for the mechanism of denaturation. Adv Protein Chem. 1970;24:1–95. [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. Nature of the early folding intermediate of ribonuclease A. Biochemistry. 1995 Mar 28;34(12):4088–4096. doi: 10.1021/bi00012a027. [DOI] [PubMed] [Google Scholar]
- Wagner G., Wüthrich K. Amide protein exchange and surface conformation of the basic pancreatic trypsin inhibitor in solution. Studies with two-dimensional nuclear magnetic resonance. J Mol Biol. 1982 Sep 15;160(2):343–361. doi: 10.1016/0022-2836(82)90180-2. [DOI] [PubMed] [Google Scholar]
- Wlodawer A., Svensson L. A., Sjölin L., Gilliland G. L. Structure of phosphate-free ribonuclease A refined at 1.26 A. Biochemistry. 1988 Apr 19;27(8):2705–2717. doi: 10.1021/bi00408a010. [DOI] [PubMed] [Google Scholar]
- Woodward C. K., Hilton B. D. Hydrogen isotope exchange kinetics of single protons in bovine pancreatic trypsin inhibitor. Biophys J. 1980 Oct;32(1):561–575. doi: 10.1016/S0006-3495(80)84990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C., Simon I., Tüchsen E. Hydrogen exchange and the dynamic structure of proteins. Mol Cell Biochem. 1982 Oct 29;48(3):135–160. doi: 10.1007/BF00421225. [DOI] [PubMed] [Google Scholar]