Abstract
Eukaryotic DNA replication initiates from multiple replication origins. To ensure each origin fires just once per cell cycle, initiation is divided into two biochemically discrete steps: the Mcm2-7 helicase is first loaded into prereplicative complexes (pre-RCs) as an inactive double hexamer by the origin recognition complex (ORC), Cdt1 and Cdc6; the helicase is then activated by a set of “firing factors.” Here, we show that plasmids containing pre-RCs assembled with purified proteins support complete and semi-conservative replication in extracts from budding yeast cells overexpressing firing factors. Replication requires cyclin-dependent kinase (CDK) and Dbf4-dependent kinase (DDK). DDK phosphorylation of Mcm2-7 does not by itself promote separation of the double hexamer, but is required for the recruitment of firing factors and replisome components in the extract. Plasmid replication does not require a functional replication origin; however, in the presence of competitor DNA and limiting ORC concentrations, replication becomes origin-dependent in this system. These experiments indicate that Mcm2-7 double hexamers can be precursors of replication and provide insight into the nature of eukaryotic DNA replication origins.
Keywords: DNA replication in vitro, DNA replication origins
See also: J Gros et al (March 2014)
Introduction
During each eukaryotic cell cycle, the entire complement of genomic DNA must be faithfully copied before chromosome segregation occurs. DNA replication involves the selection of potential initiation sites (replication origins), assembly of replication machinery at these sites and activation of this machinery. In budding yeast, origins are well-defined, specific DNA sequences, whilst in early metazoan embryos virtually any DNA sequence can support DNA replication. In mammalian cells, there is evidence that initiation is non-random, though the nature of replication origins is still the subject of intense investigation (discussed in Mechali, 2010). One of the key steps in determining sites of replication origins is the binding of the origin recognition complex (ORC). ORC, with two additional factors, Cdc6 and Cdt1, loads the Mcm2-7 replicative helicase at replication origins as a double hexamer bound around double-stranded DNA. This process, known as origin licensing or prereplicative complex (pre-RC) assembly, can only occur during late mitosis through G1 phase, when CDK activity is low (Bell & Dutta, 2002; Blow & Dutta, 2005; Masai et al, 2010; Mechali, 2010; Tanaka & Araki, 2010; Boos et al, 2012).
Upon entry into S phase, increase in the activities of CDK and DDK promotes the activation of licensed origins. These kinases, together with other firing factors, convert each inactive Mcm2-7 double hexamer into two Cdc45•Mcm2-7•GINS (CMG) complexes each containing a hexamer of Mcm2-7 which encircles ssDNA on the leading strand template (Moyer et al, 2006; Pacek et al, 2006; Ilves et al, 2010; Fu et al, 2011). Some firing factors, including Dpb11 and the key CDK substrates Sld2 and Sld3, are required for initiation but not elongation, whilst others, like Cdc45, GINS and the leading strand DNA polymerase ε, are required for initiation and then form part of the elongation machinery (Tanaka & Araki, 2010).
From the onset of S phase, through G2 and M phases, CDK plays a second crucial role in inhibiting pre-RC assembly until the next G1 phase (Diffley, 2004). This dual role for CDK is not only crucial for ensuring origins fire just once per cell cycle, but also has an important implication for the biochemical reconstitution of initiation: because origin licensing requires the absence of CDK activity but origin firing requires high CDK activity, these two reactions cannot be performed in a single, one-step reaction.
The mechanism of helicase activation and the steps involved in replisome assembly are still poorly understood in eukaryotes. The ability to reconstitute replication with purified proteins would provide a powerful approach to study this process. Pre-RC assembly has been reconstituted in vitro with purified components (Gillespie et al, 2001; Kawasaki et al, 2006; Evrin et al, 2009; Remus et al, 2009); however, helicase activation has not. Xenopus laevis egg extracts can replicate exogenous DNA (Blow & Laskey, 1986; Walter et al, 1998), and this system has been important for understanding replication mechanism and control. However, reconstitution of replication with purified proteins has thus far not been accomplished. Heller et al described a system in which pre-RCs assembled in an extract from G1-arrested Saccharomyces cerevisiae cells could be transferred to an S-phase extract and activated (Heller et al, 2011). This was an important development and has already provided insights into helicase activation. However, the relationship between the pre-RCs assembled in the G1 extract and those assembled from purified proteins is unclear. Moreover, input plasmids are only partially replicated in this system. This may be because factors required for elongation are limiting in the extract or because attachment of the input plasmid to beads somehow impedes replisome progression. Attachment of the DNA to beads allows removal of pre-RCs from the G1 extract, which contains inhibitors of origin firing including the CDK inhibitor Sic1, before transferring to the S-phase extract. In this paper, we show that pre-RCs assembled on plasmid DNA with purified proteins can effectively replace the G1 extract. Consequently, attachment to beads is not required. We show that input plasmids are replicated completely in this system, resulting in covalently closed, circular products indicating that elongation is not limiting. Whilst pre-RC assembly is strictly dependent on the presence of a functional yeast origin in G1 extracts (Seki & Diffley, 2000; Bowers et al, 2004), pre-RC assembly with purified proteins has been shown to have significantly relaxed specificity (Remus et al, 2009). Here, we show that this relaxed specificity is reflected in DNA replication: plasmids with or without a functional yeast origin replicate with similar efficiencies in this system. However, we show that origin specificity can be conferred by reducing ORC concentration and including competitor DNA during pre-RC assembly. We discuss the implications of this on our understanding of eukaryotic replication origins.
Results
DDK phosphorylation of the Mcm2-7 double hexamer with purified proteins
After reconstituting pre-RC assembly with purified proteins (Remus et al, 2009), we next set out to reconstitute the phosphorylation of this pre-RC by DDK. Recombinant DDK was expressed and purified in two different protein expression systems: baculovirus-infected Hi5 insect cells and Saccharomyces cerevisiae. DDKs from both sources were similarly efficient and were used interchangeably. Purified DDK was phosphorylated on both Dbf4 and Cdc7 subunits, as evidenced by the increase in mobility of both polypeptides in SDS–PAGE after treatment with λ protein phosphatase (Fig 1A, lanes 1 and 2). The purified, dephosphorylated DDK appears to be a more active kinase since it was better at phosphorylating a fragment of Mcm2 (Supplementary Fig S1) and resulted in more extensive phosphorylation of full-length Mcm4 and Mcm6 in the context of the pre-RC as evidenced by the more pronounced shift in SDS–PAGE (Fig 1B, compare lanes 2 and 4). Fig 1C shows that Mcm4 and 6 in the double hexamer could be extensively phosphorylated with this kinase, and the maximum amount of kinase (100 nM) in this experiment was used in all subsequent experiments.
Figure 1.

- Recombinant DDK was phosphorylated on both subunits when expressed in baculovirus-infected Hi5 insect cells (lanes 1 and 2). DDK in lane 2 was treated with lambda phosphatase. Protein samples were analysed by SDS–PAGE and Coomassie staining.
- Dephosphorylated DDK was more active than the phosphorylated form in phosphorylating Mcm2-7 complexes. Pre-RCs reconstituted with purified proteins were subjected to phosphorylation by either the phosphorylated (lanes 4 and 5) or the dephosphorylated (lanes 2 and 3) DDK. Samples were analysed by SDS–PAGE and immunoblotting of the indicated Mcm2-7 subunits.
- Mcm2-7 complexes were quantitatively phosphorylated by purified DDK in vitro. Increasing concentrations of dephosphorylated DDK (0, 25, 50, 100 nM) were used to phosphorylate a constant amount of Mcm2-7 complexes assembled into pre-RCs. Samples were analysed by SDS–PAGE and Western blotting of the indicated Mcm2-7 subunits.
- DDK phosphorylation did not result in Mcm2-7 double hexamer dissociation. Representative micrographs of Mcm2-7 double hexamers, either untreated or DDK-treated, analysed by electron microscopy after negative staining.
- 2D-class averages and surface representations of the 3D reconstruction and cylindrically averaged 3D reconstructions of untreated and DDK-treated MCM double hexamers. Contoured central sections of side views of the cylindrically averaged MCM double hexamers. Left: untreated; centre, DDK-treated; and right, superimposition of untreated and DDK-treated sections. Brackets mark the N-terminal tiers and the C-terminal AAA+-containing outer tiers of the MCM complex. The circle (dashed line) indicates the location of slight conformational changes around the pore of the C-terminal ring.
The majority of DDK phosphorylation sites in the Mcm2-7 complex are in the N-termini of Mcm2, 4 and 6 (Randell et al, 2010). Moreover, mcm mutants that bypass DDK requirement have been identified in the N-termini of Mcm4 and 5 (Hardy et al, 1997; Sheu & Stillman, 2006, 2010). Given that the two hexamers in the pre-RC interact stably via their N-termini, we considered that DDK might function by promoting separation of the Mcm2-7 double hexamer, presumably a critical step in helicase activation (see Boos et al, 2012 for discussion). To examine this, pre-RCs were assembled on 1-kb linear DNA and either mock-treated or phosphorylated with the highest amount of DDK described in Fig 1C. DNA-bound Mcm2-7 complexes were released from the beads by EcoRI digestion as previously described (Remus et al, 2009) and applied directly onto carbon-coated copper grids for single particle EM analysis after negative staining. Fig 1D shows that Mcm2-7 double hexamers were readily observed in both the treated and untreated samples and, more significantly, Mcm2-7 single hexamers were not found in either sample. This indicates that DDK phosphorylation was not sufficient to cause efficient dissociation of the Mcm2-7 double hexamers despite the extensive phosphorylation of virtually all the Mcm4 and 6. This is consistent with work from Gambus et al who showed that chromatin-bound Mcm2-7 complexes, which appeared by gel filtration to be double hexamers, were not converted to a smaller form after DDK treatment in a Xenopus egg extract (Gambus et al, 2011). To examine whether these DDK-treated Mcm2-7 double hexamers had undergone conformational change, class averages (Fig 1E, left), 3D reconstructions (Fig 1E, centre) and cylindrically averaged 3D volumes (Fig 1E, right) derived from the untreated and DDK-phosphorylated Mcm2-7 double hexamers were compared. Although relatively small data sets were used, it is clear that, aside from a potential small conformational change in the C-terminal region of the double hexamer (Fig 1E, lower panel), DDK did not induce major structural rearrangements. Thus, we have no evidence that DDK phosphorylation induces dissociation of the Mcm2-7 double hexamer.
Recruitment of firing factors from an S-Phase extract
We generated extracts from S-phase cells using a strategy similar to that described by Heller et al (2011) (see Fig 2A). Briefly, firing factors (Dpb11, Sld2, Sld3, Sld7 and Cdc45) were overexpressed from galactose-inducible promoters and cells were synchronised in G1 phase with alpha factor. After release from alpha factor at 37°C, the cdc7 temperature-sensitive mutation in the strain induced arrest of cells in an S-phase-like state (Fig 2A). Fig 2B shows that the overexpressed firing factors were indeed present at elevated levels in both the G1-and S-phase whole-cell extracts [compare lanes 1 and 2 with lanes 3 and 4—note that antibodies against Cdc45, Dpb11, Sld2 and Sld3 were unable to detect the endogenous proteins from either α factor-or nocodazole-arrested cells (Fig 2B, lanes 1 and 2) but readily detected the overproduced proteins (lanes 3 and 4)]. Both Orc6 and Sld2, two important S-CDK substrates (Nguyen et al, 2001; Masumoto et al, 2002; Wilmes et al, 2004), migrated more slowly in SDS–PAGE in the S-phase extract compared to the equivalent extract made from cells held in G1 phase. Moreover, Sic1, which is targeted for degradation by CDK phosphorylation at the end of G1 phase, was present in the G1 extract, but absent from the S-phase extract. These results suggest that cells used to make the S-phase extract were not significantly contaminated with G1 cells, and this extract contains sufficient CDK activity to maintain the phosphorylation of key CDK substrates.
Figure 2.

- Schematic diagram showing the synchronisation procedures of the S-phase whole-cell extract preparation from the strain yKO3.
- Cell lysates from the background strain W303 arrested either in G1 phase by α-factor (lane 1) or M phase by nocodazole (lane 2) and yKO3 strains arrested either in S phase (lane 3) or G1 phase by α-factor (lane 4) were analysed by SDS–PAGE and Western blotting of the indicated proteins.
- Schematic diagram showing the procedures of the replisome assembly reaction.
- DDK-dependent recruitment of all overproduced firing factors and the endogenous Psf1 protein to the reconstituted pre-RCs. Samples were analysed by SDS–PAGE and immunoblotting of the indicated proteins.
Next, we examined the recruitment of firing factors to pre-RCs in the S-phase extract. Briefly, pre-RCs were assembled from purified proteins on DNA attached to paramagnetic beads, either treated with DDK or not, and then incubated in the S-phase extract (Fig 2C). Bound proteins were identified by immunoblotting after washing the beads. As shown in Fig 2D, overproduced firing factors were recruited to the reconstituted pre-RCs in a DDK-dependent manner. Moreover, Psf1, a subunit of GINS which was not overexpressed in the extract, was also recruited specifically to the DDK-treated pre-RCs (Fig 2D, compare lanes 2 and 3). Detailed proteomic analysis of proteins bound to pre-RCs is presented in a later section.
Characterisation of DNA replication products
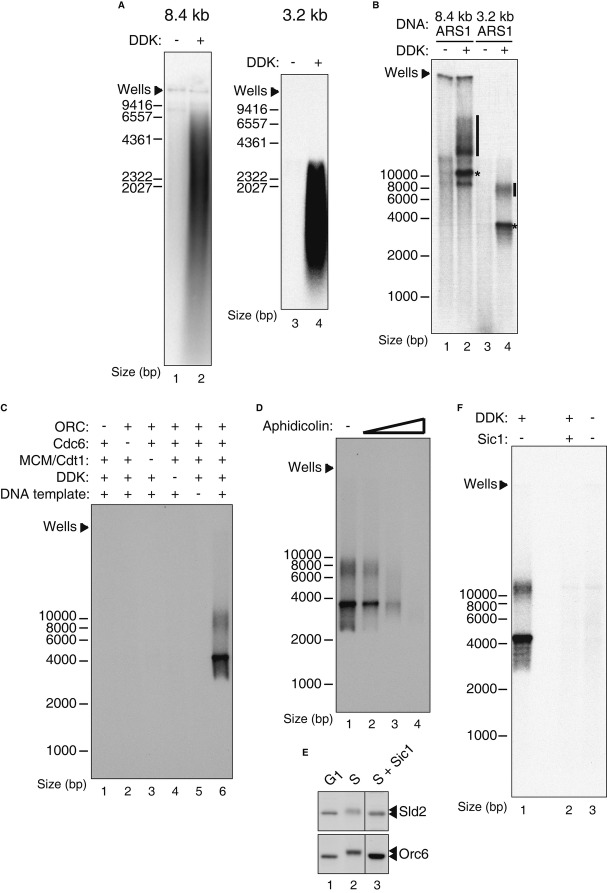
To determine whether recruitment of firing factors was an indication that these extracts could support DNA replication, pre-RCs were assembled on two different ARS1-containing plasmids, pBS/ARS1WTA (3.2 kb) (Marahrens & Stillman, 1992) and YRp14/CEN4/ARS1 (8.4 kb) (Hieter et al, 1985) attached to paramagnetic beads and incubated with the S-phase extract in the presence of nucleoside triphosphates (NTPs) and deoxynucleoside triphosphates (dNTPs) containing α32P-dCTP. Products were run on an alkaline agarose gel, and the gel was dried and subjected to autoradiography (Fig 3A). This experiment shows that DDK-dependent DNA synthesis does indeed occur in this extract and the products resolve as a smear of radioactivity (Fig 3A). As observed by Heller et al, the length of replicated products from reactions using bead-coupled DNA templates was relatively short (Heller et al, 2011) and substantial amounts of full-length products were not observed with either template plasmid. These results indicate that purified pre-RCs can replace the G1 extract for DNA replication in the system described by Heller et al.
Figure 3.
- DDK-dependent in vitro DNA synthesis. Bead-coupled DNA templates (8.4 kb: YRp14/CEN4/ARS1; 3.2 kb: pBS/ARS1 WTA) were used for pre-RC assembly with purified proteins. Reconstituted pre-RCs were then either mock-treated or treated with purified DDK and incubated with the S-phase extract and 32P-α-dCTP. Samples were analysed by denaturing agarose gel electrophoresis and autoradiography.
- In vitro DNA synthesis reactions performed with soluble plasmid DNA of the indicated sizes (*, migration position of unlabelled, relaxed plasmid; bar, replication intermediates).
- Mcm2-7 loading-, DDK-and plasmid DNA-dependent in vitro DNA synthesis. DNA synthesis reaction was performed with soluble plasmid DNA (pBS/ARS1 WTA) instead of bead-coupled DNA. Reactions were performed in the presence or absence of the indicated factors. Samples were analysed by native agarose gel electrophoresis and autoradiography.
- B-family replicative DNA polymerase-dependent in vitro DNA synthesis. DNA synthesis reactions were performed in the presence of an increasing concentration of aphidicolin (0, 0.77, 2.33, 7 μg/ml).
E, F CDK-dependent in vitro DNA synthesis. S-phase extract was either mock-treated (lane 2) or incubated with purified, recombinant Sic1 protein (320 nM) (lane 3) at 4°C for 20 min (E). Samples were analysed by SDS–PAGE and Western blotting of the indicated proteins. Mock-treated and Sic1-treated S-phase extracts from (E) were used in the DNA synthesis reactions (F). Samples were analysed by native agarose gel electrophoresis and autoradiography.
We reasoned that attachment to beads should not be required in our system since pre-RCs assembled with purified proteins should not contain S-phase inhibitors. To test this, we assembled pre-RCs in solution on these plasmids, added DDK and incubated for 30 min. The S-phase extract was then added along with nucleotides as described above, and after further 45 min, reactions were stopped and plasmid DNA was purified and subjected to neutral agarose electrophoresis. Fig 3B shows that both plasmids incorporated labelled nucleotide, and in both cases, this incorporation was DDK-dependent. Quantification of 32P incorporation indicates that the amount of DNA synthesis in these reactions corresponds to only a few per cent of the input plasmid under these conditions (Supplementary Fig S2), indicating that replication is relatively inefficient. However, most of the labelled plasmid ran as discrete bands roughly at the position of the unlabelled relaxed/nicked plasmid (asterisks in Fig 3B), suggesting that they are completely replicated. With both plasmids, there was also some incorporation into products which ran more slowly than relaxed/nicked plasmid (bars in Fig 3B), which might be theta replication intermediates (RIs) like those seen during SV40 replication, for example (Li & Kelly, 1984; Stillman et al, 1985). In Fig 3C, we examined some of the requirements for DNA synthesis. This experiment shows that generation of the putative complete products and RIs required ORC, Cdc6, Mcm2-7•Cdt1 and template DNA as well as DDK. This shows that pre-RCs assembled prior to addition of the S-phase extract are required for replication and that ORC, Cdc6 or Mcm2-7•Cdt1 from the S-phase extract cannot support replication. DNA synthesis was also completely inhibited by aphidicolin, an inhibitor of the three B-family replicative DNA polymerases (Fig 3D).
Replication initiation in vivo requires CDK, which phosphorylates the firing factors Sld2 and Sld3 and triggers origin firing (Masumoto et al, 2002; Tanaka et al, 2007; Zegerman & Diffley, 2007). Unlike DDK, which we add exogenously, CDK activity is already present and CDK targets are already phosphorylated in the S-phase extract (Fig 2B). Preincubation of recombinant Sic1 protein (Supplementary FigS3) in the extract for 20 min at 4°C resulted in an increase in the migration of both Sld2 and Orc6 in SDS–PAGE, relative to the untreated S-phase extract (Fig 3E). This increased migration was similar to that seen in the G1 extract and indicates that Orc6 and Sld2 were dephosphorylated by some phosphatases in the extract. Most importantly, the S-phase extract preincubated with purified Sic1 did not support in vitro DNA synthesis compared to mock-treated extract (Fig 3F), indicating that DNA synthesis also required CDK activity.
A time-course experiment was conducted to examine the kinetics of DNA synthesis in this system, and replicated products were analysed by electrophoresis in a native agarose gel (Fig 4A). Both the complete product and the RIs first appeared 20 min after the S-phase extract was added to the reaction (Fig 4A, lane 3). Both products, along with the topoisomers, continued to accumulate and reached maximal amount at 45 min (Fig 4A, lanes 4 and 5). At later time points, a slight decrease in product amount was observed (Fig 4A, lanes 6–8) perhaps due to degradation of the replicated products.
Figure 4.
Replicated products were full-length and covalently closed molecules.
A, B Replicated products were full-length molecules. A time-course experiment showing the accumulation of replicated products over time. Samples were either (A) analysed by native agarose gel electrophoresis, or (B) EcoRI-treated and analysed by denaturing agarose gel electrophoresis (linear template: linearised, radioactively labelled template DNA).
C Replicated products were covalently closed molecules. Replicated products were analysed by native agarose gel electrophoresis either in the absence (left panel) or in the presence (right panel) of ethidium bromide. Relaxed plasmid DNA was analysed in the same gel to visualise the effect of ethidium bromide intercalation into DNA.
To obtain more information about replication in this system, labelled DNA products were examined by electrophoresis in denaturing alkaline agarose gels. When the replicated plasmids from the time-course experiment were linearised with EcoRI and subjected to electrophoresis in an alkaline gel, the majority of products migrated at the same position as the linearised, end-labelled plasmid template (Fig 4B, compare lanes 3–8 with 11). Thus, the replicated products were predominantly the same length as the full-length template. Products that were not linearised migrated slightly faster than the linearised products (Fig 4B, compare lanes 8 and 10), consistent with the possibility that at least some full-length products were covalently closed DNA molecules which, because they are circular, run differently compared to the linear products.
Reaction products were next analysed by electrophoresis in native agarose gels either with or without the DNA-intercalating agent ethidium bromide (EtBr) to test for the presence of covalently closed circular products. Ethidium ions intercalate into DNA (Waring, 1965), resulting in a decrease in the twist of the DNA molecule. If this DNA is a covalently closed molecule, a decrease in twist must be compensated by an increase in writhe (Shimada & Yamakawa, 1985). Hence, ethidium intercalation in closed circular DNA leads to an increase in positive supercoiling (Crawford & Waring, 1967). The right side of each panel shows the position of various control plasmids in stained agarose gels. Plasmid DNA relaxed by topoisomerase I migrates as a set of topoisomers more slowly than supercoiled plasmids in neutral agarose gels lacking EtBr (Fig 4C, lanes 3 and 4), but these two plasmid forms co-migrate in gels containing EtBr (Fig 4C, lanes 7 and 8). Most of the plasmid DNA synthesised in the extract (excluding RIs) migrated at the position of relaxed or nicked plasmid with a few apparent topoisomers below in gels lacking EtBr (Fig 4C, lane 1). In gels containing EtBr, approximately half of the product continued to migrate at the position of nicked plasmid, whilst the other half co-migrated with supercoiled DNA (Fig 4C, lane 5). The appearance of this latter product is an indication that a significant fraction of the replicated plasmids is covalently closed. It is possible that some of the nicked product arose during purification of the DNA. Alternatively, it is possible that the extract is inefficient in some aspect of termination.
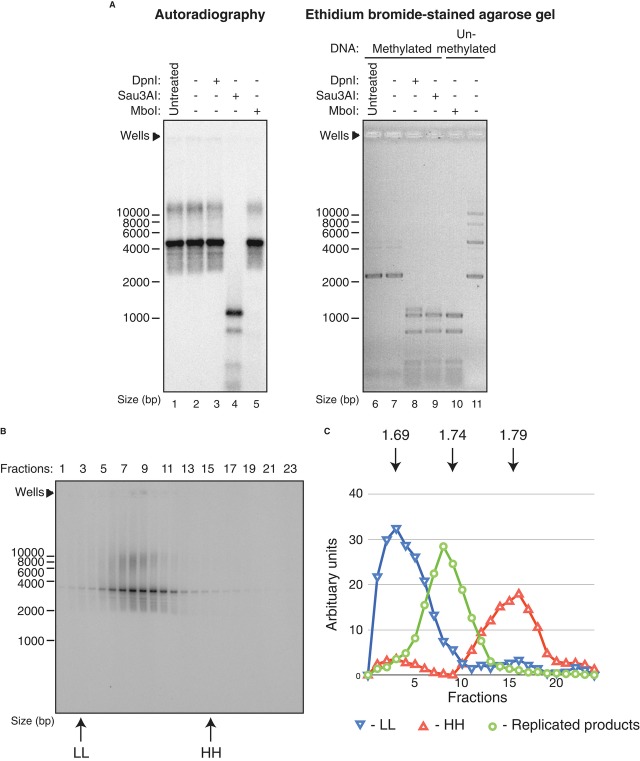
To determine whether the product plasmids have been fully replicated in a semi-conservative manner, we took two approaches. DpnI, Sau3AI and MboI all recognise and cleave the same DNA sequence (GATC), which is represented 16 times across the pBS/ARS1WTA plasmid. These enzymes are distinguished by the effects DNA methylation of adenine (GAmTC) by dam methylase has on their ability to cleave DNA: Sau3AI is insensitive to DNA methylation state, MboI is blocked by methylation on either strand, and DpnI requires methylation on both strands for full activity. Complete semi-conservative replication of our fully methylated input plasmid (as isolated from dam+ E. coli) should generate a fully hemimethylated product, which should be resistant along its entire length to both DpnI and MboI, but should remain sensitive to Sau3AI. As an internal control to show the activity of each of the enzymes, 0.5 μg of starting plasmid purified from either dam+ (DpnI, Sau3AI) or dam− (MboI) E. coli was added to each digestion reaction. The right panel of Fig 5A shows an EtBr-stained gel demonstrating that all three enzymes digested the exogenous control plasmid (Fig 5A lanes 6–11). The left panel shows the autoradiogram from the same gel. From this, it is clear that the labelled products were efficiently digested by Sau3AI (Fig 5A lane 4), but were not digested at all by DpnI or MboI (Fig 5A, lanes 3 and 5). Resistance to DpnI shows that all GATC sequences have lost full methylation; resistance to MboI shows that none of the GATC sequences have become fully unmethylated. Therefore, all of the GATC sequences in the labelled products appear to be hemimethylated, consistent with full semi-conservative replication. The lack of fully unmethylated GATC sequence also indicates that none of the replicated plasmids are the products of re-replication events.
Figure 5.
DNA synthesised in vitro were products of semi-conservative DNA replication.
A In vitro synthesised DNA products were resistant to DpnI digestion. Radioactive reaction products were mock-treated (lane 2) or treated with the indicated restriction enzymes (lanes 3–5) (left panel). Unlabelled, supercoiled plasmid DNA present in the reactions served as internal control for the activity of the restriction enzymes (right panel).
B, C Semi-conservative DNA synthesis in vitro. Radioactive reaction products were analysed by centrifugation in a CsCl gradient (B). Fractions were collected from the top of the gradient and analysed by native agarose gel electrophoresis and autoradiography (arrows indicate fractions at which control DNA peak at. LL, Light-Light; HH, Heavy-Heavy). Signal intensities of the radioactive products and control DNA were quantified and plotted in (C) to indicate the peaks of each DNA population (arrows indicate the refractive indexes of the fractions).
To more directly examine this, replication products were generated with α32P-dCTP and BrdUTP. BrdU base pairs with adenine, but is heavier than thymine, and thus generates a denser product. Replication products were subjected to equilibrium centrifugation in CsCl, and fractions from the gradient were analysed by agarose gel electrophoresis (Fig 5B; quantification of incorporation in Fig 5C). This experiment shows that the replication products have a hybrid density (1.74 g/ml) halfway between fully light DNA (LL: 1.69 g/ml) and fully substituted heavy (HH: 1.79 g/ml) DNA. Consistent with the restriction enzyme analysis above, HH DNA was not detected indicating little or no re-replication. From these experiments, we conclude that the replication products generated in this reaction are full-length molecules in which one strand has been entirely synthesised in the extract.
Protein composition of replicating molecules
To determine the components of replication complexes more completely, we used mass spectrometry to identify proteins with intensity-based absolute quantification (iBAQ) (Schwanhausser et al, 2011) to measure the relative abundance of proteins recruited to pre-RCs under a variety of conditions (Supplementary Table S1). Fig 6A–C shows identical graphs comparing iBAQ values (log10-transformed) of proteins bound to pre-RCs with (y-axis) and without (x-axis) DDK treatment, each highlighting a different subset of replication factors. Because the iBAQ values are log10-transformed, each unit on the y-axis represents a tenfold difference in abundance. Proteins present at similar levels in the two reactions lie along the diagonal line. Mcm2-7 are amongst the most abundant proteins along the diagonal, whilst the other licensing factors (ORC, Cdc6 and Cdt1) are present at approximately 10–100 times lower levels (Fig 6A). Loss of these other licensing factors is likely due to extra washes after DDK phosphorylation and after incubation with the extract. The slightly higher persistence of Cdc6 is likely due to non-specific binding to beads (Remus et al, 2009). Most importantly, the presence of licensing components along the diagonal provides an internal control, indicating that pre-RCs were loaded equally in both samples. Other proteins found along the diagonal include proteins that bind non-specifically to the DNA beads, but may also include factors from the extract that bind pre-RCs independent of DDK. Fig 6B highlights the firing factors. All of the overexpressed firing factors (Dpb11, Sld2, Sld3, Sld7 and Cdc45) as well as the two DDK subunits, which were added exogenously, are enriched in the +DDK reaction (i.e. lie above the diagonal line) and have iBAQ values just slightly below those of Mcm2-7, suggesting that they have been efficiently recruited to the loaded pre-RCs. In addition, the two largest subunits of DNA polymerase ε (Pol2 and Dpb2), which were not overexpressed, as well as the GINS subunit Sld5, were enriched in the DDK-treated sample to a similar level, suggesting that formation of a preinitiation complex (pre-IC) (Zou et al, 1997; Zou & Stillman, 2000) is not limiting in these reactions. Although some firing factors (Dpb2, Pol2, DDK, Cdc45 and Sld3) also appear to be present without DDK, because the y-axis is log10-transformed, each is enriched by at least 60-fold in the presence of DDK. Mcm10 was detected but did not appear to be enriched in the +DDK sample. Reasons for this are unclear: it may reflect DDK-independent association of Mcm10 with Mcm2-7 as seen previously (Wohlschlegel et al, 2002; Ricke & Bielinsky, 2004; van Deursen et al, 2012), or may reflect Mcm10's non-specific DNA binding activity (Robertson et al, 2008; Warren et al, 2008; Eisenberg et al, 2009). Regardless, Mcm10 is present at levels 10-100 times lower than other firing factors, suggesting that this step may be limiting in our system.
Figure 6.
Mass spectrometry analysis of protein composition of replication reactions.
A–C DDK-dependent recruitment of firing factors, RPC components and other replication factors to reconstituted pre-RCs. Note that the y-axis is log10-transformed. Thus, each unit represents a tenfold difference in abundance. Proteins with iBAQ values ≤ 3 were all assigned a value of 3 to simplify the graph.
D Enrichment of DNA-associated polymerases (Pol α and Pol δ) but not RPA under condition of replisome stalling by aphidicolin. In all cases, pre-RCs were incubated with S-phase extract as described in Materials and Methods.
Most of the other components of the replisome progression complex (RPC) (Gambus et al, 2006, 2009) were enriched in the +DDK sample (Fig 6C) including Mrc1, Tof1, Csm3, Ctf4 and the FACT subunit Pob3 as were subunits of DNA polymerase α (Pol1, Pol12, Pri1 and Pri2), DNA polymerase δ (Pol3, Pol31, Pol32), RFC (RFC1,2,3,5) and RPA (RFA1-3). The degree of enrichment of these replisome components was significantly less than the firing factors, suggesting that one or more of these components may be limiting in the extract. All four core histones (HTA1,2, HTB1,2, HHF1 and HHT1) were also partly enriched in the +DDK sample, suggesting that some replication-associated chromatin assembly may be occurring during replication.
Finally, to determine whether any replication or checkpoint proteins were enriched specifically at stalled replication forks, we compared the protein profile in two complete (+DDK, +NTPs and dNTPs) reactions, one unperturbed and one treated with a high concentration of aphidicolin, which blocks DNA synthesis (Fig 3D). Fig 6D shows that, under these conditions, very few proteins are enriched in the +aphidicolin sample (i.e. below the diagonal line). Both DNA polymerases α and δ are slightly enriched in the aphidicolin-treated sample, suggesting that stalling the polymerases may stabilise them at the fork. None of the RPA subunits are enriched under these conditions. This may suggest that significant uncoupling of unwinding and polymerisation does not occur in these extracts, in contrast to the situation in Xenopus egg extracts (Pacek & Walter, 2004; Byun et al, 2005). However, there was quite a high background of RPA binding even in the absence of DDK (Fig 6C), suggesting that most RPA binding is non-specific in these extracts. Thus, further work is required to determine whether or not unwinding and DNA synthesis are tightly coupled in this system.
Origin-dependent and independent replication
Although budding yeast uses specific DNA sequences as origins in vivo, and ORC possesses specific DNA binding activity (Bell & Stillman, 1992), even budding yeast ORC has significant non-specific DNA binding activity in vitro (Remus et al, 2009). This non-specific DNA interaction might be functional: ORC can load Mcm2-7 complexes onto DNA substrates in vitro irrespective of whether a functional replication origin is present or not (Remus et al, 2009). Alternatively, it is possible that Mcm2-7 double hexamers may assemble at non-origin sites as well as origin sites, but only Mcm2-7 double hexamers bound to origins are functional in replication. To begin to test this, we examined replication on pBS/ARS1WTA containing a wild-type ARS1 and pBS/ARS1A−B2−, the same plasmid in which two key origin elements (A and B2) have been replaced with an XhoI linker (Marahrens & Stillman, 1992; Remus et al, 2009). Fig 7A shows that both plasmids replicated equally well in a DDK-dependent manner, indicating that a functional origin is not required for replication in this system. We next tested an entirely heterologous DNA, the bacteriophage lambda chromosome. Fig 7B shows that lambda DNA replicates in a DDK-dependent manner as efficiently as the ARS1-containing plasmid. To determine whether origin-independent replication is simply because we are using excessive levels of ORC, we performed an ORC titration during the pre-RC assembly step (Fig 7C). This experiment shows that replication of the plasmid lacking a functional origin was also ORC dependent and that reducing ORC concentration reduced overall replication, but did not confer origin specificity.
Figure 7.

Origin-independent and dependent DNA replication in vitro.
A, B Replication origin was not required for in vitro DNA replication of the template DNA. Plasmid DNA containing (A) either wild-type ARS1 (WT) or mutant ARS1 (A−B2−) and (B) either wild-type ARS1 plasmid (700 ng) or lambda DNA (700 ng) were used as templates in the in vitro replication reactions. Products were run on neutral agarose gels (A) or quantified by liquid scintillation counting (B).
C Origin-independent in vitro replication was dependent on ORC. Replication reactions were performed as in (A) in the presence of an increasing concentration of ORC (0, 1.3, 4, 13, 40 nM).
D–G Competitor DNA restored origin specificity in the in vitro replication reaction. Replication reactions in (D) were performed as in (A) in the presence of an increasing concentration of the competitor DNA poly(dA-dT) (0, 0.009, 0.028, 0.083 mg/mL). The amount of incorporation was quantified and plotted in (E). Replication reactions in (F) were performed in the presence of poly(dA-dT) (0.009 mg/mL) and increasing concentrations of ORC (0, 4, 13, 40 nM). The amount of incorporation was quantified and plotted in (G).
Stricter origin dependency of Mcm2-7 loading can be enforced by including competitor DNA during pre-RC assembly (Remus et al, 2009). Therefore, we next performed a titration of the non-specific competitor, poly(dA-dT), which was used because it will not interfere with detection of 32P-dCTP incorporation into replicated products. As shown in Fig 7D and E, addition of competitor DNA inhibited the overall reaction but also generated some origin preference (compare lanes 2 and 6). We then repeated the ORC titration in the presence of an intermediate amount of competitor DNA (Fig 7F and G) and found that, at intermediate ORC levels, the origin-containing plasmid was 5–10× more efficient compared to the mutant origin plasmid. Therefore, functional yeast origins are not required for ORC-and DDK-dependent replication in vitro. Moreover, origin specificity can be achieved by manipulating levels of ORC and competitor DNA.
Discussion
We have described a soluble, cell-free DNA replication system to study yeast DNA replication. DNA replication of plasmids in extracts from S-phase cells is semi-conservative, requires preassembly of pre-RCs and results in the formation of fully replicated, covalently closed circular product. Consequently, factors required for elongation and termination do not appear to be lacking from this extract. Using a proteomic approach, all three replicative polymerases as well as virtually all of the other identified components of the RPC (Gambus et al, 2006, 2009) were found specifically associated with template DNA in a DDK-dependent manner, suggesting that our extracts assemble replisomes similar to those generated in vivo. Surprisingly, few proteins other than known replisome components and firing factors were found associated in a DDK-dependent manner (Supplementary Table S1), suggesting that most or all core factors involved in initiation and elongation have been identified. If true, it should be possible to reconstitute this process with this set of purified proteins in future.
Only a small fraction of the input plasmid is replicated in these extracts. Several results suggest that initiation occurs slowly and asynchronously in the extracts. Firstly, the accumulation of fully replicated products is relatively slow, occurring over 30–45 min, yet significant accumulation of replication intermediates does not occur during the time course. This suggests that, once initiated, elongation proceeds relatively rapidly, and thus, the rate-limiting step is initiation rather than elongation. Secondly, although the firing factors Dpb11, Sld2, Sld3 and Sld7 are not believed to play any role in elongation, nor to move with replication forks (Masumoto et al, 2000; Kanemaki & Labib, 2006), we identified them as amongst the most abundant factors associated with pre-RCs after DDK treatment and in fact are almost as abundant as pre-RCs even after 30 min in the presence of NTPs and dNTPs. This suggests that most pre-RCs are converted to a pre-IC-like state but do not proceed efficiently beyond this stage. It may be that some factors involved in the transition to the complete replisome are limiting in the extract. A candidate for such a limiting factor is Mcm10. Several recent reports have indicated that Mcm10 is important for initiation after pre-IC assembly and CMG formation, but before any significant unwinding (Heller et al, 2011; van Deursen et al, 2012; Kanke et al, 2012; Watase et al, 2012). Recruitment of Mcm10 is inefficient in our extracts and occurs at levels similar to most of the elongation factors. Whether it is Mcm10 itself or something required for Mcm10's recruitment which is limiting remains to be determined.
In bacteria and viruses, replication origins are often complex DNA sequences with precisely arranged binding sites for initiator proteins and auxiliary factors and other elements like DNA unwinding elements (DUEs) (Challberg & Kelly, 1989; Kowalski & Eddy, 1989; Stillman, 1989; Leonard & Grimwade, 2010; Skarstad & Katayama, 2013). In yeast, the requirement for multiple DNA sequence elements (Marahrens & Stillman, 1992) might have suggested a similar complex organisation; however, the enzymology of initiation is largely conserved across Eukarya, and replication in other systems, like early Xenopus embryos, does not require specific DNA sequences (Harland & Laskey, 1980). Two of the most important sequence elements in the prototypal yeast origin, ARS1, are the ARS consensus sequence and the B1 element, which both contribute to ORC binding (Rao & Stillman, 1995; Rowley et al, 1995). Moreover, these two elements are sufficient for weak origin activity in vivo (Marahrens & Stillman, 1992), and tethering mammalian ORC to an array of GAL4 DNA binding sites is sufficient to generate a mammalian replication origin (Takeda et al, 2005). This suggests that ORC binding is sufficient to determine a eukaryotic replication origin.
Our results contribute to this idea: complex functional DNA replication origins are not required for DNA replication of naked DNA in our extract system. Instead, replication competence appears to correlate with ORC binding since conditions that promote sequence-specific ORC binding (i.e. reduced ORC concentrations and the presence of competitor DNA) also promote origin dependence. Whilst yeast origin DNA sequences are not required for replication in our system, we do not rule out the possibility that ORC is binding to heterologous sequences with moderate sequence specificity. These heterologous sequences must be sufficiently simple to be present in both bacterial plasmids and lambda bacteriophage DNA. Moreover, we cannot rule out the possibility that some simple DNA sequences may contribute to events downstream of pre-RC assembly, such as origin melting. Given that the Mcm2-7 double hexamer can slide on DNA once loaded (Remus et al, 2009), it is also possible that the actual site of initiation may not correspond to the initial location of ORC binding and pre-RC assembly.
Thus far, the templates we have used for replication have been naked DNA. It is clear from previous work that ORC has important interactions with nucleosomes (Lipford & Bell, 2001; Hizume et al, 2013). Work from MacAlpine and colleagues has shown that yeast origins correlate very strongly with ORC binding sites adjacent to AT-rich sequences which exclude nucleosomes (Eaton et al, 2010). It will be important to reconstitute pre-RCs and subsequent replication on chromatinised templates in future. Regardless, we can make our system dependent on DNA sequences. This should be useful for future work aimed at mapping protein–DNA interactions, identifying initial DNA melting events and mapping the positions of replication initiation at nucleotide resolution.
Materials and Methods
Yeast strains, protein expression vectors, protein expression, antibodies and electron microsocopy
The strains and expression vectors used in this study are listed in Supplementary Materials. Detailed protein purifications, antibodies used and procedures for electron microscopy and single-particle analysis are included in the Supplementary Materials and Methods.
Preparation of S-phase whole-cell extract
For small-scale extracts, 4 L of S. cerevisiae cells (yKO3) were grown in YP-raffinose at 25°C to a density of 1 × 107 cells/ml. Protein expression was induced by adding 2% galactose for 2 h at 25°C. Cells were then arrested in G1 by α-factor for 3 h. Arrested cells were collected and resuspended in YP-galactose (prewarmed to 37°C) and α-factor for 45 min. Next, cells were released from α-factor arrest by three washes in prewarmed YP-galactose. Cells were harvested 45 min after release. For large-scale extracts, 10 L of S. cerevisiae cells (yKO3) was grown in a fermentor in YP-raffinose at 25°C to a density of 1 × 107 cells/ml. Protein expression was induced by adding 2% galactose for 2 h at 25°C. Cells were then shifted from to 37°C, grown for 5 h and harvested. Cell harvest was performed by centrifugation at 2,602 g for 15 min at 4°C. Cell pellet was washed twice with SE buffer I (50 mM Hepes-KOH pH 7.6, 0.3 M K-glutamate, 2 mM EDTA, 0.8 M sorbitol)/3 mM DTT and then resuspended in 0.3 volumes of SE buffer II (100 mM Hepes-KOH pH 7.6, 10 mM Mg(OAc)2, 0.8 M sorbitol)/1.5 M K-glutamate/5 mM DTT/protease inhibitors (Roche) and frozen dropwise in liquid nitrogen. Frozen drops of cells were crushed using a freezer mill (SPEX CertiPrep 6850 Freezer/Mill) with six cycles of 2-min crushing at a rate of 15. Frozen cell powder was transferred to a prechilled centrifuge tube and allowed to thaw completely on ice. The lysate was centrifuged for 1 h at 50,000 rpm at 4°C using a Ti 70 rotor (∼257,000 g). The clear phase was recovered and dialysed against SE buffer III (50 mM Hepes-KOH pH 7.6, 0.3 M K-glutamate, 5 mM Mg(OAc)2, 1 mM EGTA, 1 mM EDTA, 10% glycerol)/3 mM DTT/protease inhibitors (Roche) at 4°C for 3.5 h. The dialysed extract was recovered and centrifuged at 90,000 rpm for 30 min at 4°C using a TLA 100.3 rotor (∼440,000 g). The clear phase was recovered and stored in aliquots at −80°C.
Replisome assembly assay and mass spectrometry
Mcm2-7 loading was performed as described previously on 300 ng bead-coupled plasmid DNA template (8.4 kb) at 25°C for 20 min (Remus et al, 2009). Next, supernatant of the reaction was removed and the beads were resuspended in a reaction mix containing 5× bead buffer (125 mM Hepes-KOH pH 7.6, 50 mM Mg(OAc)2, 0.1% NP-40, 212.5 mM K-glutamate, 25% glycerol), 1 mM DTT, 5 mM ATP, 1 mM spermine (Sigma-Aldrich) and distilled water. The reaction was supplemented with purified DDK and incubated at 25°C with agitation for 15 min. The supernatant of the reaction was removed and beads were resuspended in a reaction mix containing 20× replication buffer (800 mM Hepes-KOH pH 7.6, 160 mM MgCl2), 1 mM DTT, 5 mM ATP, 100 μM dATP/dCTP/dTTP/dGTP (Invitrogen) and 200 μM CTP/GTP/UTP (Invitrogen), 40 mM creatine phosphate (Calbiochem) and 10 μg creatine phosphokinase (Calbiochem). Seven hundred and fifty micrograms of yKO3 S-phase extract were added last. The reaction was then incubated at 25°C with agitation for 20 min. Supernatant of the reaction was removed, followed by 2 washes with low-salt wash buffer (45 mM Hepes-KOH pH 7.6, 300 mM K-glutamate, 5 mM Mg(OAc)2, 1 mM EDTA, 1 mM, 0.02% NP-40, 10% glycerol). Next, beads were resuspended in 40 μl of 1× SDS sample buffer, boiled and analysed by SDS–PAGE and immunoblotting.
For mass spectrometry, experiments were performed as described above with the following modifications: 1,250 μg of extract was used per replisome assembly reaction; DDK phosphorylation step and extract incubation step were both extended to 30 min; and 4 replisome assembly reactions were pooled for each mass spectrometry sample. For the replisome stalling sample, aphidicolin (7 μg/ml) was added 15 min after the extract was added to the DDK reaction. Each SDS–PAGE lane was cut into eight equal-sized pieces and subjected to in-gel trypsin digestion. Peptide extracts were analysed using a nanoACQUITY UPLC (Waters Corporation) coupled to a LTQ-Orbitrap XL (Thermo Fisher Scientific) mass spectrometer via an Advion Biosciences Nanomate. Protein identification and quantification was performed using MaxQuant 1.3.0.5. (Cox & Mann, 2008). The Uniprot S. cerevisiae reference proteome was used as the search database, and the protein false discovery rate was set to 1%. Protein quantification was performed using intensity-based quantification (iBAQ).
In vitro DNA replication assay
Unless otherwise stated, Mcm2-7 loading reactions were performed either on soluble plasmid DNA templates (200 ng) or bead-coupled DNA (300 ng). Purified DDK was then added to the Mcm2-7 loading reaction and incubated at 25°C with agitation for 30 min. Next, a reaction mix containing 20× replication buffer, 1 mM DTT, 5 mM ATP, 100 μM dATP/dTTP/dGTP, 200 μM CTP/GTP/UTP, 5 μCi 32P-α-dCTP, 40 mM creatine phosphate and 10 μg creatine phosphokinase was added to the DDK reaction. Finally, 1,250 μg of yKO3 S-phase extract were added. The reaction was then incubated at 25°C with agitation for 45 min. Replication reactions were quenched with the addition of 20 mM EDTA. Proteins were denatured and digested by adding 0.5% SDS and 20 μg proteinase K into the reaction. This mixture was incubated at 37°C with agitation for 30 min, followed by a 25:24:1 phenol/chloroform/isoamyl alcohol (Invitrogen) extraction step. Unincorporated radioactive nucleotides were then removed by passing the aqueous layer through an Illustra MicroSpin S-400 HR column (GE Healthcare) according to the manufacturer's instructions. Finally, RNA was digested by incubating the sample with two units of RNase A (Sigma, molecular biology grade) at 30°C for 15 min. Samples were then analysed by native or denaturing agarose gel electrophoresis and autoradiography.
Radioactive products from the in vitro replication assay were analysed in a CsCl gradient as described previously (Walter et al, 1998). Fractions were collected from the top of the gradient after centrifugation. Non-radioactive control DNA was generated by PCR with either dTTP (Light-Light) or BrdUTP (Heavy-Heavy). The resultant DNA products were mixed and separated by CsCl gradient. Locations of the control DNA were determined by ethidium bromide staining, whereas the positions of the radioactive replicated products were detected by autoradiography.
Acknowledgments
We are grateful to Dirk Remus for communicating results prior to publication, to Karim Labib, Boris Pfander and Philip Zegerman for antibodies, to Stephen Sweet for early characterisation of DDK and to members of the Diffley laboratory for discussion. This work was funded by Cancer Research UK and by a European Research Council Advanced grant to JD (249883—EUKDNAREP) and a Boehringer Ingelheim Ph.D. Fellowship to KFO.
Author contributions
KO and JD devised all experiments. Mass spectrometry was performed and analysed by DF and AS; electron microscopy was performed and analysed by FB and EM. All authors contributed to manuscript preparation.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information
for this article is available online: http://emboj.embopress.org
References
- Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu Rev Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- Bell SP, Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat Rev Mol Cell Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopus eggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Boos D, Frigola J, Diffley JFX. Activation of the replicative DNA helicase: breaking up is hard to do. Curr Opin Cell Biol. 2012;24:423–430. doi: 10.1016/j.ceb.2012.01.011. [DOI] [PubMed] [Google Scholar]
- Bowers JL, Randell JC, Chen S, Bell SP. ATP hydrolysis by ORC catalyzes reiterative Mcm2-7 assembly at a defined origin of replication. Mol Cell. 2004;16:967–978. doi: 10.1016/j.molcel.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challberg MD, Kelly TJ. Animal virus DNA replication. Annu Rev Biochem. 1989;58:671–717. doi: 10.1146/annurev.bi.58.070189.003323. [DOI] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Crawford LV, Waring MJ. Supercoiling of polyoma virus DNA measured by its interaction with ethidium bromide. J Mol Biol. 1967;25:23–30. doi: 10.1016/0022-2836(67)90276-8. [DOI] [PubMed] [Google Scholar]
- van Deursen F, Sengupta S, De Piccoli G, Sanchez-Diaz A, Labib K. Mcm10 associates with the loaded DNA helicase at replication origins and defines a novel step in its activation. EMBO J. 2012;31:2195–2206. doi: 10.1038/emboj.2012.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JFX. Regulation of early events in chromosome replication. Curr Biol. 2004;14:R778–R786. doi: 10.1016/j.cub.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. Conserved nucleosome positioning defines replication origins. Genes Dev. 2010;24:748–753. doi: 10.1101/gad.1913210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg S, Korza G, Carson J, Liachko I, Tye BK. Novel DNA binding properties of the Mcm10 protein from Saccharomyces cerevisiae. J Biol Chem. 2009;284:25412–25420. doi: 10.1074/jbc.M109.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrin C, Clarke P, Zech J, Lurz R, Sun J, Uhle S, Li H, Stillman B, Speck C. A double-hexameric MCM2-7 complex is loaded onto origin DNA during licensing of eukaryotic DNA replication. Proc Natl Acad Sci USA. 2009;106:20240–20245. doi: 10.1073/pnas.0911500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu YV, Yardimci H, Long DT, Guainazzi A, Bermudez VP, Hurwitz J, van Oijen A, Scharer OD, Walter JC. Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell. 2011;146:931–941. doi: 10.1016/j.cell.2011.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of Cdc45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gambus A, Khoudoli GA, Jones RC, Blow JJ. MCM2-7 form double hexamers at licensed origins in Xenopus egg extract. J Biol Chem. 2011;286:11855–11864. doi: 10.1074/jbc.M110.199521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, Calzada A, Labib K. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie PJ, Li A, Blow JJ. Reconstitution of licensed replication origins on Xenopus sperm nuclei using purified proteins. BMC Biochem. 2001;2:15. doi: 10.1186/1471-2091-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy CF, Dryga O, Seematter S, Pahl PM, Sclafani RA. Mcm5/Cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc Natl Acad Sci USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harland RM, Laskey RA. Regulated replication of DNA microinjected into eggs of Xenopus laevis. Cell. 1980;21:761–771. doi: 10.1016/0092-8674(80)90439-0. [DOI] [PubMed] [Google Scholar]
- Heller RC, Kang S, Lam WM, Chen S, Chan CS, Bell SP. Eukaryotic origin-dependent DNA replication in vitro reveals sequential action of DDK and S-CDK kinases. Cell. 2011;146:80–91. doi: 10.1016/j.cell.2011.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hizume K, Yagura M, Araki H. Concerted interaction between origin recognition complex (ORC), nucleosomes and replication origin DNA ensures stable ORC-origin binding. Genes Cells. 2013;18:764–779. doi: 10.1111/gtc.12073. [DOI] [PubMed] [Google Scholar]
- Ilves I, Petojevic T, Pesavento JJ, Botchan MR. Activation of the MCM2-7 helicase by association with Cdc45 and GINS proteins. Mol Cell. 2010;37:247–258. doi: 10.1016/j.molcel.2009.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanemaki M, Labib K. Distinct roles for Sld3 and GINS during establishment and progression of eukaryotic DNA replication forks. EMBO J. 2006;25:1753–1763. doi: 10.1038/sj.emboj.7601063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanke M, Kodama Y, Takahashi TS, Nakagawa T, Masukata H. Mcm10 plays an essential role in origin DNA unwinding after loading of the CMG components. EMBO J. 2012;31:2182–2194. doi: 10.1038/emboj.2012.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki Y, Kim HD, Kojima A, Seki T, Sugino A. Reconstitution of Saccharomyces cerevisiae prereplicative complex assembly in vitro. Genes Cells. 2006;11:745–756. doi: 10.1111/j.1365-2443.2006.00975.x. [DOI] [PubMed] [Google Scholar]
- Kowalski D, Eddy MJ. The DNA unwinding element: a novel cis-acting component that facilitates opening of the Escherichia coli replication origin. EMBO J. 1989;8:4335–4344. doi: 10.1002/j.1460-2075.1989.tb08620.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard AC, Grimwade JE. Regulating DnaA complex assembly: it is time to fill the gaps. Curr Opin Microbiol. 2010;13:766–772. doi: 10.1016/j.mib.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JJ, Kelly TJ. Simian virus 40 DNA replication in vitro. Proc Natl Acad Sci USA. 1984;81:6973–6977. doi: 10.1073/pnas.81.22.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipford JR, Bell SP. Nucleosomes positioned by ORC facilitate the initiation of DNA replication. Mol Cell. 2001;7:21–30. doi: 10.1016/s1097-2765(01)00151-4. [DOI] [PubMed] [Google Scholar]
- Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Masai H, Matsumoto S, You Z, Yoshizawa-Sugata N, Oda M. Eukaryotic chromosome DNA replication: where, when, and how? Annu Rev Biochem. 2010;79:89–130. doi: 10.1146/annurev.biochem.052308.103205. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechali M. Eukaryotic DNA replication origins: many choices for appropriate answers. Nat Rev Mol Cell Biol. 2010;11:728–738. doi: 10.1038/nrm2976. [DOI] [PubMed] [Google Scholar]
- Moyer SE, Lewis PW, Botchan MR. Isolation of the Cdc45/Mcm2-7/GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc Natl Acad Sci USA. 2006;103:10236–10241. doi: 10.1073/pnas.0602400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen VQ, Co C, Li JJ. Cyclin-dependent kinases prevent DNA re-replication through multiple mechanisms. Nature. 2001;411:1068–1073. doi: 10.1038/35082600. [DOI] [PubMed] [Google Scholar]
- Pacek M, Tutter AV, Kubota Y, Takisawa H, Walter JC. Localization of MCM2-7, Cdc45, and GINS to the site of DNA unwinding during eukaryotic DNA replication. Mol Cell. 2006;21:581–587. doi: 10.1016/j.molcel.2006.01.030. [DOI] [PubMed] [Google Scholar]
- Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell JC, Fan A, Chan C, Francis LI, Heller RC, Galani K, Bell SP. Mec1 is one of multiple kinases that prime the Mcm2-7 helicase for phosphorylation by Cdc7. Mol Cell. 2010;40:353–363. doi: 10.1016/j.molcel.2010.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao H, Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc Natl Acad Sci USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remus D, Beuron F, Tolun G, Griffith JD, Morris EP, Diffley JFX. Concerted loading of Mcm2-7 double hexamers around DNA during DNA replication origin licensing. Cell. 2009;139:719–730. doi: 10.1016/j.cell.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke RM, Bielinsky AK. Mcm10 regulates the stability and chromatin association of DNA polymerase-alpha. Mol Cell. 2004;16:173–185. doi: 10.1016/j.molcel.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Robertson PD, Warren EM, Zhang H, Friedman DB, Lary JW, Cole JL, Tutter AV, Walter JC, Fanning E, Eichman BF. Domain architecture and biochemical characterization of vertebrate Mcm10. J Biol Chem. 2008;283:3338–3348. doi: 10.1074/jbc.M706267200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley A, Cocker JH, Harwood J, Diffley JF. Initiation complex assembly at budding yeast replication origins begins with the recognition of a bipartite sequence by limiting amounts of the initiator, ORC. EMBO J. 1995;14:2631–2641. doi: 10.1002/j.1460-2075.1995.tb07261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Seki T, Diffley JFX. Stepwise assembly of initiation proteins at budding yeast replication origins in vitro. Proc Natl Acad Sci USA. 2000;97:14115–14120. doi: 10.1073/pnas.97.26.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. Cdc7-Dbf4 phosphorylates MCM proteins via a docking site-mediated mechanism to promote S phase progression. Mol Cell. 2006;24:101–113. doi: 10.1016/j.molcel.2006.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheu YJ, Stillman B. The Dbf4-Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4. Nature. 2010;463:113–117. doi: 10.1038/nature08647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada J, Yamakawa H. Statistical mechanics of DNA topoisomers. The helical worm-like chain. J Mol Biol. 1985;184:319–329. doi: 10.1016/0022-2836(85)90383-3. [DOI] [PubMed] [Google Scholar]
- Skarstad K, Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol. 2013;5:a012922. doi: 10.1101/cshperspect.a012922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman B. Initiation of eukaryotic DNA replication in vitro. Ann Rev Cell Biol. 1989;5:197–245. doi: 10.1146/annurev.cb.05.110189.001213. [DOI] [PubMed] [Google Scholar]
- Stillman B, Gerard RD, Guggenheimer RA, Gluzman Y. T antigen and template requirements for SV40 DNA replication in vitro. EMBO J. 1985;4:2933–2939. doi: 10.1002/j.1460-2075.1985.tb04026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda DY, Shibata Y, Parvin JD, Dutta A. Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev. 2005;19:2827–2836. doi: 10.1101/gad.1369805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Araki H. Regulation of the initiation step of DNA replication by cyclin-dependent kinases. Chromosoma. 2010;119:565–574. doi: 10.1007/s00412-010-0291-8. [DOI] [PubMed] [Google Scholar]
- Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- Walter J, Sun L, Newport J. Regulated chromosomal DNA replication in the absence of a nucleus. Mol Cell. 1998;1:519–529. doi: 10.1016/s1097-2765(00)80052-0. [DOI] [PubMed] [Google Scholar]
- Waring MJ. Complex formation between ethidium bromide, nucleic acids. J Mol Biol. 1965;13:269–282. doi: 10.1016/s0022-2836(65)80096-1. [DOI] [PubMed] [Google Scholar]
- Warren EM, Vaithiyalingam S, Haworth J, Greer B, Bielinsky AK, Chazin WJ, Eichman BF. Structural basis for DNA binding by replication initiator Mcm10. Structure. 2008;16:1892–1901. doi: 10.1016/j.str.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watase G, Takisawa H, Kanemaki MT. Mcm10 plays a role in functioning of the eukaryotic replicative DNA helicase, Cdc45-Mcm-GINS. Curr Biol. 2012;22:343–349. doi: 10.1016/j.cub.2012.01.023. [DOI] [PubMed] [Google Scholar]
- Wilmes GM, Archambault V, Austin RJ, Jacobson MD, Bell SP, Cross FR. Interaction of the S-phase cyclin Clb5 with an ‘RXL’ docking sequence in the initiator protein Orc6 provides an origin-localized replication control switch. Genes Dev. 2004;18:981–991. doi: 10.1101/gad.1202304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel JA, Dhar SK, Prokhorova TA, Dutta A, Walter JC. Xenopus Mcm10 binds to origins of DNA replication after Mcm2-7 and stimulates origin binding of Cdc45. Mol Cell. 2002;9:233–240. doi: 10.1016/s1097-2765(02)00456-2. [DOI] [PubMed] [Google Scholar]
- Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Stillman B. Assembly of a complex containing Cdc45p, replication protein A, and Mcm2p at replication origins controlled by S-phase cyclin-dependent kinases and Cdc7p-Dbf4p kinase. Mol Cell Biol. 2000;20:3086–3096. doi: 10.1128/mcb.20.9.3086-3096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.