Abstract
In Drosophila, Dicer-1 produces microRNAs (miRNAs) from pre-miRNAs, whereas Dicer-2 generates small interfering RNAs from long double-stranded RNA (dsRNA), a process that requires ATP hydrolysis. We previously showed that inorganic phosphate inhibits Dicer-2 cleavage of pre-miRNAs, but not long dsRNAs. Here, we report that phosphate-dependent substrate discrimination by Dicer-2 reflects dsRNA substrate length. Efficient processing by Dicer-2 of short dsRNA requires a 5′ terminal phosphate and a two-nucleotide, 3′ overhang, but does not require ATP. Phosphate inhibits cleavage of such short substrates. In contrast, cleavage of longer dsRNA requires ATP but no specific end structure: phosphate does not inhibit cleavage of these substrates. Mutation of a pair of conserved arginine residues in the Dicer-2 PAZ domain blocked cleavage of short, but not long, dsRNA. We propose that inorganic phosphate occupies a PAZ domain pocket required to bind the 5′ terminal phosphate of short substrates, blocking their use and restricting pre-miRNA processing in flies to Dicer-1. Our study helps explain how a small molecule can alter the substrate specificity of a nucleic acid processing enzyme.
Keywords: Dicer, dsRNA, miRNA, phosphate, siRNA
Introduction
Unlike mammals, flies and other arthropods divide the tasks of making microRNAs (miRNAs) and small interfering RNAs (siRNAs) between Dicer-1 and Dicer-2 (Lee et al, 2004). These two paralogs likely descend from an ancestral enzyme that produced both miRNAs to regulate protein-coding genes and siRNAs to initiate an anti-viral RNAi response. The evolution of different dicer enzymes devoted to distinct small RNA pathways presumably allowed selection for adaptations that enhance each enzyme's specialized function, at the expense of the general ability to convert any double-stranded RNA (dsRNA) substrate into small silencing RNAs.
Both Dicer-1 and Dicer-2 contain an amino-terminal “helicase” domain, but only Dicer-2 hydrolyzes ATP, an activity stimulated by dsRNA (Zamore et al, 2000; Hutvágner et al, 2001; Bernstein et al, 2003; Jiang et al, 2005). Dicer-2 hydrolyzes approximately 21 ATP molecules for each siRNA duplex produced from long dsRNA, suggesting that ATP fuels the translocation of Dicer-2 along dsRNA with a one base-pair step size (Zamore et al, 2000; Nykanen et al, 2001; Liu et al, 2003; Cenik et al, 2011; Welker et al, 2011). The Dicer-2 helicase domain comprises DExDc and Helicase C domains. In contrast, Dicer-1 contains only a Helicase C domain. Consistent with the lack of a DExDc domain, Dicer-1 liberates a miRNA/miRNA* duplex from pre-miRNA without ATP (Jiang et al, 2005; Tsutsumi et al, 2011). Like all RNase III enzymes, the active sites of Dicer-1 and Dicer-2 are formed by the dimerization of two RNase III domains, which pair intramolecularly for these multi-domain proteins (Zhang et al, 2004; MacRae et al, 2006; Ye et al, 2007). The dimer interface creates two ribonuclease active sites, allowing the enzyme to make staggered breaks in the two arms of a pre-miRNA or in the two strands of dsRNA (Zhang et al, 2004; MacRae et al, 2006; Ye et al, 2007). Dicer-1 and Dicer-2 also contain a central dsRNA-binding domain (dsRBD, previously known as DUF283) and a PAZ domain. PAZ domains, which are also found in Argonaute proteins, bind the two-nucleotide, 3′ overhanging ends created when Drosha or Dicer cleave dsRNA (Cerutti et al, 2000; Lingel et al, 2003, 2004; Song et al, 2003; Yan et al, 2003; Ma et al, 2004; MacRae et al, 2006, 2007; Park et al, 2011). The PAZ domain of human Dicer has also been shown to recognize the 5′ monophosphate present on pre-miRNAs (Park et al, 2011). Finally, Dicer-1 and Dicer-2 contain a carboxy-terminal, canonical dsRBD, which is thought to enhance affinity for substrate (Provost et al, 2002; Zhang et al, 2004).
Despite the in vivo specialization of Dicer-1 and Dicer-2, purified Dicer-2 can process pre-miRNAs into miRNAs. Notably, Dicer-2 makes miRNA products that are shorter than the authentic products produced by Dicer-1 (Cenik et al, 2011). Such a difference in the length of miRNA/miRNA* duplexes could cause their inappropriate loading into Argonaute2, which favors 21-nt RNAs, rather than Argonaute1, which prefers 22-nt RNAs (Ameres et al, 2011). Moreover, for the approximately 60% of D. melanogaster miRNAs derived from the 3′ arm of their pre-miRNA, the seed sequence (positions 2–8 of a miRNA) of the Dicer-2 product would differ from that of the canonical miRNA produced by Dicer-1. Since the target specificity of a miRNA is determined mainly by its seed, the shorter miRNA products produced by Dicer-2 would regulate mRNAs different from those controlled by authentic miRNAs. Therefore, cleavage of pre-miRNA by Dicer-2 would likely have adverse consequences that need to be suppressed in vivo.
Pre-miRNA processing by Dicer-2 is inhibited both by the Dicer-2 partner protein R2D2 and by physiological concentrations of inorganic phosphate. Phosphate inhibition of Dicer-2 is dose-dependent and specific; inorganic phosphate affects neither the rate of production by Dicer-2 of siRNAs from long dsRNA nor the cleavage by Dicer-1 of pre-miRNA into authentic miRNAs, and other anions do not inhibit Dicer-2 from processing pre-miRNA (Cenik et al, 2011). What features of pre-miRNA and long dsRNA underlie the specific inhibition by inorganic phosphate of pre-miRNA processing by Dicer-2? What differences between Dicer-2 and Dicer-1 allow inorganic phosphate to inhibit pre-miRNA processing by one dicer but not the other? Why does Dicer-2 process long dsRNA efficiently, but Dicer-1 does not?
To begin to answer these questions, we measured the kinetics of substrate cleavage using purified recombinant Dicer-1 and Dicer-2, examining substrate RNAs encompassing different lengths and terminal structures, in the presence and absence of inorganic phosphate and ATP. We find that inorganic phosphate prevents Dicer-2 from cleaving short, but not long, dsRNA by inhibiting both binding and catalysis. Cleavage of short dsRNA by Dicer-2 requires an end with a 5′ monophosphate and a two-nucleotide, 3′ overhang, whereas cleavage of long dsRNA does not require a specific terminal structure. Mutation of a pair of evolutionarily conserved arginine residues in the Dicer-2 PAZ domain blocked cleavage of short dsRNA without affecting the ATP-dependent conversion of long dsRNA into siRNAs. Thus, Dicer-2 uses distinct mechanisms to recognize and process long and short RNA substrates. We propose that inorganic phosphate prevents the use of pre-miRNA substrates by occupying a binding pocket in the Dicer-2 PAZ domain that would otherwise recognize the 5′ terminal phosphate of short dsRNA substrates.
Results
Dicer-2 cleaves pre-miR-8 and pre-miR-79 into aberrant miRNAs with altered seed sequences
We previously reported that purified recombinant Dicer-2 cleaves pre-let-7 into 5′ and 3′ products that were 1 nt shorter than the authentic let-7 and let-7* species made by Dicer-1 (Cenik et al, 2011). Because mature let-7 resides on the 5′ arm of pre-let-7, the shorter RNA product generated by Dicer-2 contains the same seed sequence as let-7 produced by Dicer-1. miR-8 and miR-79 reside on the pre-miRNA 3′ arm. For these miRNAs, purified Dicer-2 generated products 1–2 nt shorter than those made by Dicer-1 (Supplementary Fig S1). Consequently, the seed sequences of miR-8 and miR-79 produced by Dicer-2 were shifted relative to those made by Dicer-1; such aberrant seeds are predicted to target a set of mRNAs different from those regulated by the authentic miR-8 and miR-79 produced by Dicer-1 (Lewis et al, 2003, 2005; Stark et al, 2003; Brennecke et al, 2005).
Dicer-2 does not produce aberrant miRNAs in vivo
To test if Dicer-2 produces aberrant miRNAs in vivo, we sequenced 18–29-nt RNA from the ovaries of control (w1118) and dicer-2 heterozygous (dicer-2L811fsx/CyO) and homozygous (dicer-2L811fsx) mutant flies (Supplementary Tables S1, S2 and S3). The most abundant isoforms of miR-8 (23 nt) and miR-79 (22 nt) in the dicer-2L811fsx homozygous and heterozygous mutants and control ovaries (Fig1A) corresponded to the products made by purified Dicer-1, but not Dicer-2 (Supplementary Fig S1). In fact, no miRNA changed significantly (Bonferroni-corrected P-value < 0.05) in its abundance or length in dicer-2L811fsx homozygous and heterozygous mutants compared with the control, except miR-7 whose mean length changed from 23.5 to 23.4 nt in the dicer-2L811fsx homozygous mutant (Bonferroni-corrected P-value = 0.004; Fig1B and C). We conclude that there is no biologically meaningful change in miRNA abundance or length in the absence of Dicer-2. As expected (Kawamura et al, 2008; Okamura et al, 2008a,b), the abundance of endo-siRNAs was significantly decreased in the dicer-2L811fsx homozygous mutant flies (Supplementary Table S3).
Figure 1.
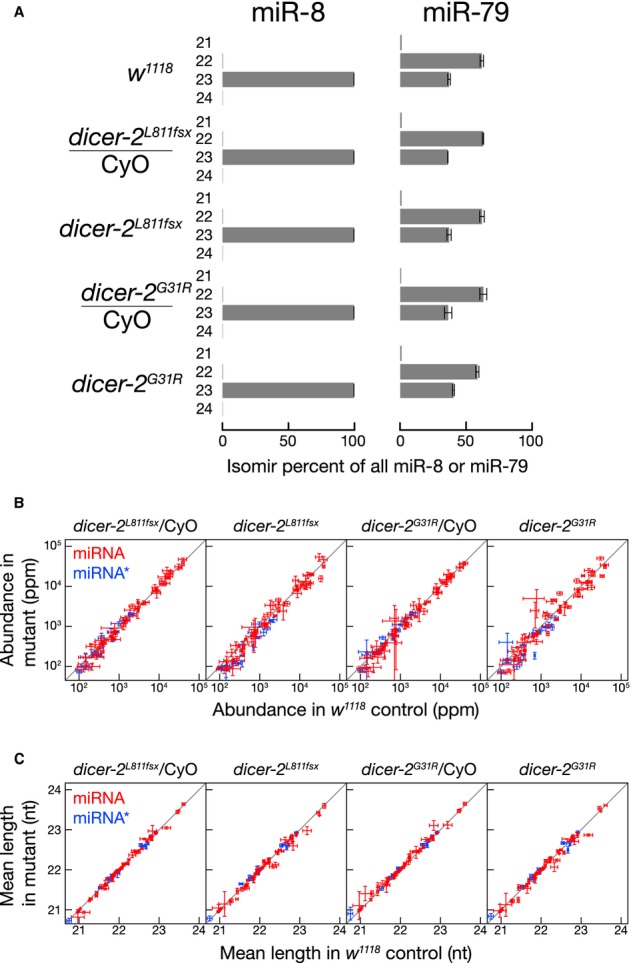
- A Isoform distribution of miR-8 and miR-79 in wild-type, heterozygous, and dicer-2 mutant fly ovaries.
- B, C Normalized number of reads (ppm) (B) or mean length (C) for miRNA (red) and miRNA* (blue) compared among control w1118, heterozygous or homozygous dicer-2L811fsx or dicer-2G31R mutant fly ovaries. Only miRNAs and miRNA* strands whose abundance was >100 ppm were analyzed in (B) and (C). Data are mean ± s.d. for three biologically independent replicates.
The Dicer-2 G31R mutation blocks siRNA production in vivo (Lee et al, 2004; Pham et al, 2004) and is predicted to inhibit the binding of ATP to the helicase domain. Dicer-2G31R does not efficiently hydrolyze ATP or process long dsRNA, but retains the ability to cleave pre-miRNA (Cenik et al, 2011) and collaborate with its partner, R2D2, in loading small RNA duplexes into Ago2 (Pham et al, 2004; Forstemann et al, 2007). As in dicer-2L811fsx, both the abundance and mean length of each miRNA were essentially unchanged in the dicer-2G31R heterozygous and homozygous mutant flies (Fig1B and C). In contrast, the dicer-2G31R mutation reduced endo-siRNAs compared with the controls (P-value = 0.003, 0.03, 0.0008 for esi-1.1, esi-1.2, and esi-2.1, respectively; Supplementary Table S3). We conclude that in vivo Dicer-2 produces siRNAs from long dsRNA, but not miRNAs from pre-miRNAs.
Inorganic phosphate and ATP act on Dicer-2 independently
Physiological concentrations of inorganic phosphate (25 mM) inhibit Dicer-2, but not Dicer-1, from cleaving pre-let-7, whereas ATP is required for the production of siRNAs from long dsRNA but not for the cleavage of pre-let-7 by Dicer-2 in the absence of inorganic phosphate (Fig2A; Cenik et al, 2011). To test whether phosphate binds to the ATP-binding site of Dicer-2, we measured the effect of inorganic phosphate on pre-let-7 processing by Dicer-2 in the absence and presence of 1 mM ATP, a concentration greater than the approximately 14 μM KMATP of Dicer-2 (Cenik et al, 2011). ATP had no measurable effect on the ability of phosphate to inhibit Dicer-2 cleavage of pre-let-7, pre-miR-8 or pre-miR-79 (Fig2A). Moreover, phosphate inhibited cleavage of pre-miR-8 by Dicer-2G31R in both the presence and absence of ATP (Fig2B). Phosphate did not significantly (P-value >> 0.05) change the rate of wild-type Dicer-1 cleavage for any of these pre-miRNAs (Fig2A). Thus, for Dicer-2, the inhibition of pre-miRNA cleavage by phosphate is independent from the stimulation of long-dsRNA processing by ATP.
Figure 2.
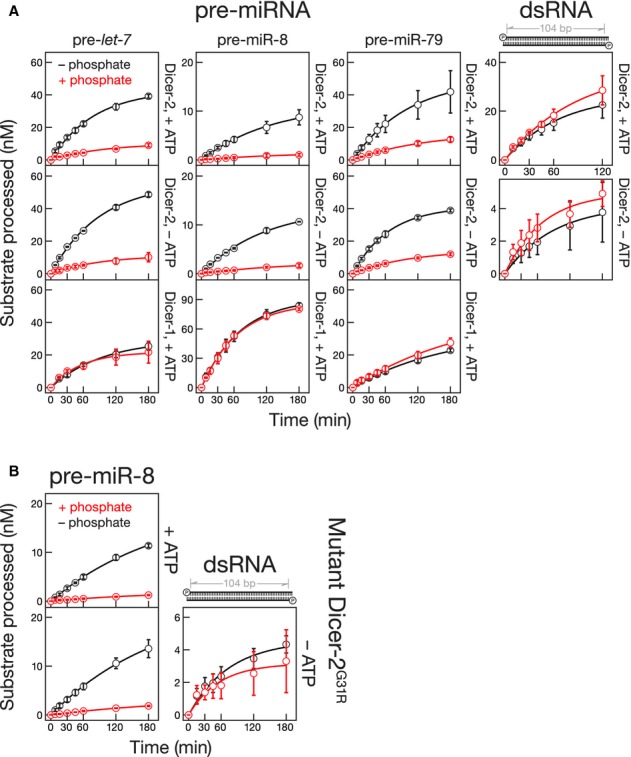
- Pre-miRNAs (100 nM, 5′ 32P-radiolabeled) or a 104-bp dsRNA with a two-nucleotide, 3′ overhanging end (100 nM, uniformly 32P-radiolabeled) were incubated with Dicer-1 (2 nM) or Dicer-2 (8 nM) with (red) or without (black) 25 mM inorganic phosphate. For 104-bp dsRNA −ATP, 12 nM Dicer-2 was used.
- Pre-miR-8 (100 nM, 5′ 32P-radiolabeled) or a 104-bp dsRNA with a two-nucleotide, 3′ overhanging end (100 nM, uniformly 32P-radiolabeled) was incubated with Dicer-2G31R (8 nM for +ATP and 12 nM for −ATP) with (red) or without (black) 25 mM inorganic phosphate. Data are mean ± s.d. for three independent trials.
Long dsRNA, but not pre-miRNA, stimulates ATP hydrolysis by Dicer-2 (Cenik et al, 2011). Perhaps ATP allows Dicer-2 to overcome the inhibitory effect of phosphate for cleavage of long dsRNA? In the presence of ATP, inorganic phosphate did not inhibit cleavage of 5′ monophosphorylated 106- or 104-bp dsRNAs bearing either a blunt or 3′ overhanging end (Fig2A; Cenik et al, 2011). To test whether the stimulation of long-dsRNA processing by ATP enables Dicer-2 to escape inhibition by inorganic phosphate, we measured the effect of phosphate on processing of long dsRNA in the absence of ATP. Without ATP, the rate of siRNA production from long dsRNA was reduced, but remained unaffected by phosphate (Fig2A). We obtained similar results using the Dicer-2G31R mutant (Fig2B), which does not hydrolyze ATP in the presence of long dsRNA (Cenik et al, 2011). We conclude that stimulation by ATP of dsRNA cleavage cannot explain why inorganic phosphate does not inhibit the conversion of long dsRNA into siRNAs by Dicer-2.
Phosphate inhibits Dicer-2 binding to pre-miRNA
To understand how inorganic phosphate inhibits processing of short dsRNAs by Dicer-2, we tested whether phosphate impedes binding of such substrates to the enzyme. We used 302-nm UV cross-linking to examine the binding of Dicer-2 to pre-let-7 and 30- and 104-bp dsRNA bearing a 5′ monophosphate and two-nucleotide, 3′ overhang (Fig3; Supplementary Fig S2). All substrates contained a 5′-iodouridine at the penultimate position. Pre-let-7 and the 30-bp dsRNA were 5′ 32P-radiolabeled; one of the strands of the 104-bp-long dsRNA was 32P-radiolabeled at the phosphodiester bond linking nucleotides 36 and 37. Inorganic phosphate inhibited binding of Dicer-2 to pre-let-7 and 30-bp dsRNA. In contrast, inorganic phosphate did not affect binding of Dicer-2 to the long dsRNA. Dicer-1 cross linked inefficiently to the RNAs bearing a 5′-iodouridine at the penultimate position, so we used 254-nm UV non-site-specific cross-linking to monitor the binding of Dicer-1 to unmodified pre-let-7. Inorganic phosphate had no detectable effect on the binding of Dicer-1 to pre-let-7. We conclude that inorganic phosphate inhibits Dicer-2 binding to pre-miRNAs and short 30-bp dsRNA.
Figure 3.
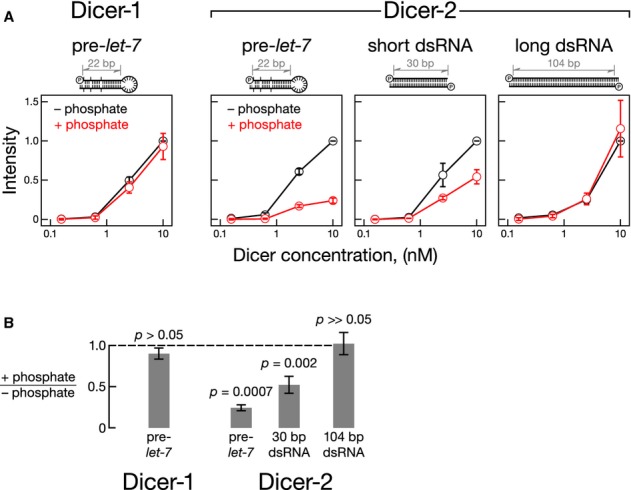
- For Dicer-1, 32P-radiolabeled pre-let-7 (30 pM) was incubated with Dicer with or without 25 mM phosphate for 20 min at 4°C, then irradiated with 254-nm light to cross link bound substrate to enzyme. For Dicer-2, 32P-radiolabeled RNA (30 pM) containing 5-iodouridine as the penultimate nucleotide was incubated with Dicer with or without 25 mM phosphate for 20 min at 4°C, then irradiated with 302-nm light. Samples were resolved by SDS-PAGE. Mean signal intensity for 10 nM Dicer proteins without inorganic phosphate was arbitrarily set to one.
- The ratio of substrate cross-linked to 10 nM Dicer-1 or Dicer-2 with and without inorganic phosphate is shown. Data are mean ± s.d. for three independent trials.
Dicer-2 prefers short 5′ monophosphorylated dsRNAs with two-nucleotide, 3′ overhangs
Our data suggest that Dicer-2 binds pre-miRNA and long dsRNA by distinct mechanisms. Considering the similar chemical structures of a 5′ monophosphate and inorganic phosphate, we sought to test whether a 5′ monophosphate was required for Dicer-1 or Dicer-2 to process short substrates such as pre-let-7 (Fig4). We prepared pre-let-7 bearing either a 5′ monophosphate or a 5′ hydroxyl group. Dicer-1 cleaved both forms of pre-let-7 with similar efficiencies. Inorganic phosphate had no detectable effect on the rate of cleavage by Dicer-1 of either 5′ monophosphorylated or 5′ hydroxy pre-let-7. In contrast, Dicer-2 efficiently cleaved 5′ monophosphorylated, but not 5′ hydroxy, pre-let-7. Similarly, the rate of cleavage by Dicer-2 of 5′ monophosphorylated pre-miR-87 was 2.7-fold faster than that for 5′ hydroxy pre-miR-87, whereas Dicer-1 processed the two substrates with similar efficiency. Inorganic phosphate inhibited processing by Dicer-2, but not by Dicer-1, of both types of pre-miR-87 substrates. The rate of cleavage by Dicer-2 of 5′ monophosphorylated pre-miR-307a was 43-fold faster than that for 5′ hydroxy pre-miR-307a; cleavage was inhibited by inorganic phosphate for both terminal structures (Supplementary Fig S3).
Figure 4.
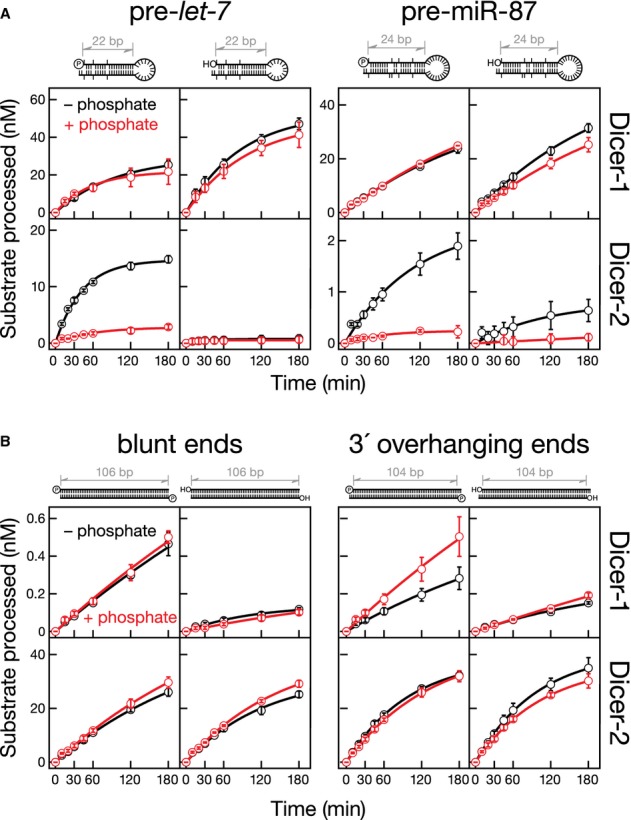
- Uniformly 32P-radiolabeled pre-miRNAs (100 nM) bearing a 5′ monophosphate or hydroxyl group were incubated with Dicer-1 (2 nM) or Dicer-2 (8 nM) with (red) or without (black) 25 mM inorganic phosphate.
- Uniformly 32P-radiolabeled 104- or 106-bp dsRNA (100 nM) bearing a 5′ monophosphate or hydroxyl group and either a blunt or two-nucleotide, 3′ overhang end were incubated with Dicer-1 or Dicer-2 (8 nM) with (red) or without (black) 25 mM inorganic phosphate. Data are mean ± s.d. for three independent trials.
Terminal structure had no detectable effect on the processing of long dsRNA by Dicer-2. We compared uniformly 32P-radiolabeled 104- or 106-bp dsRNAs bearing (i) 5′ monophosphate, blunt ends, (ii) 5′ monophosphate, two-nucleotide, 3′ overhanging ends, (iii) 5′ hydroxyl, blunt ends, and (iv) 5′ hydroxyl, two-nucleotide, 3′ overhanging ends (Fig4). Dicer-2 cleaved all four long dsRNAs with similar rates.
Even in the presence of ATP, production of the first siRNA from the end of a long dsRNA is rate-determining for Dicer-2 (Cenik et al, 2011). Thus, subsequent production of siRNAs from the interior of a long dsRNA appears to proceed at the same rate as production of the first, terminal siRNA. All the long dsRNA substrates used here were diced at the similar rates (Fig4), supporting the idea that production of the first siRNA limits the rate of producing internal siRNAs from substrates with all possible termini (Cenik et al, 2011). Consistent with this observation, we were unable to detect intermediates for any long dsRNA, irrespective of its terminal structure.
Although Dicer-1 favored RNAs with 5′ monophosphate ends over 5′ hydroxyl ends, the cleavage activity was low for all four long dsRNAs, consistent with previous observations that Dicer-1 does not appreciably cleave long dsRNA (Cenik et al, 2011). Inorganic phosphate had no significant inhibitory effect on the rate of cleavage of any of the four long dsRNAs by Dicer-1 or Dicer-2.
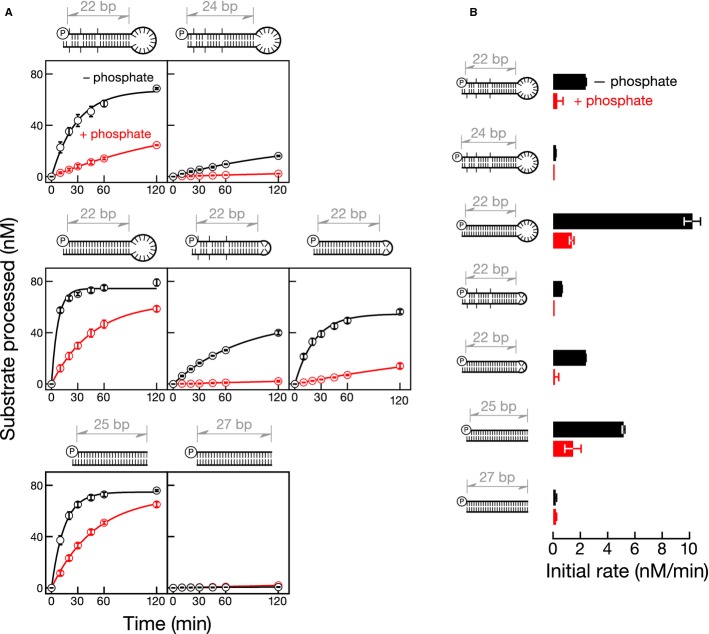
In addition to a 5′ monophosphate, pre-miRNAs bear two-nucleotide, 3′ overhangs, contain mismatches in the stem, and include a long terminal loop. What pre-miRNA feature(s) does Dicer-2 recognize? To begin to answer this question, we compared pre-let-7 to six pre-let-7 variants: (i) pre-let-7 with a blunt end; (ii) pre-let-7 with a perfectly base-paired stem; (iii) pre-let-7 with a tetraloop in place of the wild-type 14-nt terminal loop; (iv) pre-let-7 with both a perfectly base-paired stem and a tetraloop; (v) a fully paired 25-bp dsRNA with a two-nucleotide, 3′ overhang; and (vi) a fully paired 27-bp dsRNA with a blunt end (Fig5).
Figure 5.
- 5′ 32P-radiolabeled pre-let-7 variants or short dsRNAs (100 nM) were incubated with Dicer-2 (7 nM) with (red) or without (black) 25 mM inorganic phosphate.
- Initial rates determined from the data in (A). Gray denotes deoxynucleotides in short dsRNAs. Data are mean ± s.d. for three independent trials.
Pre-let-7 with a blunt end was cleaved approximately 12-fold more slowly than the wild-type pre-let-7, indicating that Dicer-2 prefers a two-nucleotide, 3′ overhang to a blunt end. Pre-let-7 with a perfectly base paired stem was cleaved by Dicer-2 more efficiently than wild-type pre-let-7, indicating that Dicer-2 prefers a perfectly base-paired stem to a stem containing mismatches. In contrast, pre-let-7 with a tetraloop was cleaved less efficiently than the wild-type pre-let-7, revealing that Dicer-2 prefers the natural terminal loop to the compact tetraloop. Consistent with these findings, pre-let-7 with both a perfectly base-paired stem and a tetraloop was cleaved at a rate intermediate to the two pre-let-7 variants, like that observed for wild-type pre-let-7.
Similarly, a fully paired 25-bp dsRNA bearing a two-nucleotide, 3′ overhang was processed more efficiently than the corresponding 27-bp dsRNA with a blunt end [For each of the two dsRNAs, one end contained two deoxy-nucleotides to block access to Dicer-2 (Rose et al, 2005; Cenik et al, 2011)]. We conclude that in processing short dsRNAs, Dicer-2 prefers a two-nucleotide, 3′ overhang, while it exhibits no preference for end structure when cleaving long dsRNA in the presence of ATP (Fig4).
A Short Length is the signature feature of dsRNAs whose processing by Dicer-2 is inhibited by phosphate
What pre-miRNA feature(s) are required for inorganic phosphate to inhibit Dicer-2 cleavage? We tested the effect of phosphate on Dicer-2 cleavage for each of the pre-let-7 variants (Fig5). Inorganic phosphate inhibited cleavage of all substrates tested (cleavage of the blunt, fully paired 27-bp dsRNA was too slow to permit comparison of rates with and without phosphate). Together with our finding that cleavage of long dsRNAs by Dicer-2 was unaffected by phosphate (Figs2A and 4B), these data suggest that substrate length determines whether phosphate inhibits its cleavage by Dicer-2: cleavage of short, but not long, dsRNAs is inhibited by phosphate. Using UV cross-linking, we tested the effect of inorganic phosphate on the binding of Dicer-2 to short, 30-bp dsRNA bearing a 5′ monophosphate and a two-nucleotide, 3′ overhang end (Fig3; Supplementary Fig 2). Inorganic phosphate inhibited binding, supporting our hypothesis that substrate length determines whether phosphate inhibits its cleavage by Dicer-2.
Next, we explored the relationship between substrate length and the requirement for specific terminal features, enhancement by ATP, or inhibition by inorganic phosphate. We assayed Dicer-2 processing—with and without ATP and in the presence or absence of phosphate—for sixteen different RNAs (Fig6; Supplementary Fig S4): 30-, 38-, 52-, and 73-bp dsRNAs bearing (i) a 5′ monophosphate and two-nucleotide, 3′ overhang; (ii) a 5′ hydroxyl group and two-nucleotide, 3′ overhang; (iii) a 5′ monophosphate with a blunt end; or (iv) a 5′ hydroxyl group with a blunt end. For each, the access of Dicer-2 to one end of the dsRNA was blocked by including two deoxynucleotides (Rose et al, 2005; Cenik et al, 2011). Our data show that the shorter the substrate, the more Dicer-2 prefers a 5′ monophosphate to a 5′ hydroxyl group and a two-nucleotide, 3′ overhang to a blunt end. Conversely, the longer the substrate, the greater the requirement for ATP for efficient cleavage by Dicer-2 and the less the susceptibility to inhibition by phosphate. The transition between “long” substrates—whose processing required ATP but whose end structure mattered little if at all—and “short” substrates—for which a 5′ phosphate and two-nucleotide, 3′ overhang was essential but ATP was dispensable—occurred around 38 bp. We note that the median length of the pre-miRNA stem of the 60 most abundant miRNAs in flies is 26 bp.
Figure 6.
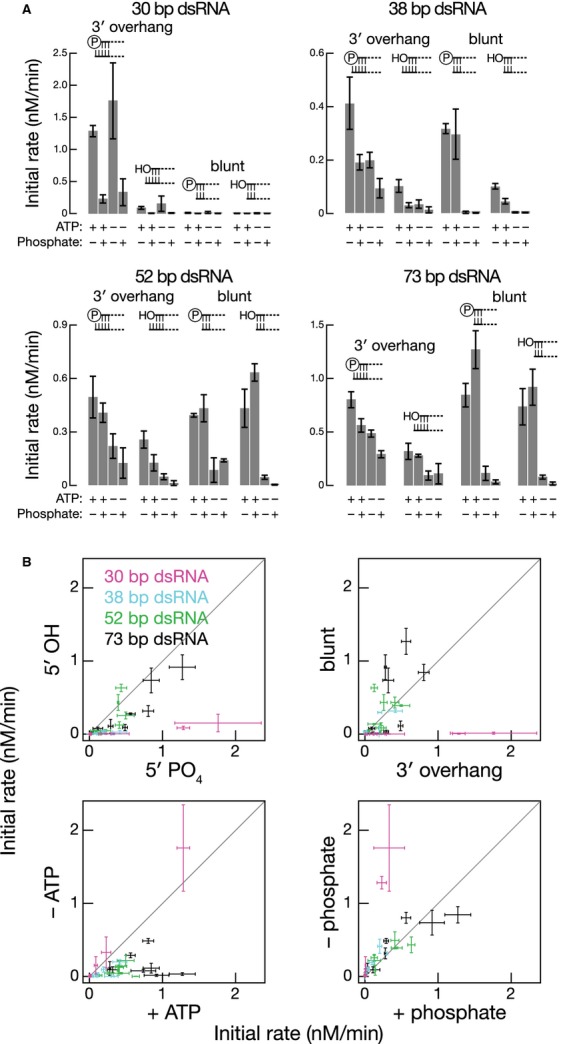
- Initial rates determined from the time course in Supplementary Fig S4.
- Initial rates were compared between 5′ monophosphorylated and hydroxyl ends (upper left panel), between blunt ends and two-nucleotide 3′ overhangs (upper right panel), between the presence or absence of ATP (lower left panel), and between the presence or absence of inorganic phosphate (lower right panel). Data are mean ± s.d. for three independent trials.
Inorganic phosphate increases KM and decreases kcat of Dicer-2 for short, but not long, dsRNA
To better understand how inorganic phosphate inhibits cleavage by Dicer-2 of short but not long dsRNA, we analyzed the kinetics of cleavage by Dicer-2 using three substrates bearing a 5′ monophosphorylated, two-nucleotide, 3′ overhanging end: pre-let-7 and 30- and 104-bp dsRNAs (Table1; Supplementary Fig S5). For pre-let-7, phosphate increased KM (1.5 ± 0.5 μM versus 3.2 ± 1.0 μM; P-value = 0.04) and reduced kcat (2.0 ± 0.5 min versus 0.6 ± 0.2 min; P-value = 0.03). Similarly, phosphate increased KM for the 30-bp dsRNA from 1.1 ± 0.3 μM to 2.7 ± 0.6 μM (P-value = 0.04) and decreased kcat from 2.7 ± 0.3 μM to 2.0 ± 0.2 min (P-value = 0.03). Thus, for these two short RNA substrates inorganic phosphate increased the substrate concentration at which Dicer-2 was half-maximally active and decreased the rate of enzyme turnover. Overall, inorganic phosphate decreased enzyme efficiency (kcat/KM) >7-fold for pre-let-7 and >3-fold for the 30-bp dsRNA. Because KM = (kcat + koff)/kon, an increase in KM accompanied by a decrease in kcat requires either that phosphate increases koff or decreases kon or both, consistent with our result that inorganic phosphate inhibited binding of Dicer-2 to pre-let-7 and 30-bp dsRNA (Fig3; Supplementary Fig S2). In contrast, inorganic phosphate had no measurable affect on KM or kcat for the 104-bp dsRNA: KM values were 42 ± 10 nM and 41 ± 5 nM (P-value >> 0.05) and kcat values were 0.086 ± 0.008 min and 0.090 ± 0.008 min (P-value >> 0.05), without and with phosphate, respectively.
Table 1.
Kinetic analysis of Dicer-2 processing in the presence and absence of 25 mM phosphate. Data are mean ± standard deviation for three independent trials
| Substrate | Enzyme | KM (nM) | Change in KM | kcat (min) | Change in kcat | kcat/KM (nM/min) | Change in kcat/KM |
|---|---|---|---|---|---|---|---|
| pre-let-7 | Dicer-2 | 1500 ± 500 | 1.0 | 2.0 ± 0.5 | 1.0 | 0.0014 ± 0.0001 | 1.0 |
| Dicer-2 + Phosphate | 3200 ± 1000 | 2.1 | 0.6 ± 0.2 | 0.31 | 0.00020 ± 0.00003 | 0.14 | |
| 30-bp dsRNA | Dicer-2 | 1100 ± 300 | 1.0 | 2.7 ± 0.3 | 1.0 | 0.0026 ± 0.0003 | 1.0 |
| Dicer-2 + Phosphate | 2700 ± 600 | 2.5 | 2.0 ± 0.2 | 0.73 | 0.00077 ± 0.00012 | 0.29 | |
| 104-bp dsRNA | Dicer-2 | 42 ± 10 | 1.0 | 0.086 ± 0.008 | 1.0 | 0.0021 ± 0.0003 | 1.0 |
| Dicer-2 + Phosphate | 41 ± 5 | 0.98 | 0.090 ± 0.008 | 1.0 | 0.0022 ± 0.0001 | 1.0 |
Mutation of the Dicer-2 PAZ domain disrupts cleavage of short but not long dsRNA
The Platform and PAZ domains of human Dicer form a pocket that binds the 5′ phosphate of a pre-miRNA (Park et al, 2011). Interestingly, in the three-dimensional structure of this region of human Dicer contains a phosphate molecule chelated by the side chains of three evolutionarily conserved arginine residues (Park et al, 2011). The corresponding region of Dicer-2 in flies and other arthropods has diverged considerably from Dicer-1 and lacks these three conserved arginine residues, leading to the suggestion that Dicer-2 does not recognize 5′ phosphates on its substrates (Park et al, 2011).
Our data suggest that Dicer-2 does, in fact, bind 5′-terminally phosphorylated ends and raises the possibility that an alternative set of positively charged residues in the Dicer-2 PAZ domain interacts with phosphate. Supporting this idea, a pair of PAZ domain arginine residues, R943 and R965, are evolutionarily conserved among arthropod Dicer-2 orthologs (Fig7A). Mutation of these conserved arginines to alanine (Dicer-2R943A,R956A) blocked cleavage of a 30-bp dsRNA bearing 5′ monophosphorylated, two-nucleotide, 3′ overhanging end, but not processing of a 104-bp dsRNA (Fig7B). In contrast, mutation of the helicase domain (Dicer-2G31R) reduced the rate of processing of the 104-bp dsRNA, but had no effect on cleavage of the 30-bp dsRNA. Cleavage of this short, 5′ phosphorylated substrate by mutant Dicer-2G31R was inhibited by inorganic phosphate.
Figure 7.
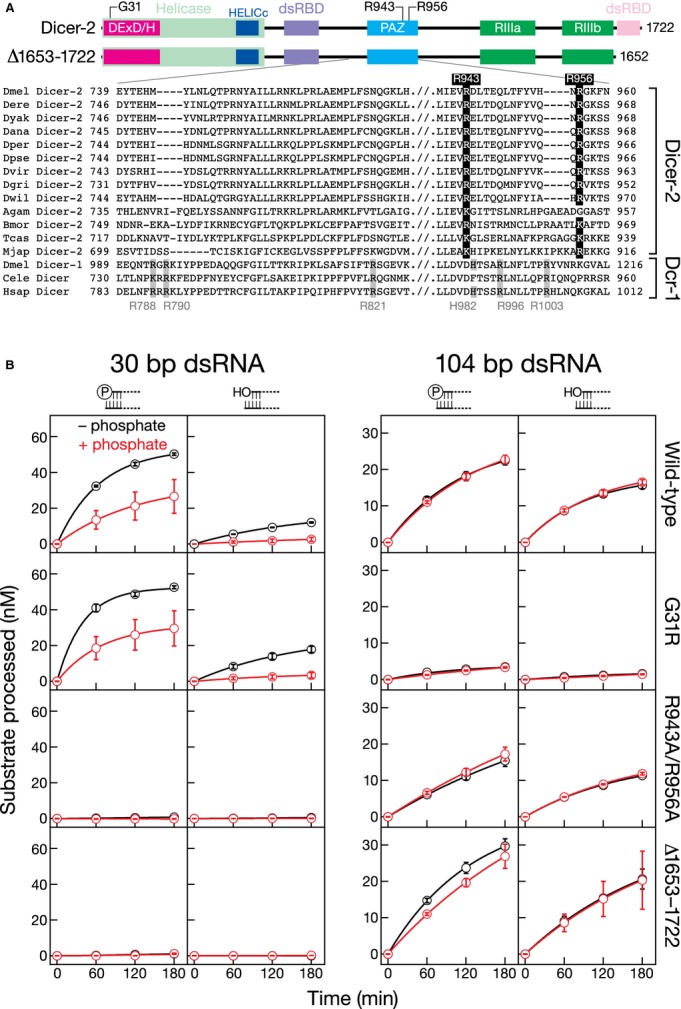
- The domains of Drosophila melanogaster Dicer-2 and alignment of the PAZ domain sequence from Dicers from various arthropods, C. elegans, and human. In the alignment, the bottom three Dicer-1-related enzymes cleave pre-miRNA to produce miRNA in vivo. The residues that compose the 5′ monophosphate-binding pocket in human Dicer are highlighted. Legend: DExD/H, DExD/DExH box helicase domain; HELICc, Helicase conserved C-terminal domain; PAZ, PAZ domain; RIIIa and RIIIb, Ribonuclease III domain; dsRBD, dsRNA-binding domain.
- 5′ 32P-radiolabeled 30-bp dsRNA (100 nM) or 104-bp dsRNA (100 nM uniformly 32P-radiolabeled), with a 5′ monophosphate or hydroxyl end with a two-nucleotide, 3′ overhanging end, was incubated with the wild-type and mutant Dicer-2 (6 nM) and 1 mM ATP in the presence (red) or absence (black) of 25 mM inorganic phosphate. Data are mean ± s.d. for three independent trials.
We also measured dsRNA processing by a Dicer-2 mutant in which the C-terminal dsRNA-binding domain was truncated (Dicer-2Δ1653–1722, Fig7). Like Dicer-2R943A,R956A, Dicer-2Δ1653-1722 efficiently cleaved long dsRNA, but not short. We were unable to detect intermediate cleavage products of the long dsRNAs in the presence or absence of inorganic phosphate. Together, these three mutants identify distinct domains of Dicer-2 that are differentially required for processing long and short dsRNA. Our data suggest that processing of long dsRNA requires ATP binding and hydrolysis by the helicase domain, whereas processing of short dsRNA requires recognition of a 5′ phosphate by the PAZ domain and binding of dsRNA by the carboxy-terminal dsRNA-binding domain.
Discussion
Drosophila Dicer-2 uses distinct mechanisms to recognize short and long dsRNA (Fig6). Our data suggest that the boundary between short and long dsRNAs is >30 bp but <38 bp, although more extensive work will be required to know if the transition between “short” and “long” occurs sharply or gradually over dsRNA length. To efficiently cleave short dsRNA, Dicer-2 requires a 5′ monophosphorylated, two-nucleotide 3′ overhanging end, but does not require ATP. In contrast, to efficiently cleave long dsRNA, Dicer-2 requires ATP, but not a specific terminal RNA structure.
We propose a model in which the PAZ domain of Dicer-2 forms binding pockets for the 5′ monophosphate and the two-nucleotide, 3′ overhang end of the dsRNA substrate, as suggested for human Dicer (Park et al, 2011). For short dsRNA substrates to be efficiently bound and cleaved by Dicer-2, the RNA needs to have a 5′ monophosphate and a two-nucleotide, 3′ overhang, each recognized by binding pockets. The substrate also needs to be bound by the C-terminal dsRNA binding domain. In contrast, long dsRNA does not need to have a specific terminal structure to be efficiently recognized. Nor does long dsRNA need the C-terminal dsRNA binding domain, likely because long dsRNA can interact with the helicase domain and/or the central dsRNA binding domain (Lau et al, 2012).
In our model, an inorganic phosphate molecule occupies the same binding pocket as the 5′ monophosphorylated end of the dsRNA substrate (Fig8). The bound inorganic phosphate serves to block binding of the 5′ monophosphorylated end of a short dsRNA or pre-miRNA (Fig3 and Table1). In the presence of inorganic phosphate, binding of short dsRNA or pre-miRNA to Dicer-2 may require distorting the substrate or the enzyme, explaining the decreased kcat we observed for such substrates in the presence of inorganic phosphate. Our data suggest that the bound inorganic phosphate also blocks binding of a short dsRNA with a 5′ hydroxyl end, perhaps because a phosphate oxygen and the terminal hydroxyl group occupy the same portion of the phosphate binding pocket (Figs4 and 7B).
Figure 8.
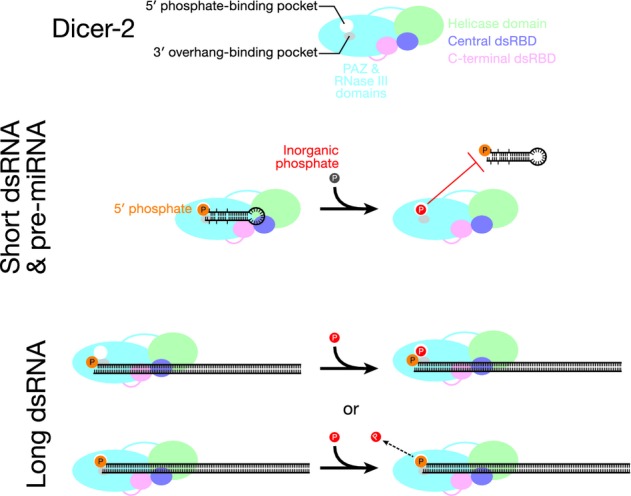
A model for the inhibition by inorganic phosphate of Dicer-2 processing of short, but not long dsRNA
We hypothesize that inorganic phosphate occupies the same binding pocket in the PAZ domain of Dicer-2 as the 5′ monophosphorylated end of a dsRNA substrate and blocks binding of the 5′-monophosphorylated end of a short dsRNA or pre-miRNA. Because long dsRNA can be recognized by the helicase domain and/or the central dsRNA-binding domain, binding of long dsRNA is not inhibited when inorganic phosphate occupies the PAZ domain. Alternatively, the initial high affinity recognition of long dsRNA by the helicase or central dsRNA binding-domain might allow the end of a long dsRNA to displace the inorganic phosphate from its binding pocket. The model aligns well with the previous structural model of Dicer-2 (Lau et al, 2012).
Because long dsRNA can be recognized by the helicase domain and/or the central dsRNA binding domain, its binding is not inhibited by inorganic phosphate occupying the PAZ domain. Alternatively, the initial high affinity recognition by the helicase domain or the central dsRNA binding domain might allow the long dsRNA to displace a bound inorganic phosphate from its binding pocket.
Point mutations in a pair of highly conserved arginine residues in the PAZ domain impair processing of short, but not long, dsRNA (Fig7). Such mutations likely disrupt the binding pocket for a 5′ terminal monophosphate. The finding that Dicer-2 recognizes the 5′ phosphate on short dsRNA substrates such as pre-miRNAs is consistent with such phosphate recognition being present in the ancestral dicer from which Dicer-1 and Dicer-2 descend. Specialization of the two arthropod dicers has allowed phosphate recognition to become dispensable for long dsRNA processing by Dicer-2. Mutation of the Dicer-2 helicase domain converts Dicer-2 from an enzyme optimized for the processive production of siRNAs from long dsRNA into a Dicer-1-like protein that generates miRNAs from pre-miRNAs. Our data help explain the different evolutionary trajectories of these paralogous dsRNA-endonucleases, whose PAZ and helicase domains have diverged, restricting them to produce distinct classes of small silencing RNAs. Our study also helps explain how a small molecule can alter the substrate specificity of a nucleic acid processing enzyme.
Why does Dicer-2 retain a binding pocket for a 5′ monophosphorylated dsRNA end although it is dispensable for processing long dsRNA? Dicer-2 acts not only to cleave long dsRNA into siRNAs, but also to load siRNA duplexes into Argonaute2 (Liu et al, 2003; Tomari et al, 2004). We speculate that the 5′ phosphate-binding pocket functions during the loading of an siRNA duplex into Argonaute2.
We previously found that the Dicer-2 partner protein R2D2, but not the alternative partner Loquacious-PD, inhibits Dicer-2 from processing pre-miRNA (Cenik et al, 2011). Dicer-2 binds R2D2 via its helicase domain (Hartig and Forstemann, 2011 and Nishida et al, 2013). The inhibitory effects of R2D2 and inorganic phosphate are additive, suggesting that they act independently and consistent with the idea that inorganic phosphate binds to the Dicer-2 PAZ domain rather than the helicase domain. Dicer-2 achieves the highest substrate specificity in the presence of both R2D2 and inorganic phosphate (Cenik et al, 2011), which we propose is the in vivo situation.
Our finding that a small molecule—phosphate—can alter the function of Dicer-2 is encouraging for developing a small molecule drug targeting mammalian Dicer. Such drugs might inhibit or enhance production of miRNA, thereby promoting or inhibiting oncogenesis (Bernstein et al, 2003; Srikantan et al, 2011). Destruction of toxic Alu RNA by mammalian Dicer helps prevent age-related macular degeneration (Kaneko et al, 2011; Tarallo et al, 2012); drugs that enhance this activity might prove useful in managing this disease.
Materials and Methods
Small RNA libraries
Small RNA libraries were prepared as described (Fukunaga et al, 2012). Ovaries were dissected from 3–5-day-old females. Total RNA was isolated using the mirVana kit (Ambion, Life Technologies, Grand Island, NY, USA). Supplementary Tables S1 and S2 summarize sequencing statistics.
RNA substrates
RNA substrates for dicing were prepared by chemical synthesis or transcription with T7 RNA polymerase. The sequences of 104- and 106-bp dsRNA were as described (Cenik et al, 2011). Supplementary Table S4 lists the sequences of other RNAs. The 5′ triphosphate was converted to a hydroxyl group with alkaline phosphatase (NEB, Ipswich, MA, USA) and then to a 5′ monophosphate using T4 polynucleotide kinase (NEB). RNA was 5′ 32P-radiolabeled RNA using γ-32P ATP (6, 000 Ci/mmol; PerkinElmer, Waltham, MA, USA) and T4 polynucleotide kinase. Site-specifically 32P-radiolabeled RNA was prepared by DNA-splinted ligation (Moore & Sharp, 1993; Moore & Query, 2000). Uniformly 32P-radiolabeled RNA was prepared by T7 RNA polymerase transcription in the presence of α-32P UTP (800 Ci/mmol; PerkinElmer). All RNAs were gel-purified.
Dicing assays
Recombinant Dicer-1 and Dicer-2 were expressed and purified, and dicing reactions were performed at 25°C and analyzed as described (Cenik et al, 2011; Fukunaga et al, 2012). Two different preparations of recombinant wild-type Dicer-2 were used in this study: preparation 1 was used in Fig5, and preparation 2 was used for all other experiments. Mutations in Dicer-2 were introduced by PCR. Membranes or dried gels were exposed to image plates and analyzed with an FLA-9000 and ImageGauge 3.0 software (Fujifilm, Tokyo, Japan).
To determine rates of reaction, substrate processed versus time was fit to y = y0 + A(1 – e−kt), where dy/dt = Ake−kt. When t = 0, dy/dt = Ak and k gives the initial rate of reaction (Lu & Fei, 2003). In Supplementary Fig S5, data were fit to the Michaelis-Menten scheme using Visual Enzymics 2008 (Softzymics, Princeton, NJ, USA) for Igor Pro 6.31 (WaveMetrics, Lake Oswego, OR, USA).
Binding assays
To measure Dicer-2 binding, 30 pM 32P-radiolabeled substrate RNA was incubated with Dicer-2, then cross linked for 2 min on ice with 302-nm light (UVM-57, UVP, Upland, CA, USA) through the lid of a polystyrene microtiter plate. The light source was placed upside-down directly on the lid. For Dicer-1 binding, 30 pM 32P-radiolabeled substrate RNA was incubated with Dicer-1, then the samples, in a polystyrene microtiter plate without the lid, were cross linked for 2 min on ice with 254-nm light (Spectroline ENF-240C). The light source was placed directly on top of the microtiter plate. Binding was for 20 min at 4°C with 5 mM EDTA to prevent substrate cleavage. After cross linking, the samples were resolved by SDS-PAGE. Gels were dried and exposed to image plates and analyzed with an FLA-9000 and ImageGauge 3.0 software (Fujifilm).
Statistical analyses
The two-tailed Student's t-test was used for all statistical analyses.
Accession numbers
The SRA accession number for the small RNA libraries reported in this paper is SRX337129.
Acknowledgments
We thank Richard Carthew and the Bloomington Stock Center for fly strains, Alicia Boucher and Cindy Tipping for help with fly husbandry; and members of the Zamore laboratory for help, discussions, advice, and comments on the manuscript. We thank the UMMS Deep Sequencing Core for technical assistance. This work was supported in part by grants from the National Institutes of Health to PDZ (GM62862 and GM65236) and a JSPS Research Fellowship for Research Abroad and a Charles A. King Trust Postdoctoral Fellowship to RF.
Author contributions
RF and PDZ conceived and designed the experiments. RF and CC performed the experiments and analyzed the data. RF and BWH performed bioinformatics analysis. RF and PDZ wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting Information
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Legends
Review Process File
References
- Ameres SL, Hung JH, Xu J, Weng Z, Zamore PD. Target RNA-directed tailing and trimming purifies the sorting of endo-siRNAs between the two Drosophila Argonaute proteins. RNA. 2011;17:54–63. doi: 10.1261/rna.2498411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik ES, Fukunaga R, Lu G, Dutcher R, Wang Y, Tanaka Hall TM, Zamore PD. Phosphate and R2D2 restrict the substrate specificity of Dicer-2, an ATP-Driven Ribonuclease. Mol Cell. 2011;42:172–184. doi: 10.1016/j.molcel.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L, Mian N, Bateman A. Domains in gene silencing and cell differentiation proteins: the novel PAZ domain and redefinition of the Piwi domain. Trends Biochem Sci. 2000;25:481–482. doi: 10.1016/s0968-0004(00)01641-8. [DOI] [PubMed] [Google Scholar]
- Forstemann K, Horwich MD, Wee L, Tomari Y, Zamore PD. Drosophila microRNAs are sorted into functionally distinct Argonaute complexes after production by Dicer-1. Cell. 2007;130:287–297. doi: 10.1016/j.cell.2007.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukunaga R, Han BW, Hung JH, Xu J, Weng Z, Zamore PD. Dicer partner proteins tune the length of mature miRNAs in flies and mammals. Cell. 2012;151:533–546. doi: 10.1016/j.cell.2012.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartig JV, Förstemann K. Loqs-PD and R2D2 define independent pathways for RISC generation in Drosophila. Nucleic Acids Res. 2011;39:3836–3851. doi: 10.1093/nar/gkq1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvágner G, McLachlan J, Pasquinelli AE, Balint É, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ye X, Liu X, Fincher L, McKearin D, Liu Q. Dicer-1 and R3D1-L catalyze microRNA maturation in Drosophila. Genes Dev. 2005;19:1674–1679. doi: 10.1101/gad.1334005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko H, Dridi S, Tarallo V, Gelfand BD, Fowler BJ, Cho WG, Kleinman ME, Ponicsan SL, Hauswirth WW, Chiodo VA, Kariko K, Yoo JW, Lee DK, Hadziahmetovic M, Song Y, Misra S, Chaudhuri G, Buaas FW, Braun RE, Hinton DR, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura Y, Saito K, Kin T, Ono Y, Asai K, Sunohara T, Okada TN, Siomi MC, Siomi H. Drosophila endogenous small RNAs bind to Argonaute2 in somatic cells. Nature. 2008;453:793–797. doi: 10.1038/nature06938. [DOI] [PubMed] [Google Scholar]
- Lau PW, Guiley KZ, De N, Potter CS, Carragher B, MacRae IJ. The molecular architecture of human Dicer. Nat Struct Mol Biol. 2012;19:436–440. doi: 10.1038/nsmb.2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature. 2003;426:465–469. doi: 10.1038/nature02123. [DOI] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, Sattler M. Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat Struct Mol Biol. 2004;11:576–577. doi: 10.1038/nsmb777. [DOI] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X. R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science. 2003;301:1921–1925. doi: 10.1126/science.1088710. [DOI] [PubMed] [Google Scholar]
- Lu WP, Fei L. A logarithmic approximation to initial rates of enzyme reactions. Anal Biochem. 2003;316:58–65. doi: 10.1016/s0003-2697(03)00034-4. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, Patel DJ. Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature. 2004;429:318–322. doi: 10.1038/nature02519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- MacRae IJ, Zhou K, Li F, Repic A, Brooks AN, Cande WZ, Adams PD, Doudna JA. Structural basis for double-stranded RNA processing by Dicer. Science. 2006;311:195–198. doi: 10.1126/science.1121638. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC. Joining of RNAs by splinted ligation. Methods Enzymol. 2000;317:109–123. doi: 10.1016/s0076-6879(00)17009-0. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Nishida KMA, Miyoshi K, Ogino A, Miyoshi T, Siomi H, Siomi MS. Roles of R2D2, a cytoplasmic D2 body component, in the endogenous siRNA pathway in Drosophila. Mol Cell. 2003;49:680–691. doi: 10.1016/j.molcel.2012.12.024. [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Okamura K, Balla S, Martin R, Liu N, Lai EC. Two distinct mechanisms generate endogenous siRNAs from bidirectional transcription in Drosophila melanogaster. Nat Struct Mol Biol. 2008a;15:581–590. doi: 10.1038/nsmb.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008b;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature. 2011;475:201–205. doi: 10.1038/nature10198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham JW, Pellino JL, Lee YS, Carthew RW, Sontheimer EJ. A Dicer-2-dependent 80s complex cleaves targeted mRNAs during RNAi in Drosophila. Cell. 2004;117:83–94. doi: 10.1016/s0092-8674(04)00258-2. [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O. Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J. 2002;21:5864–5874. doi: 10.1093/emboj/cdf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose SD, Kim DH, Amarzguioui M, Heidel JD, Collingwood MA, Davis ME, Rossi JJ, Behlke MA. Functional polarity is introduced by Dicer processing of short substrate RNAs. Nucleic Acids Res. 2005;33:4140–4156. doi: 10.1093/nar/gki732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Liu J, Tolia NH, Schneiderman J, Smith SK, Martienssen RA, Hannon GJ, Joshua-Tor L. The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nat Struct Biol. 2003;10:1026–1032. doi: 10.1038/nsb1016. [DOI] [PubMed] [Google Scholar]
- Srikantan S, Marasa BS, Becker KG, Gorospe M, Abdelmohsen K. Paradoxical microRNAs: individual gene repressors, global translation enhancers. Cell Cycle. 2011;10:751–759. doi: 10.4161/cc.10.5.14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarallo V, Hirano Y, Gelfand BD, Dridi S, Kerur N, Kim Y, Cho WG, Kaneko H, Fowler BJ, Bogdanovich S, Albuquerque RJ, Hauswirth WW, Chiodo VA, Kugel JF, Goodrich JA, Ponicsan SL, Chaudhuri G, Murphy MP, Dunaief JL, Ambati BK, et al. DICER1 loss and Alu RNA induce age-related macular degeneration via the NLRP3 inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomari Y, Matranga C, Haley B, Martinez N, Zamore PD. A protein sensor for siRNA asymmetry. Science. 2004;306:1377–1380. doi: 10.1126/science.1102755. [DOI] [PubMed] [Google Scholar]
- Tsutsumi A, Kawamata T, Izumi N, Seitz H, Tomari Y. Recognition of the pre-miRNA structure by Drosophila Dicer-1. Nat Struct Mol Biol. 2011;18:1153–1158. doi: 10.1038/nsmb.2125. [DOI] [PubMed] [Google Scholar]
- Welker NC, Maity TS, Ye X, Aruscavage PJ, Krauchuk AA, Liu Q, Bass BL. Dicer's helicase domain discriminates dsRNA termini to promote an altered reaction mode. Mol Cell. 2011;41:589–599. doi: 10.1016/j.molcel.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan KS, Yan S, Farooq A, Han A, Zeng L, Zhou MM. Structure and conserved RNA binding of the PAZ domain. Nature. 2003;426:468–474. doi: 10.1038/nature02129. [DOI] [PubMed] [Google Scholar]
- Ye X, Paroo Z, Liu Q. Functional anatomy of the Drosophila microRNA-generating enzyme. J Biol Chem. 2007;282:28373–28378. doi: 10.1074/jbc.M705208200. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Jaskiewicz L, Westhof E, Filipowicz W. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Figure S5
Supplementary Table S1
Supplementary Table S2
Supplementary Table S3
Supplementary Table S4
Supplementary Legends
Review Process File