Abstract
Following translation termination, ribosomal subunits dissociate to become available for subsequent rounds of protein synthesis. In many translation-inhibiting stress conditions, e.g. glucose starvation in yeast, free ribosomal subunits reassociate to form a large pool of non-translating 80S ribosomes stabilized by the ‘clamping’ Stm1 factor. The subunits of these inactive ribosomes need to be mobilized for translation restart upon stress relief. The Dom34-Hbs1 complex, together with the Rli1 NTPase (also known as ABCE1), have been shown to split ribosomes stuck on mRNAs in the context of RNA quality control mechanisms. Here, using in vitro and in vivo methods, we report a new role for the Dom34-Hbs1 complex and Rli1 in dissociating inactive ribosomes, thereby facilitating translation restart in yeast recovering from glucose starvation stress. Interestingly, we found that this new role is not restricted to stress conditions, indicating that in growing yeast there is a dynamic pool of inactive ribosomes that needs to be split by Dom34-Hbs1 and Rli1 to participate in protein synthesis. We propose that this provides a new level of translation regulation.
Keywords: glucose-starvation, ribosome recycling, Rli1, Stm1, translation initiation
Introduction
The production of proteins by ribosomes can be divided into four stages that together form the translation cycle: initiation, elongation, termination and recycling (Krebs et al, 2011). Eukaryotic translation initiation requires separate 40S and 60S ribosomal subunits, which assemble on the initiation codons of messenger RNAs (mRNA) to form the actively translating 80S ribosome. Ribosomal subunits available for initiation result from ribosome recycling, which occurs after, and is tightly coupled to, translation termination. Termination and recycling are triggered when a translating ribosome encounters a termination codon. At this point, the termination factors eRF1 and eRF3, together with the NTPase Rli1 (also known as ABCE1), catalyze peptide release and subsequent ribosome dissociation (Pisarev et al, 2010; Shoemaker & Green, 2011).
Many stress conditions cause a global shut-down of translation, allowing cells to economically use limited metabolic resources to only produce proteins important for adaptation to the changing environment. Ribosomal subunits that are released through recycling may not engage in new rounds of protein synthesis, but instead associate to form a large pool of non-translating, inactive ribosomes (Nielsen et al, 1981; Tzamarias et al, 1989; Ashe et al, 2000, 2001; Montero-Lomeli et al, 2002; Uesono & Toh, 2002; Krokowski et al, 2011). Formation of these inactive ribosomes may protect ribosomal subunits from damage and/or degradation. Moreover, upon stress relief, these inactive ribosomes can be easily and economically mobilized without a requirement for ribosome biogenesis. Upon prolonged stress, ribosomes may eventually be degraded by ribophagy to provide cells with energy and nutrients (Kraft et al, 2008).
In bacteria, it is well described that stress-induced factors bind to ‘hibernating’ ribosomes and induce the formation of ribosome dimers (70S + 70S). The binding sites of the stress-induced factors overlap with those of mRNA and transfer RNA (tRNA) thus inhibiting normal ribosome activities (Polikanov et al, 2012).
Eukaryotic hibernating ribosomes may in some organisms also form dimers (Krokowski et al, 2011), but mostly accumulate as inactive 80S monomers. This was shown for example to occur in mammalian cells upon serum depletion (Nielsen et al, 1981), in yeast and mammalian cells after amino acid shortage (Tzamarias et al, 1989; Krokowski et al, 2011), and in yeast during osmotic stress (Uesono & Toh, 2002), lithium-induced stress (Montero-Lomeli et al, 2002) and after exposure to fusel alcohols (Ashe et al, 2001). The most detailed example stems probably from the analysis of glucose starvation in the yeast Saccharomyces cerevisiae. This condition leads to the accumulation of 80S ribosomes (Ashe et al, 2000) that contain the protein Stm1 in a conformation that clamps the ribosomal subunits together (Ben-Shem et al, 2011). This structure is incompatible with translation as Stm1 occupies part of the mRNA channel. Consistent with this structural observation, Stm1 was identified as a ribosome-binding factor (Inada et al, 2002; Van Dyke et al, 2006) and has translation-inhibiting activity in vitro (Balagopal & Parker, 2011). In addition, Stm1 was shown to enhance recovery following nutritional stress (Van Dyke et al, 2006), having a positive effect on the number of ribosomes preserved during nutrient deprivation (Van Dyke et al, 2013).
Mobilization of inactive ribosomes, which allows a rapid restart of translation upon stress relief (Ashe et al, 2000) requires dissociation, making their subunits available for initiation. We wondered whether this process, in analogy to ribosome recycling after termination, depends on recycling factor activity.
In addition to normal termination and recycling factors that are thought to function on stop codons, recent studies in yeast and mammalian systems identified Dom34 (Pelota in humans) and the GTPase Hbs1, forming a complex structurally similar to eRF1 and eRF3 (Chen et al, 2010; van den Elzen et al, 2010; Kobayashi et al, 2010), that together with Rli1 similarly promote subunit dissociation. Interestingly, however, these factors appear to function on mRNA-bound ribosomes in a codon-independent manner (Shoemaker et al, 2010; Shoemaker & Green, 2011) or to promote subunit splitting on completely empty ribosomes (Pisareva et al, 2011). Current models suggest that the Dom34-Hbs1 complex binds to the ribosomal A site, followed by GTP hydrolysis, dissociation of Hbs1 and accommodation of Dom34 in the ribosome. Rli1 then binds and induces ATP-dependent subunit dissociation (Shoemaker & Green, 2011). CryoEM structures provide clear support for eRF3/eRF1/Rli1 and Hbs1/Dom34/Rli1 playing related roles in ribosome recycling (Becker et al, 2011, 2012).
In addition to these biochemical insights, Dom34 and Hbs1 were shown in genetic experiments to be important for RNA quality control in No-Go decay (NGD) targeting aberrant mRNAs (Doma & Parker, 2006)) and in non-functional 18S-rRNA decay targeting defective or incompletely matured 40S ribosomal subunits (Cole et al, 2009; Soudet et al, 2010). In NGD, the Dom34-Hbs1 complex may use its dissociation activity to release ribosomes that are stalled at the 3′ end of mRNAs lacking a termination codon (Tsuboi et al, 2012). Recent reports also suggest that the Dom34-Hbs1 complex and Rli1 mediate dissociation of pre-40S and 60S subunits in a quality control step during ribosome maturation (Lebaron et al, 2012; Strunk et al, 2012). Most of these processes involve the recognition of ribosomes stalled on an mRNA during translation.
We report here a new function for Dom34-Hbs1. We observe that Dom34-Hbs1 stimulates the dissociation of non-translating ribosomes that accumulate upon glucose starvation in yeast. The biological relevance for this activity is seen in the dependence on these proteins of translational recovery in yeast cells after glucose deprivation. We further extended these ideas and show that Dom34-Hbs1 mediated dissociation of non-translating ribosomes can stimulate translation even in non-stressed conditions.
Results
The Dom34-Hbs1 complex stimulates restart of translation after glucose starvation
When the yeast S. cerevisiae is exposed to media lacking glucose for as little as 10 min, a change in the polysome profile occurs that is characteristic of translation inhibition: the polysome levels drop and ribosomes accumulate in a large 80S peak (Ashe et al, 2000). These 80S ribosomes are known to be inactive, bound by Stm1 in a conformation incompatible with translation (Ben-Shem et al, 2011). Glucose addition leads to a rapid recovery of translation, characterized by the reappearance of polysomes and a decrease in the 80S peak (Ashe et al, 2000). These observations were reproduced in our hands with strong translational recovery detectable 5 min after glucose addition (Fig 1A).
Figure 1.
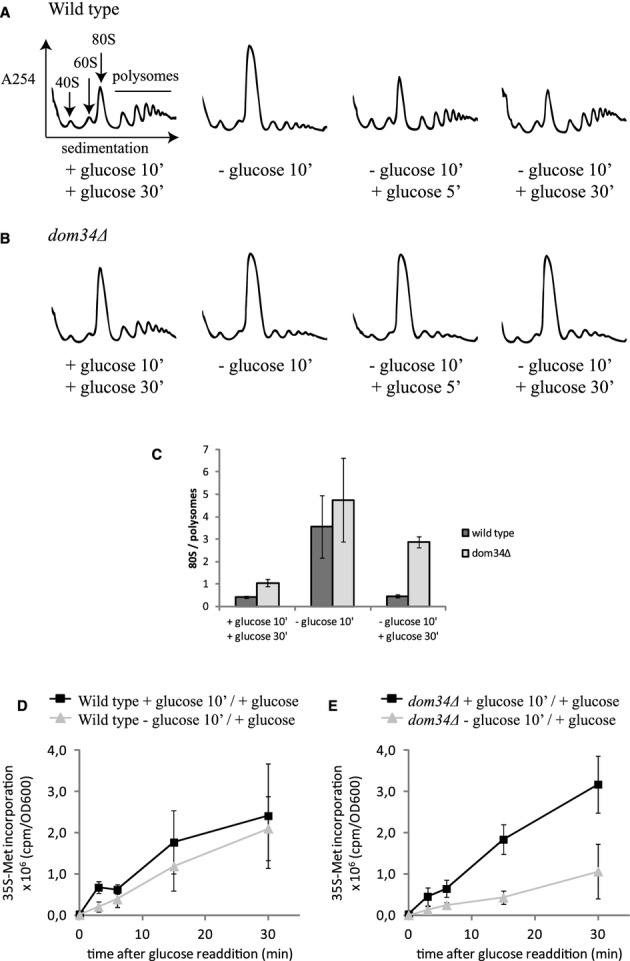
Dom34 stimulates restart of translation in yeast recovering from glucose starvation stress.
A, B Dom34 stimulates the rapid reappearance of polysomes in cells recovering from glucose starvation stress. Polysome profiles of wild-type (A) or dom34Δ (B) yeast grown in glucose-rich medium (left graph), after 10 min of glucose starvation (second graph), and 5 and 30 min after glucose addition (third and fourth graph) at 16°C.
C Quantification of 80S over polysome ratios, corresponding to the first, second and fourth graphs in (A) and (B). Means ± standard deviations of two independent experiments are shown.
D, E Dom34 stimulates protein production in cells recovering from glucose starvation stress. Wild-type (D) and dom34Δ (E) yeast depleted of glucose or grown in glucose-rich medium for 10 min at 16°C was resuspended in glucose-rich medium containing 35S-methionine, followed by incubation at 16°C. 35S-methionine incorporation was measured at the indicated time points. Means ± standard deviations of 3 independent experiments are shown.
The process of translation initiation depends on the activities of separate ribosomal subunits. As such, the restart of translation in yeast recovering from glucose deprivation must depend on the dissociation of the large pool of inactive, Stm1-bound 80S ribosomes. Since human or yeast Dom34-Hbs1 complex, together with Rli1, can very effectively dissociate ribosomal subunits assembled with or without an mRNA in vitro (Shoemaker et al, 2010; Pisareva et al, 2011; Shoemaker & Green, 2011), we hypothesized that these factors might also be needed to split Stm1-bound ribosomes in vivo. To test this possibility, we monitored the polysome profiles of isogenic wild-type, dom34Δ or hbs1Δ strains in glucose-depleted conditions and after glucose addition. In contrast to the wild-type strain, we found that in the absence of Dom34 or Hbs1, the 80S peak did not diminish and the polysomes did not increase after the addition of glucose. A small delay in recovery of translation was observed in mutant strains at 30°C, but the effect was much more prominent at reduced temperatures (16°C; Fig 1A, B and C; see also Fig 4A) where ribosomal subunit dissociation is likely to be energetically more demanding.
Figure 4.
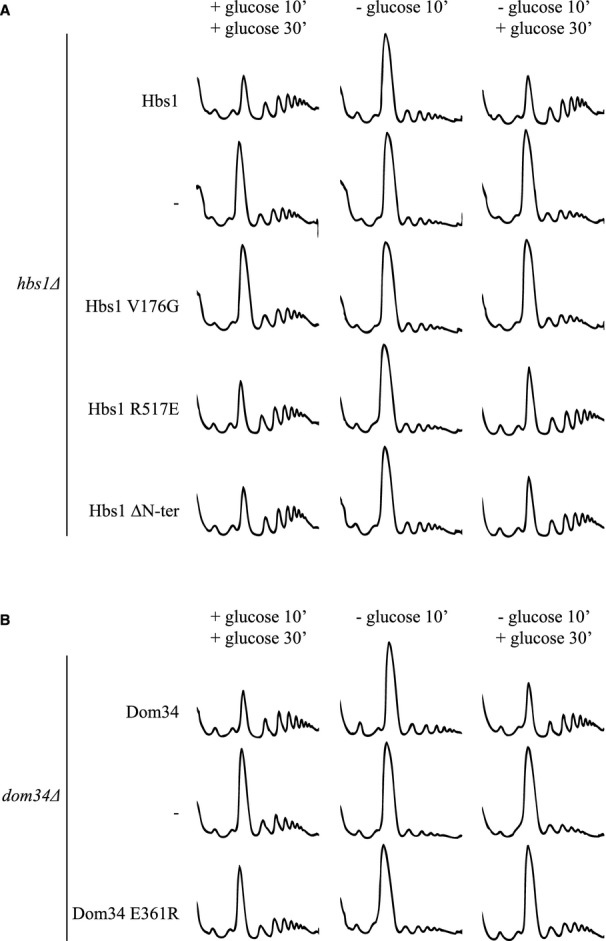
Restart of translation after glucose depletion stress requires Hbs1 GTPase activity but not Dom34-Hbs1 interaction or the Hbs1 N-terminus.
A, B Polysome profiles of hbs1Δ (A) or dom34 Δ (B) yeast transformed with plasmid expressing the indicated mutants, grown in glucose-rich medium (left graph), exposed to glucose starvation (middle graph) and after glucose readdition (right graph) at 16°C.
To further evaluate the role of Dom34-Hbs1 in promoting the restart of translation after glucose starvation, we monitored overall 35S-Met incorporation as a measure of protein synthesis in wild-type and mutant cells. While in wild-type cells, protein synthesis is equivalent in unstarved cells or cells recovering from starvation (Fig 1D), we see that protein synthesis was decreased in dom34Δ cells recovering from glucose starvation compared to unstarved cells (Fig 1E). We also addressed the importance of Dom34-dependent restart of translation for recovery of yeast growth. Yeast lacking Dom34 recovering from glucose starvation consistently displayed a delay in growth compared to a non-starved isogenic strain. In wild-type yeast, growth was practically indistinguishable between yeast recovering from glucose starvation and yeast that was not starved. (Supplementary Fig S1). These results indicate that the Dom34-Hbs1 complex is involved in the restart of translation in yeast recovering from glucose starvation.
Dom34-Hbs1 and Rli1 dissociate inactive ribosomes that accumulate in glucose-starved yeast
Because of its ribosome dissociating activity in vitro (Shoemaker et al, 2010; Pisareva et al, 2011; Shoemaker & Green, 2011) it is likely that Dom34-Hbs1 stimulates restart of translation by splitting inactive 80S ribosomes that accumulate during glucose starvation. These inactive ribosomes differ from known Dom34-Hbs1 substrates in that they contain the protein Stm1 in a conformation that clamps the subunits together (Ben-Shem et al, 2011). We performed biochemical recycling assays to test whether the Dom34-Hbs1 complex could act on these Stm1-bound inactive ribosomes.
In in vitro recycling assays, ribosomes are radioactively labeled and then incubated with various factors (Dom34, Hbs1, Rli1). The complexes are then analyzed on sucrose gradients or on native gels to determine the level of ‘splitting’ (Shoemaker et al, 2010; Shoemaker & Green, 2011). The protein Tif6, an initiation factor that binds to the subunit interface on the 60S subunit, is included in all experiments to trap ribosomes that undergo stimulated dissociation. For the experiments described here, ribosomes were non-specifically radiolabeled using casein kinase II (Shoemaker et al, 2010).
In a first experiment, ‘inactive’ ribosomes isolated from glucose-starved yeast were incubated with various combinations of Dom34, Hbs1 and Rli1 (in addition to Tif6) and the samples were analyzed on a sucrose gradient. We see that after 15 min of incubation, all 80S ribosomes were dissociated into separate subunits when all three factors were added (Fig 2A). The elimination of either Rli1 or Dom34 significantly diminished the amount of dissociation observed, though not completely (Fig 2A).
Figure 2.
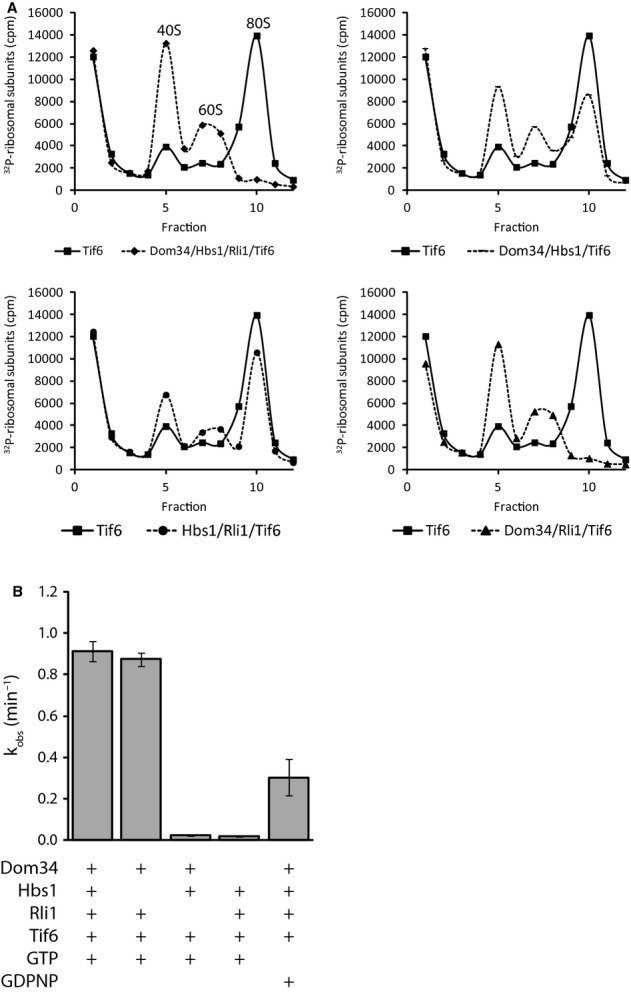
- Dom34-Hbs1 and Rli1 dissociate ribosomes from glucose-starved yeast in vitro. 32P-labeled 80S ribosomes purified from glucose-starved yeast were incubated with the indicated proteins in the presence of ATP and GTP or GDPNP. After 15 min of incubation dissociation was monitored by sucrose density gradient centrifugation and scintillation counting of collected fractions.
- Observed rate constants were determined by monitoring the fraction of dissociated ribosomes in (A) over time on a native gel system (see Supplementary Fig S2). Rate constants were normalized for endpoints. Means ± standard deviations of three independent experiments are shown.
In order to more precisely define the efficiencies of these splitting reactions, we used native gel electrophoresis (Supplementary Fig S2) to analyze equivalent experimental samples over time to determine relative rate constants. The rates that we measured for Dom34/Hbs1/Rli1-mediated splitting of Stm1-bound ribosomes is ˜0.91 min−1 which is similar to the rate we previously observed (˜1.6 min−1) for elongating ribosomes bound to mRNA and peptidyl-tRNA (Shoemaker & Green, 2011). In the absence of Dom34, the rate of dissociation decreased by ˜50-fold, whereas absence of Rli1 caused a reduction of ˜40-fold. In the absence of in vivo data indicating that Rli1 is involved in dissociating inactive ribosomes (RLI1 is an essential gene involved in other important processes) these results strongly support its participation in this process. Similar to the results of the sucrose gradient analysis (Fig 2A) and previous studies (Pisareva et al, 2011; Shoemaker & Green, 2011), elimination of Hbs1 had little effect on the observed rate of the splitting reaction, though blocking Hbs1 GTPase activity by the inclusion of a non-hydrolyzable GTP analog (GDPNP) diminished the rate of splitting by 3-fold (Fig 2B) (see Discussion).
Overall, these data demonstrate that Stm1-bound 80S ribosomes from glucose-starved yeast are good substrates for Dom34/Hbs1/Rli1-mediated subunit splitting in vitro. Indeed, while these samples were prepared at different times from different yeast cultures, the similarity in these values with earlier measurements for related but distinct complexes suggests that these ribosome complexes are equivalent targets for Dom34/Hbs1/Rli1-mediated recycling. We note that because the rates are measured with saturating amounts of Dom34/Hbs1/Rli1, the measured values are rate constants, thus allowing more readily for longitudinal comparisons to be made.
Deletion of STM1 suppresses the requirement for Dom34-Hbs1 to restart translation in vivo
We reasoned that if the Dom34-Hbs1 complex stimulates restart of translation by dissociating Stm1-bound 80S ribosomes, weakening subunit interactions might reduce the requirement for the Dom34-Hbs1 complex. We tested this hypothesis by comparing the translation recovery of strains following glucose starvation and the readdition of glucose in stm1Δ and stm1Δdom34Δ strains. In Fig 3B, we see that deletion of STM1 rescued the dom34Δ recovery-deficient phenotype (Fig 3B and C). Importantly, deletion of STM1 alone had no effect on translation inhibition or translation recovery (compare Figs 3A and 1A, see also Fig 3C).
Figure 3.
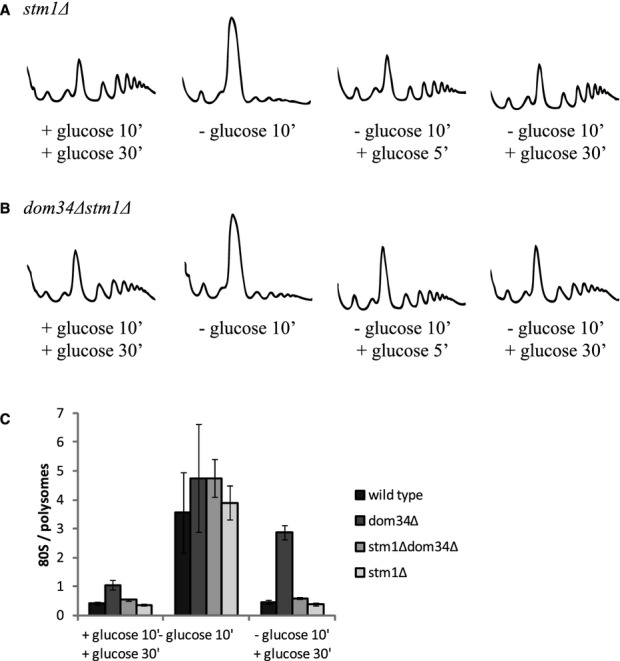
Weakening ribosome subunit interaction reduces the need for Dom34 during restart of translation after glucose starvation stress.
A, B Polysome profiles of stm1Δ (A) and dom34Δstm1Δ (B) yeast grown in glucose-rich medium (left graph), exposed to glucose starvation (second graph), and 5 and 30 min after glucose readdition (third and fourth graphs) at 16°C.
C Quantification of 80S over polysome ratios, corresponding to the first, second and fourth graphs in (A) and (B) as well as in Fig 1A and B. Means ± standard deviations of two independent experiments are shown.
The observation that the weakening of ribosomal subunit interactions reduces the requirement for Dom34 for recovery from starvation supports a model where Dom34/Hbs1/Rli1 promotes the dissociation of ribosomes in vivo.
Functional requirements of Dom34-Hbs1 for translational reactivation
Our biochemical assay indicated that Hbs1 was not essential for the dissociation of inactive, Stm1-bound ribosomes in vitro (Fig 2B). To test the role of Hbs1 in vivo, we compared polysome profiles from a hbs1Δ mutant strain carrying the wild-type HBS1 gene on a plasmid or an empty vector. Analysis of polysomes at different time points during a glucose starvation/recovery experiment indicated that Hbs1, like Dom34, is required for optimal translational restart (Fig 4A, Supplementary Fig S3A).
We next explored the requirement of several functional regions of the Dom34-Hbs1 complex for this process. Hbs1 is a GTPase belonging to the eEF-1α-like family of GTPases (Wallrapp et al, 1998; Atkinson et al, 2008), which includes the termination factor eRF3 and the elongation factor eEF-1α. Both of these factors function essentially to deliver their cargo, eRF1 and aminoacyl-tRNA, respectively, to the ribosomal A site. We asked whether the GTPase activity of Hbs1 is required for efficient restart of translation in vivo and found that the GTP binding-defective Hbs1 mutant V176G (van den Elzen et al, 2010) did not promote effective recovery from glucose starvation (Fig 4A, Supplementary Fig S3A).
We next probed the importance of the interaction between Dom34 and Hbs1 for the recovery from glucose starvation. The interface of these two proteins is comprised of contacts between several different regions in multiple domains of each protein (Chen et al, 2010; van den Elzen et al, 2010; Kobayashi et al, 2010) where interaction-defective mutants have been previously characterized (van den Elzen et al, 2010). The Hbs1 R517E mutant was shown by two-hybrid analysis to bind poorly to Dom34 (van den Elzen et al, 2010). Interestingly, this mutation did not affect the restart of translation following glucose starvation, indicating that a stable Dom34-Hbs1 interaction is not needed for this function (Fig 4A, Supplementary Fig S3A). In parallel, we used the Dom34 E361R mutant that similarly blocks formation of the Dom34-Hbs1 complex (van den Elzen et al, 2010), but in this case, the mutation diminished the recovery of cells from glucose starvation at least partly (Fig 4B, Supplementary Fig S3B). This asymmetric requirement for the interaction surfaces of Hbs1 and Dom34 suggests that Dom34 E361 may be important for other functions in addition to its interaction with Hbs1 (see Discussion).
Members of the family of eEF1α-like GTPases are highly similar with regard to their C-terminal domains, but differ in their N-terminal length and amino-acid sequence (Inagaki & Ford Doolittle, 2000); the function of the N-terminus of Hbs1 is not known. Cryo-EM analysis of the Dom34-Hbs1 complex bound to an 80S ribosome revealed that it is located proximal to the mRNA entry channel (Becker et al, 2011). When this cryoEM structure was aligned with the high-resolution crystal structure of the ribosome from glucose-depleted yeast (Ben-Shem et al, 2011), we found that the N-terminus of Hbs1 would be in close contact with a portion of Stm1 located in the mRNA channel (Supplementary Fig S4). We therefore asked whether the N-terminus of Hbs1 plays a role in stimulating translation recovery after glucose depletion. Deletion of N-terminal amino acids 2–149 (mutant Hbs1 ΔN-ter) did not reduce the efficiency of translation re-initiation (Fig 4A, Supplementary Fig S3A).
The Dom34-Hbs1 complex stimulates translation in non stress-related conditions
In non-stressed conditions, the polysome profiles of yeast lacking functional Dom34 or Hbs1 show elevated 80S peaks, which, especially at low temperatures, are combined with reduced levels of polysomes (compare the polysome profiles on the left in Figs 1A, B and 4A, B; see also Bhattacharya et al, 2010; Carr-Schmid et al, 2002). This observation suggests that even in non-stressed conditions, there may be inactive 80S ribosomes that depend on Dom34-Hbs1 and Rli1-mediated dissociation for their subunits to become available for translation initiation. The higher 80S peak could be due to some amount of Stm1-bound ribosomes, of empty ribosomes lacking Stm1, or of mRNA-bound ribosomes that result, for example, from a Dom34-Hbs1-dependent defect in late translation initiation or early elongation. To distinguish between these possibilities, we analyzed the polysome profiles of wild-type and dom34Δ strains in high-and low-salt sucrose gradients. High-salt treatment is known to dissociate non-translating but not mRNA-bound 80S ribosomes (Martin & Hartwell, 1970; Zylber & Penman, 1970). As we see in Fig 5A and C, in low-salt conditions the 80S peak was higher for the dom34Δ strain compared to wild-type, resulting in an 80S/ polysome ratio that is significantly higher. The 80S peaks were small and of equivalent size for the wild-type and dom34Δ strains when analyzed in high-salt conditions (Fig 5B and C). These data indicate that in the absence of Dom34, primarily non-mRNA-bound 80S ribosomes accumulate.
Figure 5.
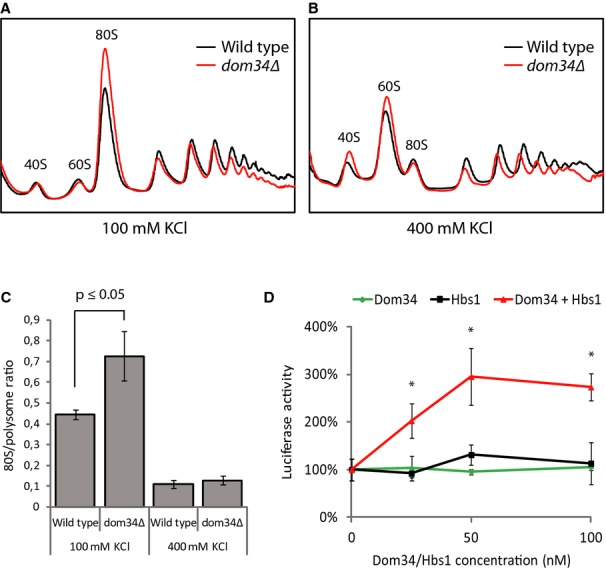
The Dom34-Hbs1 complex stimulates translation in non-stress-related conditions.
A, B Inactive 80S ribosomes accumulate in dom34Δ yeast. Polysome profiles were obtained from wild-type and dom34Δ yeast in low-salt (100 mM KCl) (A) and high-salt (400 mM KCl) (B) conditions. Yeast strains were grown at 30°C.
C Quantification of 80S over polysome ratios corresponding to (A) and (B). Means ± standard deviations of three independent experiments are shown. The indicated significance level was calculated using a Mann–Whitney U-test.
D Dom34-Hbs1 stimulates translation by ribosomes that were not exposed to starvation stress. A firefly luciferase mRNA was translated for 1 h in cell extract obtained from a dom34Δhbs1Δ strain, after which luciferase activity was measured. Addition of increasing amounts of recombinant Dom34-Hbs1 complex, but not of Hbs1 or Dom34 alone, stimulated luciferase production. Means and SDs of 3 independent experiments are shown. * indicates that at these time points the result obtained in presence of both Dom34 and Hbs1 significantly differ (P < 0.001) from the result obtained adding Dom34 or Hbs1 alone (Student's t-test).
This observation supports a potential role for Dom34-Hbs1 and Rli1 in dissociating mRNA-free 80S ribosomes even in actively growing cells. If this is true, then the Dom34-Hbs1 complex would likely stimulate translation even in non-stressed conditions. To test this hypothesis, we used an in vitro translation assay where a synthetic mRNA encoding the firefly luciferase was incubated in cellular extract from a dom34Δhbs1Δ strain, and varying amounts of recombinant Dom34 and Hbs1 were added. Luciferase activity measurements were used to monitor translation. We see that the addition of increasing concentrations of the Dom34-Hbs1 complex stimulated the translation of a firefly luciferase reporter mRNA up to 3-fold, whereas addition of Dom34 or Hbs1 alone did not (Fig 5D).
Together these results support the idea that the Dom34-Hbs1 complex generally stimulates translation in cells, stressed or non-stressed, by facilitating the dissociation of mRNA-free 80S ribosomes into their constituent 40S and 60S subunits.
Discussion
Currently the Dom34-Hbs1 complex is considered a central player in co-translational quality control on RNAs that cause translating ribosomes to stall (Graille & Seraphin, 2012; Shoemaker & Green, 2012). Dom34-Hbs1 stimulates degradation of such mRNAs and rRNAs (Doma & Parker, 2006; Cole et al, 2009), most likely by facilitating the removal of stalled ribosomes from mRNAs (Tsuboi et al, 2012). Here we show that the Dom34-Hbs1 complex is a key player in the quick recovery of cells from stress and also stimulates translation under non-stress conditions. These observations expand the biochemical and physiological roles of Dom34-Hbs1 in the cell because every inactive 80S ribosome becomes a potential substrate for this complex.
Dom34-Hbs1 dissociates inactive ribosomes, promoting recovery after stress
Our data show that the Dom34-Hbs1 complex is critical for the restart of translation in yeast recovering from glucose starvation. Two independent lines of evidence provide support for the idea that this stimulation depends on Dom34-Hbs1 dissociating inactive ribosomes, liberating subunits for new rounds of translation initiation. First, we showed that inactive ribosomes from glucose-depleted yeast are biochemical substrates of the complex (Fig 2). Second, deletion of Stm1, that stabilizes ribosomal subunit interaction (Correia et al, 2004; Ben-Shem et al, 2011) and therefore antagonizes dissociation, abolishes the need for the Dom34-Hbs1 complex for recovery.
Surprisingly we found that Hbs1 did not increase the rate of ribosome splitting in the in vitro reactions (Fig 2). On the other hand, our data clearly show that the factor is essential for efficient recovery of translation after glucose starvation in vivo (Fig 4A). The lack of effect of Hbs1 may have arisen because of the 8-fold excess of Dom34 over ribosome in our in vitro reaction. Indeed, in similar ribosome splitting assays, eRF3 becomes essential only when eRF1 is present in substoichiometric levels relative to ribosomes (Eyler et al, 2013). Preliminary experiments suggest that this is not the case, however. An alternative possibility is that in vivo certain factors are present, that negatively affect ribosome dissociation, therefore rendering the counteracting action of Hbs1 more important. These may be factors interacting with ribosomes or with Dom34. Some of these factors (e.g., tRNAs, translation elongation factors, or ribosome-associated proteins) may have been present in previous ribosomes splitting assays explaining why the stimulatory effect of Hbs1 could be observed in other conditions (Pisareva et al, 2011; Shoemaker & Green, 2011). Detailed biochemical analysis of this complex system will be required to explain the requirement of Hbs1 for ribosome splitting in vivo.
The GTPase activity of Hbs1 was previously shown to be important for all of the protein's identified functions including RNA quality control (van den Elzen et al, 2010; Kobayashi et al, 2010), complementation of a growth defect in a rps30aΔhbs1Δ or a rps28aΔhbs1Δ strain (Carr-Schmid et al, 2002; van den Elzen et al, 2010) and Dom34-Hbs1-Rli1 mediated dissociation of ribosomes (Shoemaker et al, 2010; Pisareva et al, 2011; Shoemaker & Green, 2011). Our data here are consistent with these earlier observations. First, GTPase defective Hbs1 variants were unable to function in the recovery of cells from glucose starvation. Second, the substitution of GDPNP for GTP in the in vitro subunit-splitting assays resulted in an overall inhibition of the reaction. The latter observation may seem surprising, considering that Hbs1 does not stimulate ribosome splitting in vitro. It has been reported that blocking Hbs1 GTPase activity in presence of GDPNP locks both Dom34 and Hbs1 on mRNA-bound ribosome substrates (Shoemaker & Green, 2011). As Hbs1 and Rli1 binding sites on the ribosome overlap (Becker et al, 2011, 2012) this should result in an inhibition of Rli1 binding. These mechanistic details suggest how blocking the GTPase activity of Hbs1 decreases ribosome splitting even if the reaction proceeds in its absence.
We further tested the importance of the interface between Dom34 and Hbs1 for promoting glucose starvation recovery. Here, we found that in absence of a stable Dom34-Hbs1 interaction translational recovery occurs efficiently. As both Dom34 and Hbs1 are required for ribosome dissociation in vivo, these data suggest that their interaction may be stabilized in vivo, e.g. in the ribosomal context or by another, unknown factor. We were somewhat surprised to see that, whereas mutating the Hbs1 interface had little impact on the in vivo phenotype, the Dom34 interface was more important for its function. Mutating the Dom34 interface, or its E361 residue, may interfere with Dom34 function in a way other than by disrupting the interaction with Hbs1. For instance, Dom34 must interact with Rli1 and changes conformation while bound to the ribosome (Becker et al, 2012). Dom34 E361 may at some stage during recycling interact with Rli1 or the ribosomes. Consistently, we note that mutation of the Dom34-Hbs1 interaction surface had a similar asymmetric impact on non-functional 18S rRNA decay (van den Elzen et al, 2010).
A general role of Dom34 –Hbs1 in modulating translation by controlling ribosomal subunit availability
Beyond its role in stress recovery, we observed that Dom34-Hbs1-mediated dissociation of inactive ribosomes can more broadly function to stimulate translation initiation. In the absence of Dom34 and/or Hbs1, polysome profiles have consistently been found to have elevated 80S peaks (Fig 5A; see also Bhattacharya et al, 2010; Carr-Schmid et al, 2002; and compare leftmost profiles in Figs 1A, B and 5A), which we found to be caused by accumulation of inactive ribosomes not bound to mRNA templates (Fig 5A and B). We show here that even in non-stressed conditions, Dom34-Hbs1 appears to broadly stimulate translation efficiency by making subunits available for new rounds of protein synthesis (Fig 5). This observation is consistent with the fact that depletion of orthologs of Rli1—which acts together with Dom34-Hbs1 to dissociate inactive ribosomes—similarly results in accumulation of 80S ribosomes and decreased levels of polysomes in yeast, human and Drosophila cells (Dong et al, 2004; Chen et al, 2006; Andersen & Leevers, 2007). Additionally, in a strain with impaired initiation (inhibition of eIF2), deletion of Dom34 or Hbs1 results in a synthetic growth defect (Carr-Schmid et al, 2002) that might suggest that these factors work in a common pathway. Finally, both Dom34 and Hbs1 are important for normal growth of yeast strains with reduced amounts of 40S subunits (Carr-Schmid et al, 2002; Bhattacharya et al, 2010; van den Elzen et al, 2010) likely because these strains, when they lack Dom34-Hbs1 complex, have too few ribosomes available to function.
During glucose deprivation, inactive ribosomes contain Stm1 in a conformation that inhibits translation and stabilizes subunit interaction (Ben-Shem et al, 2011). It is not clear whether this mechanism of ribosome inhibition is broadly used in response to stress, or whether it is used in a wide variety of physiological conditions. Our results show that in non-stressed conditions, deletion of Stm1 reduces the elevated 80S peak that forms in the absence of Dom34 to almost wild-type levels (compare leftmost polysome profiles in Figs 1B and 3B). This suggests that the translation inhibiting conformation of Stm1 is present in a large fraction of inactive 80S ribosomes, even in non-stressed cells. A role for Stm1 in antagonizing Dom34-Hbs1-mediated dissociation of inactive 80S ribosomes likely explains why overexpression of Stm1 in dom34Δ yeast causes a growth defect (Balagopal & Parker, 2011).
Our work shows that Dom34-Hbs1-mediated subunit dissociation is critical in the recovery of yeast cells from glucose starvation. Our data further suggest that Dom34-Hbs1 plays a similar role in non-stressed cells, dissociating unproductive empty 80S ribosomes so that normal translation initiation can occur. These observations provide insights into a novel general mechanism for the control of translation wherein ribosomes are stored in an unproductive state (either with Stm1 bound or simply not containing an mRNA) that is readily reversed by the activities of Dom34, Hbs1 and Rli1.
We emphasize that this mechanism is likely widely used by cells to dissociate various ribosome complexes to maintain an active supply of ribosomal subunits. Indeed, a general shut down of translation is a hallmark of a cell's response to many stress conditions including nutrient depletion, temperature shock, hypoxia and DNA damage (Spriggs et al, 2010). Moreover, Dom34 and Hbs1 are conserved proteins: Dom34 has orthologs in eukaryotes and archaea (Eberhart & Wasserman, 1995; Ragan et al, 1996). And, whereas Hbs1 has orthologs only in eukaryotes (Wallrapp et al, 1998; Inagaki & Ford Doolittle, 2000), the function is filled even in archaea by the related protein aEF1α (Kobayashi et al, 2010; Saito et al, 2010).
Materials and Methods
Yeast strains, media and plasmids
Yeast strains and plasmids are listed in Supplementary Table S1. Yeast strains, derivatives of BMA64, were constructed by standard methods. The plasmid pBS4415 was constructed by inverse PCR on a pBS3614 template (van den Elzen et al, 2010) using oligonucleotides 5′-ATATCATGAGGTTTCTTTGGTTTCATCTCGATAGTCAATAGTTGTCG-3′ and 5′-CCAAAGAAACCTCATGATATTTCTGCATTTGTTAAATCTGCCTTAC-3′ and was verified by sequencing.
Glucose-rich and glucose-depleted media were YPDA and YPA (for strains without plasmids) or CSM-Ura 2% glucose and CSM-Ura without glucose (for strains containing Dom34 or Hbs1 encoding plasmids) respectively.
Glucose starvation and re-addition
Yeast was grown at 30°C at 170 rpm to an OD600 of 0.6, then shifted to 16°C for 2 h. The culture was then split into multiple 100-ml cultures that were pelleted at 5400 × g for 6 min at 16°C, resuspended in 100 ml of media (precooled at 16°C) without or with 2% glucose and incubated at 16°C for 10 min at 170 rpm. Cells were pelleted, resuspended in 100 ml media with glucose and incubated at 16°C at 170 rpm for the indicated times.
Polysome analysis
At the indicated times after glucose depletion or glucose addition cycloheximide was added (100 μg/ml final concentration) and cells were pelleted at 5,400 × g for 6 min at 4°C. Cells were washed and then lysed at 4°C in lysis buffer (10 mM Tris-Cl pH 7.5; 100 mM KCl; 5 mM MgCl2; 6 mM β-mercaptoethanol; 100 μg/ml cycloheximide) or in lysis buffer containing 400 mM KCl (Fig 4B) by 5 cycles of 1 min vortexing followed by 1 min incubation on ice, in presence of glass beads. Nine OD260 units of lysate were loaded on a 7–47% sucrose gradient in lysis buffer, or lysis buffer containing 400 mM KCl (Fig 4B). After a 14 h spin at 16,900 rpm in an SW41 rotor (Beckman Coulter, Brea, CA, USA), absorbance (254 nm) was measured on an ISCO Teledyne Foxy Jr. fraction collector.
35S-methionine incorporation
Yeast was grown in CSM-Met containing 2% glucose, shifted to 16°C, split into 8-ml cultures and resuspended in 8 ml CSM-Met with or without 2% glucose for 10 min as described above. Then cells were resuspended in 8 ml CSM-Met 2% glucose (16°C) containing 4 μl 35S-methionine (1,175 Ci/mmol, 5 mCi/0.49 ml, Perkin Elmer, Waltham, MA, USA) and incubated at 16°C. At the indicated time points 1-ml samples were taken and 35S-methionine incorporation was measured as described (Ashe et al, 2000).
In vitro ribosome dissociation
80S ribosomes purified from glucose-depleted yeast were kindly provided by S. Melnikov and Dr. Marat Yusupov. 100 pmol ribosomes were 32P-labeled using 500 U casein kinase II (NEB) and 32P γ-ATP in the manufacturer's recommended buffer, then pelleted through a 600 μl 1.1 M sucrose cushion in buffer E (20 mM Tris-Cl pH 7.5, 2.5 mM Mg(OAc)2, 100 mM KOAc pH 7.6, 2 mM DTT, 0.25 mM spermidine) at 75,000 rpm 1 h 4°C in a MLA-130 rotor (rcf max = 34.0 × 104 g) followed by resuspension in buffer E. 6.25 pmol ribosomes were incubated in 25 μl buffer E containing 1 mM GTP or GDPNP and 1 mM ATP at 26°C for 15 min with 50 pmol Dom34, 50 pmol Hbs1, 50 pmol Rli1 and 625 pmol Tif6, purified as described previously (Shoemaker et al, 2010; Shoemaker & Green, 2011). Dissociation was analyzed by centrifugation through a 10–30% sucrose gradient in buffer E at 38,500 rpm for 3.5 h at 4°C in a SW41 rotor (rcf max = 26.4 × 104 g). Fractions were counted in Bio Safe II scintillation fluid. Kinetic analysis was performed by loading 2 μl fractions of the reactions on a 3% acrylamide gel in THEM buffer (34 mM Tris base, 57 mM Hepes, 0.1 mM EDTA, 2.5 mM MgCl2) (Acker et al, 2007) at indicated time points, running the gel in THEM buffer at 12 W at 4°C. Gels were dried and quantified using a Typhoon 9410 phosphoimager and ImageQuantTL (GE Healthcare Life Sciences, Little Chalfont, UK). The fraction of dissociated ribosomes was plotted against time and, using KaleidaGraph (Synergy Software, Reading, PA, USA) for curve fitting, rate constants were determined.
In vitro translation
Translational extracts were prepared from a dom34Δhbs1Δ strain (BSY2550) essentially as described (Tuite & Plesset, 1986). A synthetic firefly luciferase-A(50) mRNA (Gallie et al, 1991) was incubated in this extract supplemented with recombinant Dom34 and/or Hbs1 and luciferase activity was assayed. Translation conditions have been described by Tharun et al (Tarun & Sachs, 1995; see Supplementary data for details).
Acknowledgments
Authors are grateful to S. Melnikov and M. Yusupov for purified ribosomes and discussions, F. Wyers and F. Lacroute for the stm1Δ strain, M. Gas Lopez for the hbs1ΔN-ter (2-149)-PROTEIN A construct, and our team members as well as A. Ben-Shem for discussions. We thank the IGBMC mass-spectrometry platforms for their help in the early part of this project and IGBMC (Institut de Génétique et de Biologie Moléculaire et Cellulaire) for assistance. This work was supported by grants from the CERBM-IGBMC, the Ligue Contre le Cancer (Equipe Labellisée 2011), the CNRS and the Agence Nationale de la Recherche (ANR 11 BSV8 009 02) to BS. A.M.G.v.d.E. was supported by predoctoral fellowships from Université de Strasbourg, the Fondation pour la Recherche Médicale and an EMBO short-term fellowship (ASTF 162-2013).
Author contributions
AMGvdE performed all the in vivo experiments, as well as the in vitro translation, with input from BS. In vitro analyses of ribosome dissociation were performed by AS and AMGvdE with input from RG and BS. BS supervised the project. AMGvdE, AS, RG and BS wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://emboj.embopress.org
References
- Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–145. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- Andersen DS, Leevers SJ. The essential Drosophila ATP-binding cassette domain protein, pixie, binds the 40 S ribosome in an ATP-dependent manner and is required for translation initiation. J Biol Chem. 2007;282:14752–14760. doi: 10.1074/jbc.M701361200. [DOI] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, Slaven JW, De Long SK, Ibrahimo S, Sachs AB. A novel eIF2B-dependent mechanism of translational control in yeast as a response to fusel alcohols. EMBO J. 2001;20:6464–6474. doi: 10.1093/emboj/20.22.6464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson GC, Baldauf SL, Hauryliuk V. Evolution of nonstop, no-go and nonsense-mediated mRNA decay and their termination factor-derived components. BMC Evol Biol. 2008;8:290. doi: 10.1186/1471-2148-8-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R. Stm1 modulates translation after 80S formation in Saccharomyces cerevisiae. RNA. 2011;17:835–842. doi: 10.1261/rna.2677311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. doi: 10.1038/nsmb.2057. [DOI] [PubMed] [Google Scholar]
- Becker T, Franckenberg S, Wickles S, Shoemaker CJ, Anger AM, Armache JP, Sieber H, Ungewickell C, Berninghausen O, Daberkow I, Karcher A, Thomm M, Hopfner KP, Green R, Beckmann R. Structural basis of highly conserved ribosome recycling in eukaryotes and archaea. Nature. 2012;482:501–506. doi: 10.1038/nature10829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shem A, Garreau de Loubresse N, Melnikov S, Jenner L, Yusupova G, Yusupov M. The structure of the eukaryotic ribosome at 3.0 A resolution. Science. 2011;334:1524–1529. doi: 10.1126/science.1212642. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, McIntosh KB, Willis IM, Warner JR. Why Dom34 stimulates growth of cells with defects of 40S ribosomal subunit biosynthesis. Mol Cell Biol. 2010;30:5562–5571. doi: 10.1128/MCB.00618-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr-Schmid A, Pfund C, Craig EA, Kinzy TG. Novel G-protein complex whose requirement is linked to the translational status of the cell. Mol Cell Biol. 2002;22:2564–2574. doi: 10.1128/MCB.22.8.2564-2574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Muhlrad D, Hauryliuk V, Cheng Z, Lim MK, Shyp V, Parker R, Song H. Structure of the Dom34-Hbs1 complex and implications for no-go decay. Nat Struct Mol Biol. 2010;17:1233–1240. doi: 10.1038/nsmb.1922. [DOI] [PubMed] [Google Scholar]
- Chen ZQ, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The essential vertebrate ABCE1 protein interacts with eukaryotic initiation factors. J Biol Chem. 2006;281:7452–7457. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- Cole SE, LaRiviere FJ, Merrikh CN, Moore MJ. A convergence of rRNA and mRNA quality control pathways revealed by mechanistic analysis of nonfunctional rRNA decay. Mol Cell. 2009;34:440–450. doi: 10.1016/j.molcel.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia H, Medina R, Hernandez A, Bustamante E, Chakraburtty K, Herrera F. Similarity between the association factor of ribosomal subunits and the protein Stm1p from Saccharomyces cerevisiae. Mem Inst Oswaldo Cruz. 2004;99:733–737. doi: 10.1590/s0074-02762004000700012. [DOI] [PubMed] [Google Scholar]
- Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The essential ATP-binding cassette protein RLI1 functions in translation by promoting preinitiation complex assembly. J Biol Chem. 2004;279:42157–42168. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Wasserman SA. The pelota locus encodes a protein required for meiotic cell division: an analysis of G2/M arrest in Drosophila spermatogenesis. Development. 1995;121:3477–3486. doi: 10.1242/dev.121.10.3477. [DOI] [PubMed] [Google Scholar]
- van den Elzen AM, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Seraphin B, Graille M. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol. 2010;17:1446–1452. doi: 10.1038/nsmb.1963. [DOI] [PubMed] [Google Scholar]
- Eyler DE, Wehner KA, Green R. Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1. J Biol Chem. 2013;288:29530–29538. doi: 10.1074/jbc.M113.487090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR, Feder JN, Schimke RT, Walbot V. Post-transcriptional regulation in higher eukaryotes: the role of the reporter gene in controlling expression. Mol Gen Genet. 1991;228:258–264. doi: 10.1007/BF00282474. [DOI] [PubMed] [Google Scholar]
- Graille M, Seraphin B. Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat Rev Mol Cell Biol. 2012;13:727–735. doi: 10.1038/nrm3457. [DOI] [PubMed] [Google Scholar]
- Inada T, Winstall E, Tarun SZ, Jr, Yates JR, 3rd, Schieltz D, Sachs AB. One-step affinity purification of the yeast ribosome and its associated proteins and mRNAs. RNA. 2002;8:948–958. doi: 10.1017/s1355838202026018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inagaki Y, Ford Doolittle W. Evolution of the eukaryotic translation termination system: origins of release factors. Mol Biol Evol. 2000;17:882–889. doi: 10.1093/oxfordjournals.molbev.a026368. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1alpha complex. Proc Natl Acad Sci USA. 2010;107:17575–17579. doi: 10.1073/pnas.1009598107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Deplazes A, Sohrmann M, Peter M. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat Cell Biol. 2008;10:602–610. doi: 10.1038/ncb1723. [DOI] [PubMed] [Google Scholar]
- Krebs JE, Goldstein ES, Kilpatrick ST. Lewin's Genes X. Sudbury, MA: Jones and Barlett Publishers; 2011. [Google Scholar]
- Krokowski D, Gaccioli F, Majumder M, Mullins MR, Yuan CL, Papadopoulou B, Merrick WC, Komar AA, Taylor D, Hatzoglou M. Characterization of hibernating ribosomes in mammalian cells. Cell Cycle. 2011;10:2691–2702. doi: 10.4161/cc.10.16.16844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebaron S, Schneider C, van Nues RW, Swiatkowska A, Walsh D, Bottcher B, Granneman S, Watkins NJ, Tollervey D. Proofreading of pre-40S ribosome maturation by a translation initiation factor and 60S subunits. Nat Struct Mol Biol. 2012;19:744–753. doi: 10.1038/nsmb.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TE, Hartwell LH. Resistance of active yeast ribosomes to dissociation by KCl. J Biol Chem. 1970;245:1504–1506. [PubMed] [Google Scholar]
- Montero-Lomeli M, Morais BL, Figueiredo DL, Neto DC, Martins JR, Masuda CA. The initiation factor eIF4A is involved in the response to lithium stress in Saccharomyces cerevisiae. J Biol Chem. 2002;277:21542–21548. doi: 10.1074/jbc.M201977200. [DOI] [PubMed] [Google Scholar]
- Nielsen PJ, Duncan R, McConkey EH. Phosphorylation of ribosomal protein S6. Relationship to protein synthesis in HeLa cells. Eur J Biochem. 1981;120:523–527. doi: 10.1111/j.1432-1033.1981.tb05731.x. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Skabkin MA, Hellen CU, Pestova TV, Pisarev AV. Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J. 2011;30:1804–1817. doi: 10.1038/emboj.2011.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336:915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragan MA, Logsdon JM, Jr, Sensen CW, Charlebois RL, Doolittle WF. An archaebacterial homolog of pelota, a meiotic cell division protein in eukaryotes. FEMS Microbiol Lett. 1996;144:151–155. doi: 10.1111/j.1574-6968.1996.tb08522.x. [DOI] [PubMed] [Google Scholar]
- Saito K, Kobayashi K, Wada M, Kikuno I, Takusagawa A, Mochizuki M, Uchiumi T, Ishitani R, Nureki O, Ito K. Omnipotent role of archaeal elongation factor 1 alpha (EF1alpha in translational elongation and termination, and quality control of protein synthesis. Proc Natl Acad Sci USA. 2010;107:19242–19247. doi: 10.1073/pnas.1009599107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Eyler DE, Green R. Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science. 2010;330:369–372. doi: 10.1126/science.1192430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci USA. 2011;108:E1392–E1398. doi: 10.1073/pnas.1113956108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker CJ, Green R. Translation drives mRNA quality control. Nat Struct Mol Biol. 2012;19:594–601. doi: 10.1038/nsmb.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Gelugne JP, Belhabich-Baumas K, Caizergues-Ferrer M, Mougin A. Immature small ribosomal subunits can engage in translation initiation in Saccharomyces cerevisiae. EMBO J. 2010;29:80–92. doi: 10.1038/emboj.2009.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–237. doi: 10.1016/j.molcel.2010.09.028. [DOI] [PubMed] [Google Scholar]
- Strunk BS, Novak MN, Young CL, Karbstein K. A translation-like cycle is a quality control checkpoint for maturing 40S ribosome subunits. Cell. 2012;150:111–121. doi: 10.1016/j.cell.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarun SZ, Jr, Sachs AB. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 1995;9:2997–3007. doi: 10.1101/gad.9.23.2997. [DOI] [PubMed] [Google Scholar]
- Tsuboi T, Kuroha K, Kudo K, Makino S, Inoue E, Kashima I, Inada T. Dom34:hbs1 plays a general role in quality-control systems by dissociation of a stalled ribosome at the 3′ end of aberrant mRNA. Mol Cell. 2012;46:518–529. doi: 10.1016/j.molcel.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Tuite MF, Plesset J. mRNA-dependent yeast cell-free translation systems: theory and practice. Yeast. 1986;2:35–52. doi: 10.1002/yea.320020103. [DOI] [PubMed] [Google Scholar]
- Tzamarias D, Roussou I, Thireos G. Coupling of GCN4 mRNA translational activation with decreased rates of polypeptide chain initiation. Cell. 1989;57:947–954. doi: 10.1016/0092-8674(89)90333-4. [DOI] [PubMed] [Google Scholar]
- Uesono Y, Toh EA. Transient inhibition of translation initiation by osmotic stress. J Biol Chem. 2002;277:13848–13855. doi: 10.1074/jbc.M108848200. [DOI] [PubMed] [Google Scholar]
- Van Dyke N, Baby J, Van Dyke MW. Stm1p, a ribosome-associated protein, is important for protein synthesis in Saccharomyces cerevisiae under nutritional stress conditions. J Mol Biol. 2006;358:1023–1031. doi: 10.1016/j.jmb.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Van Dyke N, Chanchorn E, Van Dyke MW. The Saccharomyces cerevisiae protein Stm1p facilitates ribosome preservation during quiescence. Biochem Biophys Res Commun. 2013;430:745–750. doi: 10.1016/j.bbrc.2012.11.078. [DOI] [PubMed] [Google Scholar]
- Wallrapp C, Verrier SB, Zhouravleva G, Philippe H, Philippe M, Gress TM, Jean-Jean O. The product of the mammalian orthologue of the Saccharomyces cerevisiae HBS1 gene is phylogenetically related to eukaryotic release factor 3 (eRF3) but does not carry eRF3-like activity. FEBS Lett. 1998;440:387–392. doi: 10.1016/s0014-5793(98)01492-6. [DOI] [PubMed] [Google Scholar]
- Zylber EA, Penman S. The effect of high ionic strength on monomers, polyribosomes, and puromycin-treated polyribosomes. Biochim Biophys Acta. 1970;204:221–229. doi: 10.1016/0005-2787(70)90505-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


