Abstract
Epigenetic modifications of stem cell genome including DNA methylation and histone modifications are critical for the regulation of stem cell gene expression and maintenance of stem cell pool and their differentiation. Although the importance of epigenetic modifications specifically DNA methylation to adult hematopoietic stem cells (HSC) has been established, the identity of specific modulators and precise mechanism of integration of methylation events remain to be uncovered. In this issue, Shuai and colleagues identify the SUMO E3 ligase PIAS1 (protein inhibitor of activated STAT1) as a key regulator of DNA methylation of HSC required for their maintenance and lineage commitment (Liu et al, 2014).
See also: B Liu et al (December 2013)
The methylation of cytosine residues in the CpG dinucleotide is essential for mammalian development and maintenance of genomic integrity. DNA methylation has deep influence on gene expression involved in key physiological processes, including the silencing of germline-specific genes and of transposable elements. Patterns of DNA methylation are established during early embryogenesis and gametogenesis and subsequently maintained in somatic cells where DNA methylation is dynamically modulated during cell differentiation (Smith & Meissner, 2013). These patterns are tissue-specific and perturbed in several human diseases including cancer and disorders of imprinting. Dnmt3a and Dnmt3b are de novo methyltransferases that catalyze remethylation to establish methylation patterns in post-implantation mouse embryos and in germ cells while DNMT1 is responsible for maintenance of methylation after cell division, although the view that there is a clear dichotomy between the functions of the two types of enzymes may be changing (Jones & Liang, 2009).
DNA methylation is critical for the maintenance of hematopoietic stem cells and for their lineage commitment and stepwise differentiation (Broske et al, 2009; Challen et al, 2012; Tadokoro et al, 2007; Trowbridge et al, 2009). Specifically, DNMT1 is essential for hematopoietic stem cell self-renewal and homeostasis as well as for myeloid versus lymphoid fate choice (Broske et al, 2009; Trowbridge et al, 2009). While DNMT3a or DNMT3b were initially thought to have no effect on HSC (Tadokoro et al, 2007), their combined function appeared critical for HSC maintenance but not HSC differentiation. Subsequently, DNMT3a was found to be essential for HSC differentiation (Challen et al, 2012). With the importance of DNA methylation for the regulation of transcriptional programs in hematopoietic stem cell maintenance and lineage commitment being established (Broske et al, 2009; Challen et al, 2012; Tadokoro et al, 2007; Trowbridge et al, 2009), the molecular players that control DNA methylation and precise steps that integrate DNA methylation events remain poorly defined. One of the few recently identified components is SATB1 (special AT-rich sequence-binding protein 1), a global chromatin remodeler essential for HSC DNA methylation, self-renewal and repression of lineage commitment (Will et al, 2013).
In this issue of The EMBO Journal, Shuai and colleagues identify PIAS1, a SUMO E3 ligase as a new and essential piece of the DNA methylation scheme in hematopoietic stem cells (Liu et al, 2014). The authors had previously shown that PIAS1 is an epigenetic modifier critical for T-cell differentiation (Liu et al, 2010). PIAS1 restricts the differentiation of natural regulatory T (reg) cells by recruiting DNMT3a and DNMT3b to Foxp3 promoter and maintaining Foxp3 promoter at a repressive chromatin state.
Here they show that PIAS1-/- mice also present global defects in hematopoiesis that go beyond their previous phenotypic characterization (Liu et al, 2010). They find that while there are only minor perturbations in PIAS1-deficient circulating and bone marrow lineage-restricted cells, the numbers of lineage committed common lymphoid progenitors (CLP)s and megakaryocyte-erythroid progenitors (MEP)s are significantly altered. These abnormalities are associated with an increase in the long-term hematopoietic stem cell (LT-HSC) population. To further delineate their finding, they analyze the cell cycle and cell death in hematopoietic stem and progenitor cell populations. These analyses reveal that cell death is the major mediator of impaired CLP and B-cell production in PIAS1-deficient bone marrow indicating that PIAS1 is crucial for cell survival in lymphoid cells. They further elucidate PIAS1 as major contributor to the dormancy of LT-HSC. Loss of PIAS1 profoundly compromises competitive multi-lineage repopulation capability of HSC and their self-renewing potential. Further to this role in HSC-maintenance, PIAS1 is required for HSC differentiation as illustrated by compromised B-cell generation and differentiation at multiple stages as well as defective T-cell generation in PIAS1-deficient mice. It turns out that PIAS1 is critical to balance HSC differentiation of either myeloid or lymphoid cells since myelo-erythroid-specific genes are found highly reactivated in LT-HSC and in CLP lymphoid progenitors that lack PIAS1.
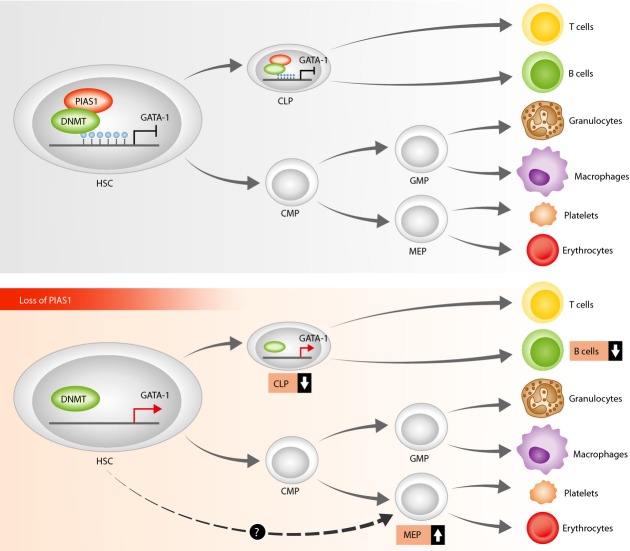
Among the highly perturbed genes, increased expression of megakaryocyte-erythroid factor GATA-1 and its downstream targets in CLP lymphoid progenitors is notable. The data are in agreement with a model in which PIAS1 is required for the suppression of myelo-erythroid genes in LT-HSC and in lymphoid progenitors and thus for proper expression of lineage-specific genes in these populations. Interestingly, the authors show that PIAS1 directly occupies GATA-1 promoter in primitive multipotential hematopoietic cells in vivo. In addition they provide evidence supporting a function for PIAS1 in mediating methylation of GATA-1 promoter as bisulfite sequencing of CpG sites are hypermethylated in wild-type but not in PIAS1-deficient LT-HSC and CLP lymphoid progenitor cells. GATA-1 promoter methylation may be mediated by DNMT3a, since DNMT3a is found at GATA-1 promoters in vivo in bone marrow cells (Fig 1). The presented data suggest that PIAS1 directly recruits DNMT3a to the GATA-1 promoter as endogenous PIAS1 and DNMT3a interact in bone marrow cells and binding of DNMT3a to GATA1 promoter is compromised in PIAS1-deficient tissue. These data are intriguing as the hematopoietic phenotype of PIAS1-deficient mice resembles more closely DNMT1- rather than DNMT3a-deficient mice (Challen et al, 2012; Trowbridge et al, 2009). In particular, the enrichment in LT-HSC and the significant decrease in CLP observed in PIAS1-deficient mice are highly reminiscent of DNMT1-deficient mice that exhibit similar but more pronounced hematopoietic defects. In contrast to DNMT1-deficient mice (Trowbridge et al, 2009), PIAS1-deficient mice did not exhibit any abnormalities within the microenvironment, overall suggesting that the observed defects are intrinsic and/or autonomous to HSC. It remains to be seen whether PIAS1 shows any interactions with DNMT1 in addition to DNMT3a during HSC differentiation. In addition it will be very interesting to see whether the enzymatic activity of PIAS1 contributes to these interactions.
Figure 1.
Model: PIAS1 controls hematopoietic stem cell homeostasis by mediating methylation of GATA-1 promoter.
Upper panel: by recruiting the DNMT3a methyltransferase to GATA-1 promoter in HSC (hematopoietic stem cells) and in common lymphoid progenitors (CLP), PIAS1 participates in silencing myelo-erythroid genes in these cells. Lower panel: Loss of PIAS1 results in decreased HSC quiescence and self-renewal, reduced generation of lymphoid progenitors, reduced B-cell production and increased production of MEP (megakaryocyte-erythroid progenitors), associated with aberrant expression of myelo-erythroid and B cell-specific genes in HSC and lymphoid progenitors. Future investigations including elucidation of steps leading to the increased MEP from PIAS-/- HSC (dashed arrow) will contribute to delineating HSC path to lineage determination. CMP: common myeloid progenitor.
In light of previous implications for regulated DNA methylation in the context of hematological malignancies and leukemias/myelodysplasias in particular (Broske et al, 2009; Ley et al, 2010; Yamashita et al, 2010; Yan et al, 2011), these novel data raise the possibility that alterations in expression and/or abnormal function of PIAS1 may be associated with hematological malignancies.
In summary, the new findings by Liu et al identify PIAS1 as a critical regulator of epigenetic silencing of myelo-erythroid genes in HSC and in lymphoid progenitors. By a mechanism that may involve the recruitment of DNMTs to particular promoters, PIAS1 participates in the intricate balance between HSC maintenance and subsequent lineage commitment.
Conflict of interest
The author declares that she has no conflict of interest.
References
- Broske AM, Vockentanz L, Kharazi S, Huska MR, Mancini E, Scheller M, Kuhl C, Enns A, Prinz M, Jaenisch R, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid 147 restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Tahk S, Yee KM, Fan G, Shuai K. The ligase PIAS1 restricts natural regulatory T cell differentiation by epigenetic repression. Science. 2010;330:521–525. doi: 10.1126/science.1193787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Yee K, Tahk S, Mackie R, Hsu C, Shuai K. PIAS1 SUMO ligase regulates the self-renewal and differentiation of hematopoietic stem cells. EMBO J. 2014;33:101–113. doi: 10.1002/embj.201283326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will B, Vogler TO, Bartholdy B, Garrett-Bakelman F, Mayer J, Barreyro L, Pandolfi A, Todorova TI, Okoye-Okafor UC, Stanley RF, et al. Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nat Immunol. 2013;14:437–445. doi: 10.1038/ni.2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita Y, Yuan J, Suetake I, Suzuki H, Ishikawa Y, Choi YL, Ueno T, Soda M, Hamada T, Haruta H, et al. Array-based genomic resequencing of human leukemia. Oncogene. 2010;29:3723–3731. doi: 10.1038/onc.2010.117. [DOI] [PubMed] [Google Scholar]
- Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]