Abstract
The role of MYC proteins in somatic stem and progenitor cells during development is poorly understood. We have taken advantage of a chick in vivo model to examine their role in progenitor cells of the developing neural tube. Our results show that depletion of endogenous MYC in radial glial precursors (RGPs) is incompatible with differentiation and conversely, that overexpression of MYC induces neurogenesis independently of premature or upregulated expression of proneural gene programs. Unexpectedly, the neurogenic function of MYC depends on the integrity of the polarized neural tissue, in contrast to the situation in dissociated RGPs where MYC is mitogenic. Within the polarized RGPs of the neural tube, MYC drives differentiation by inhibiting Notch signaling and by increasing neurogenic cell division, eventually resulting in a depletion of progenitor cells. These results reveal an unexpected role of MYC in the control of stemness versus differentiation of neural stem cells in vivo.
Keywords: Asymmetric division, differentiation, MYC, neural progenitor, Notch
Introduction
The MYC proteins are basic helix-loop-helix/leucine zipper (bHLH-Zip) transcriptional regulators that control a variety of normal cellular functions 1. MYC family members are expressed in multiple organs and, in particular, have been demonstrated to be critical during development of the nervous system 2–4. Mice lacking mycn show general malformations in the central and peripheral nervous systems 5, 6. c-myc-deficient mice demonstrate severe problems in neural tube closure, heart development, and vasculogenesis 7, 8, presenting a phenotype partly similar to mycn-deficient mice. The robust mitogenic effect of MYC in vitro and the reduction in dividing progenitor cells in mutant mice has led to the conclusion that MYC is critical for proliferation 9–11 and that the reduction in differentiated neuronal cell types is secondary 10, 12. Several studies suggest that the neuronal deficits that occur upon MYC deletion are due to insufficient proliferation of neuronal progenitors before they undergo neurogenesis and/or premature differentiation 10–16.
Here, we demonstrate a new proneurogenic function of MYC in embryonic neural stem cells in vivo. Unexpectedly, we found that neurogenic differentiation was markedly perturbed by functional knockdown of MYC. In contrast to the mitogenic MYC activity in vitro, elevated levels of MYC proteins in vivo promote neurogenic cell divisions of radial glial precursors (RGPs) and their cell cycle exit, leading to increased generation of neurons in the developing neural tube.
Results and Discussion
c-MYC and MYCN are mutually exclusively expressed during neural tube development
The temporal and spatial distribution of cells expressing c-myc and mycn mRNAs was analyzed during chick neural tube development (Supplementary Fig S1A–P). We observed mycn expression in neural progenitor cells within the neural tube and in the developing dorsal root ganglia (DRG) by in situ hybridization (ISH) (Supplementary Fig S1A–H). Importantly, mycn mRNA was detected in the ventricular zone (VZ), which is populated by Sox2-expressing RGPs (Supplementary Fig S1Q–S). In contrast, c-myc expression was found in Sox2-negative differentiating neurons of the neural tube and in DRGs (Supplementary Fig S1I–P; T–V). Consistently, a number of c-myc-expressing cells stained positive for the neuronal markers Islet 1 (Isl1) and NeuroM (Supplementary Fig S1Z–ε), whereas mycn-expressing cells were found to be Isl1-negative (Supplementary Fig S1W–Y). Thus, c-myc and mycn are expressed in a mutually exclusive pattern during chicken neural development where mycn is mainly expressed in progenitor cells and c-myc in differentiating neurons.
MYC proteins control the balance between radial glial precursor cells and differentiated neurons
We addressed whether MYC proteins regulate the fate of RGPs in vivo by selective downregulation of MYCN or c-MYC expression using siRNA. The efficiency of downregulation was confirmed by ISH 36 h after electroporation (Supplementary Fig S2). Interestingly, we found that downregulation of MYCN expression resulted in a compensatory ectopic upregulation of c-myc in the ventricular zone (VZ) cells where mycn normally is expressed (Supplementary Fig S2D–F). Therefore, we combined siRNAs against c-MYC and MYCN to downregulate both proteins in loss-of-function experiments (Fig 1A–H). Strikingly, this resulted in a significant reduction in the number of NeuN;GFP double positive (NeuN+; GFP+) differentiated neurons at E4 (Fig 1C, F, G) without affecting proliferation as assessed by EdU incorporation (Fig 1H, Supplementary Fig S2α–β).
Figure 1.

Loss- and gain-of-function experiments reveal a role of MYC in neurogenesis.
A-H Downregulation of c-myc and mycn in the chick neural tube by siRNAs. Transfection of scrambled control siRNA (A-C) or siRNAs against both c-MYC and MYCN (D-F). Transversal chick sections at E4 analyzed for the expression of c-myc (A, D) or mycn (B, E) by in situ hybridization (ISH). NeuN was revealed by immunostaining (C, F). Quantification of the proportion of NeuN+;GFP+ cells in the population of targeted (GFP+) cells (% NeuN+ cells) at E4 (G) and of EdU+;GFP+ cells in the population of targeted (GFP+) cells (% EdU+ cells) at E3 (H), respectively. Data are represented as mean ± s.e.m. of n = 4–8 embryos. ***P < 0.001, t-test.
I Schematic presentation of the MYC and MYCΔC proteins. TAD = transcriptional activation domain; I-III = conserved regions, so called Myc boxes I-III; bHLH-Zip = the basic region helix-loop-helix/leucine zipper.
J-S Overexpression of MYC proteins and their truncated forms in the chick neural tube at E4. Vector control (J, K), c-MYC (L, M), c-MYCΔC (N, O), MYCN (P, Q), or MYCNΔC (R, S) constructs, respectively. ISH revealed expression of GFP (J), c-myc (L, N), and mycn (P, R) at E4, together with immunostaining for NeuN (K, M, O, Q, S).
T Quantification of the proportion of NeuN+;GFP+ cells in the population of targeted (GFP+) cells (% NeuN+) at E4. Data are represented as mean ± s.e.m. of n = 2–7 embryos. *P < 0.01, **P < 0.005, ***P < 0.001, t-test. Scale bar, 50 μm.
Next, we ectopically expressed c-MYC or MYCN and examined neurogenesis after 48 h (E4). Nearly all c-MYC and MYCN over-expressing cells, as revealed by GFP, translocated into the mantle zone of the neural tube and differentiated into neurons, as revealed by staining for neuronal class III β3-tubulin (Tuj1) and NeuN (Fig 1J–M, P-Q, T; Supplementary Fig S3D–M). Strikingly, differentiated neurons never appeared in ectopic locations or prematurely in the VZ. Importantly, we did not observe any differences in distribution or differentiation of cells overexpressing c-MYC or MYCN at E3 compared to control (Supplementary Fig S3A–C, E, G). However, at E4, the increase in the number of differentiated neurons was paralleled by a depletion of MYC-overexpressing cells in the VZ (Fig 1K, M, Q; Supplementary Fig S3I-J, L).
To address whether MYC's effect on neurogenesis requires DNA binding, we generated C-terminal truncated versions lacking the bHLH-Zip of both c-MYC and MYCN (Fig 1I). In contrast to full-length MYC proteins, expression of the MYCΔC mutants led to a marked reduction in differentiated neurons (Fig 1N-O, R-S, T; Supplementary Fig S3F, K, H, M). This was similar to the observed effect with siRNA knockdown of both proteins (Fig 1A–G), indicating that the MYCΔC proteins act in a dominant negative fashion. We suggest that the effect of the mutant proteins is due to squelching of important co-factors, thus inhibiting endogenous MYC proteins to function efficiently in mediating neurogenesis. Importantly, these data show that the neurogenic capability of MYC is mediated by its DNA-binding function.
We next assessed possible effects on apoptosis due to MYC overexpression by TUNEL assays on neural tube sections at E3 and E4 (Supplementary Fig S4). Both c-MYC and MYCN induced apoptosis in targeted as well as in neighboring cells consistent with a recent study 17. Importantly, the apoptosis was neither specific for the Tuj1-negative progenitor population nor for the differentiated Tuj1+ cells. On the other hand, no significant increase in apoptosis was observed despite blocking the proneurogenic activity of endogenous MYC using MYCΔC constructs (Supplementary Fig S4C, H, E, J, K–M). Thus, the opposite phenotypes resulting from overexpressing the wild-type MYC versus the MYCΔC constructs in the neural tube in vivo cannot be explained by the apoptotic activity of MYC.
We then asked whether the increased neurogenesis is caused by premature expression of proneural transcriptional factors, which previously have been shown to promote premature differentiation when mis-expressed 18,19, 20. However, examination of ngn1, ngn2, neurod1, neurod2, or cash1 levels in c-MYC/MYCN and c-MYCΔC/MYCNΔC-transfected neural tubes did not reveal any expressional changes 24 h (Fig 2A–H; Supplementary Fig S3N–S, T-θ), suggesting that MYC proteins do not directly target the expression of these proneural genes.
Figure 2.
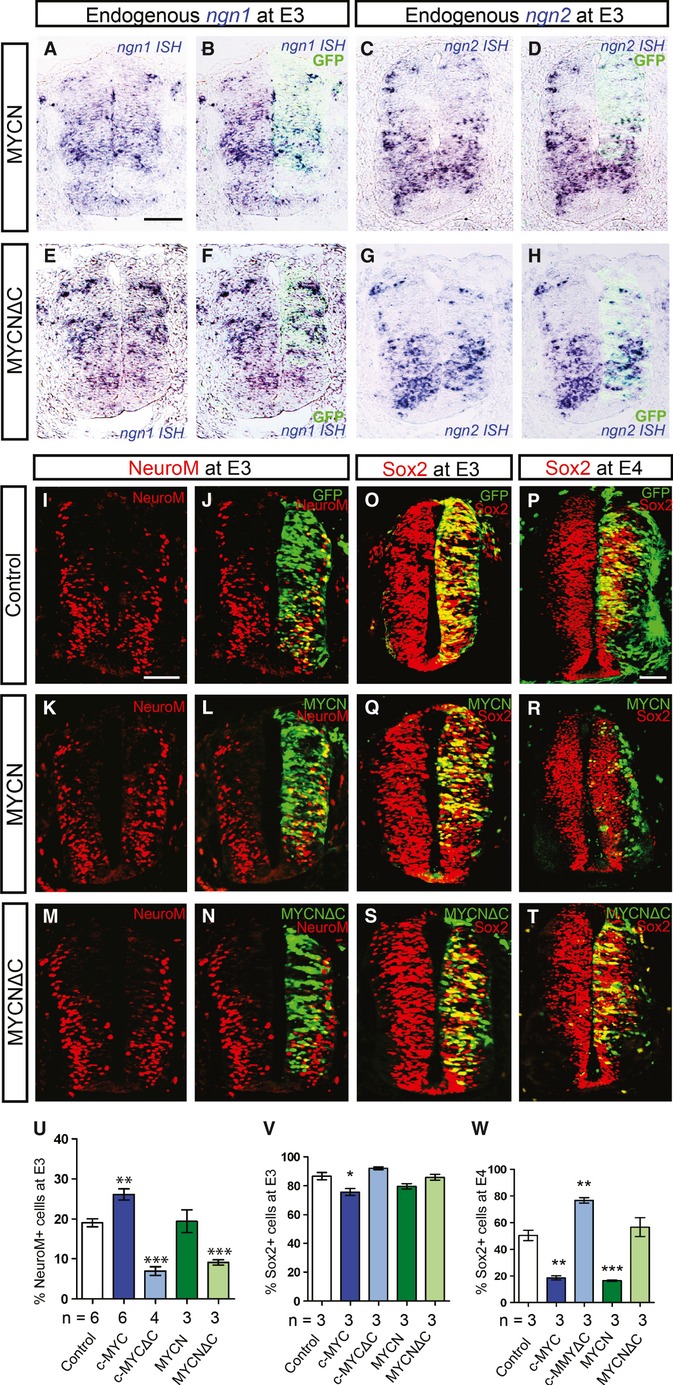
Overexpression of MYCN does not lead to the induction of proneural genes.
A-H Overexpression of MYCN or MYCNΔC followed by analysis of expression of ngn-1 (A, B, E, F) and ngn-2 (C, D, G, H) by ISH at E3. Control panels for this experiment are shown in Supplementary Fig S3 (N, Q).
I-W Endogenous levels of NeuroM (I-N) and Sox2 (O-T) as revealed by immunostaining. Control conditions and overexpression of MYCN or MYCNΔC at E3 (I-N, O, Q, S) and at E4 (P, R, T). Transfected cells were traced by GFP (J, L, N, O-T). The proportion of NeuroM+;GFP+ cells in the population of targeted (GFP+) cells was quantified (% NeuroM cells) at E3 (U), and the proportion of Sox2+;GFP+ cells in the population of targeted (GFP+) cells was quantified (% Sox2+ cells) at E3 (V) and E4 (W), respectively. Data are represented as mean ± s.e.m. of n = 3–6 embryos. **P < 0.005, ***P < 0.001, t-test. Scale bars, 50 μm.
We next analyzed the effects of MYC on the expression of NeuroM, a marker of ongoing neuronal differentiation, that specifically localizes to the intermediate differentiation zone (IZ) of the developing neural tube 21. Interestingly, we did not find any premature or ectopic induction of NeuroM by MYC (Fig 2I–L, U; Supplementary Fig S3ι–κ). Still, we observed a local transient increase in the number of NeuroM+ cells in the IZ upon MYC overexpression (Fig 2I–L, U; Supplementary Fig S3ι–κ). Conversely, the proportion of NeuroM+ cells in the IZ targeted with c-MYCΔC or MYCNΔC was significantly decreased (Fig 2M-N, U; Supplementary Fig S3λ). Taken together, our data suggest that MYC overexpression does not directly regulate NeuroM expression but rather influences the number of cells entering a transient NeuroM+ neurogenic stage between E3 and E4.
In addition, we did not find any significant changes in the numbers of Sox2+;GFP+ progenitor cells upon c-MYC or MYCN overexpression at E3, while at E4 the pool of double positive RGPs was almost completely exhausted (Fig 2O-R,V, W; Supplementary Fig S3μ–ρ). Furthermore, after c-MYCΔC overexpression, all transfected cells stayed Sox2+ in the VZ at E4, while no changes in Sox2 expression were observed at E3 (Fig 2S-T, V, W; Supplementary Fig S3ο-ς). These results indicate that MYC activity does not directly affect Sox2 expression. Similarly, Isl1+ and Brn3a+ neurons appear in normal amounts upon MYC overexpression (Supplementary Fig S3 σ-φ). Thus, specific populations of interneurons (Brn3a+) or motor neurons (Isl1+) were not affected by MYC overexpression. Collectively, our data show that neurogenesis is enhanced by MYC and that the mechanism does not involve any precautious or ectopic repression of stemness as defined by Sox2 expression or by activation of proneural gene programs.
Effects of MYC on neuronal progenitor cell proliferation depend on tissue integrity
It is well known that MYC proteins promote proliferation in cell culture (7). Given our unexpected results on MYC proteins as proneurogenic factors in vivo, we next analyzed whether MYC proteins would change the rate of proliferation of RGPs in vivo and in vitro. To this end, electroporated embryos were either kept to develop further in the eggs (E4 in vivo) or collected and dissociated into single-cell suspensions and allowed to grow in culture (E4 in vitro) (Fig 3A). At E3, we did not detect any significant differences in the number of proliferating cells between control, c-MYC-, or c-MYCΔC-expressing RGPs (Fig 3B, D, F, K), suggesting that MYC does not act as a direct mitogenic factor in RGPs in vivo. However, a significant reduction in proliferating RGPs was observed in c-MYC+; GFP+-overexpressing cells in E4 embryos (Fig 3C, E, G, L). This effect is likely secondary to the depletion of Sox2+ RGPs (see Fig 2V-W). In contrast, MYC+/GFP+ cells grown in vitro displayed a marked increase in proliferation at E4 as determined by FACS analysis (Fig 3H-J, M). Overexpression of MYC caused only mild or no effect on the length of cell cycle phases in vivo (Fig 3N). Hence, these results demonstrate opposing effects on proliferation depending on tissue integrity upon MYC overexpression. In line with our finding, MYC-induced cell cycle re-initiation has previously been shown to be blocked by epithelial cell architecture in organotypic cultures of mammary epithelial cells in vitro 22. The relationship between MYC proteins and cell integrity/polarity is particularly interesting since deregulation of polarity pathways has been shown to promote neoplastic growth in mammals 23. Our results suggest that a loss of cell polarity may not only fuel tumor development by a proposed mechanism of disrupting morphogenesis and inhibition of cell death 24 but might also unleash MYC-driven proliferation.
Figure 3.
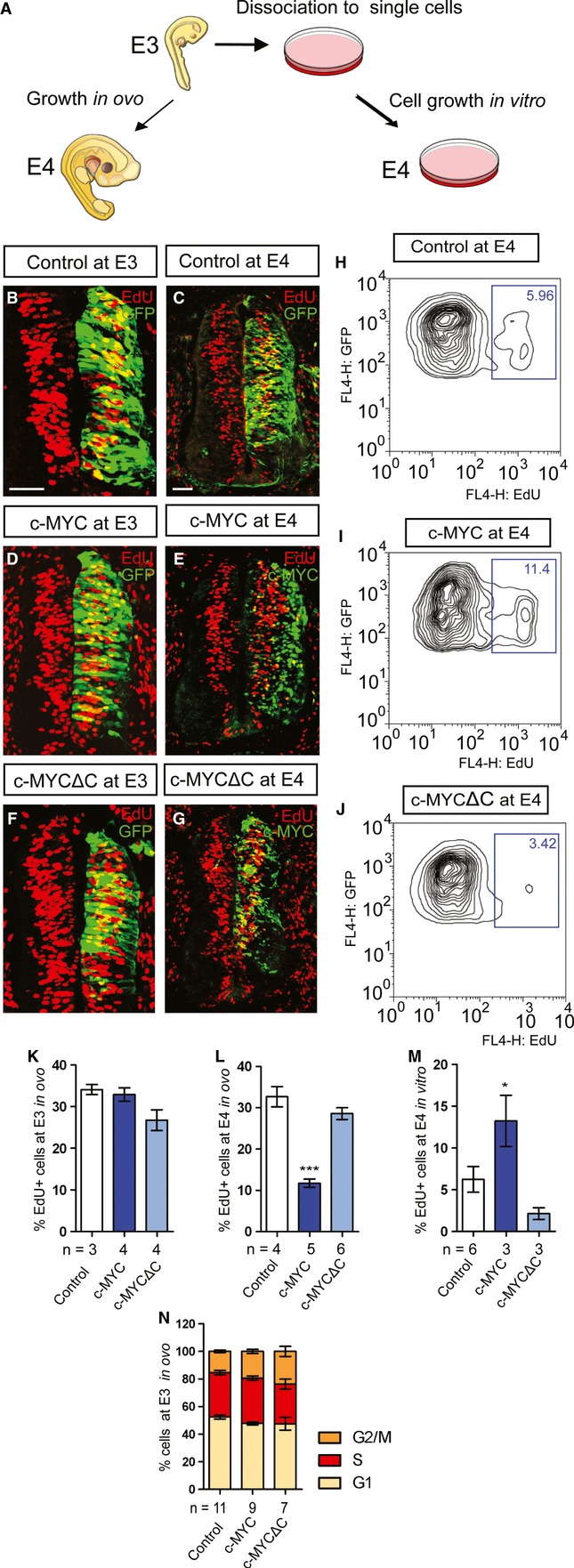
Loss of neural tissue integrity results in changes in the phenotypic outcome of MYC overexpression.
A Schematic representation of the experimental setup. Embryos at E3 (24 h post electroporation) were either allowed to grow in ovo until E4 or dissociated to single cells and cultivated in vitro for 24 h.
B-G Proliferation in ovo was analyzed with a 2-h EdU pulse in cells transfected with vector control (B, C), c-MYC (D, E), or c-MYCΔC (F, G) constructs at E3 (B, D, F) and E4 (C, E, G). Scale bars, 50 μm.
H-J FACS analysis of cells cultivated in vitro following a 2-h EdU pulse at E4 in cells transfected with vector control (H), c-MYC (I), or c-MYCΔC (J) constructs as indicated.
K-M Quantification of the proportion of EdU+;GFP+ cells in the population of targeted (GFP+) cells (% EdU cells) at E3 (K) and after 24 h (E4) either in ovo (L) or in vitro (M). Data are represented as mean ± s.e.m. of n = 3–6 embryos. *P < 0.05, ***P < 0.001, t-test.
N Distribution of cell cycle phases at E3 as analyzed by FACS. Data are represented as mean ± s.e.m. of n = 7–11 embryos.
c-MYC acts as a repressor of the Notch pathway
Notch signaling plays an important role for cell fate leading to an increase in planar cell divisions, thus preventing cell cycle exit and neuronal differentiation in the neural tube 25–27. We therefore examined whether MYC-induced neuronal differentiation involves Notch function. Interestingly, we found that notch1 expression was downregulated in response to both MYCN and c-MYC overexpression at stage E3 (Fig 4A–C). Consistently, expression of the Notch-regulated genes hes1 and hes5 was also reduced in the presence of ectopic c-MYC or MYCN (Fig 4D–I). In addition, real-time PCR showed a significant in vivo downregulation of notch1 24 h postelectroporation of MYC, while no significant difference in notch1 levels was observed in dissociated and cultured cells (Fig 4J-K).
Figure 4.
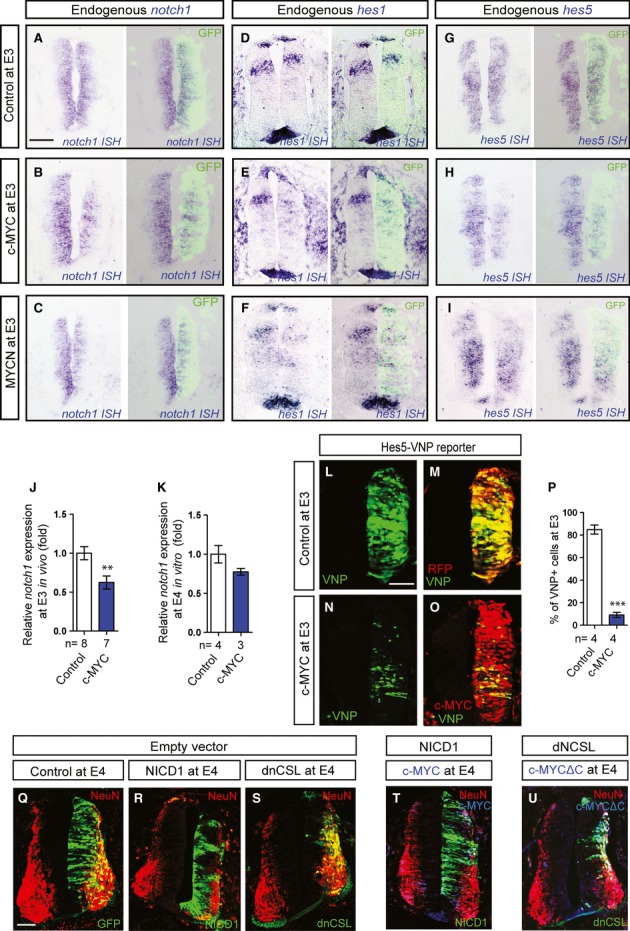
MYC and Notch Signaling are Connected and Influence the Fate of RGPs.
A-I Effects of elevated levels of c-MYC or MYCN on notch1 (A-C), hes1 (D-F), or hes5 (G-I) expression. Electroporation of vector control (A, D, G), c-MYC (B, E, H), or MYCN (C, F, I) constructs. Expression of notch1, hes1, and hes5 was analyzed by ISH at E3 (A-I).
J-K Relative notch1 expression as quantified by qPCR in targeted cells collected from embryos at E3 (J) and following dissociation and incubation for 24 h in vitro (K). Data are represented as mean ± s.e.m. of n = 3–8 embryos. *P < 0.05, t-test.
L-P Co-transfection of the pHes5-VNP reporter with vector control (L,M) or the c-MYC construct (N,O). Transfection of the vector control was revealed by RFP (M), and c-MYC was detected by immunostaining (O). Quantification of the proportion of VNP+;RFP+ cells in the population of targeted (RFP+) cells (% VNP+ cells) at E3 is shown in (P). Data are represented as mean ± s.e.m. of n = 4 embryos, ***P < 0.001, t-test.
Q-U Effects on neurogenesis of NICD1 and dnCSL expression alone and in combination with c-MYC or c-MYCΔC as indicated. Scale bars, 50 μm.
To analyze the effect of MYC proteins on Notch signaling, we co-expressed c-MYC together with a real-time reporter of Notch activity (pHes5-VNP) 27. As shown in Fig 4L-P, Notch signaling was significantly decreased upon c-MYC overexpression. Consistent with these results, stemness was strongly maintained and differentiation was prevented by overexpression of the Notch1 intracellular domain (NICD1), whereas as expected, overexpression of the Notch antagonist dnCSL 28, 29 promoted neurogenesis (Fig 4Q-S). Importantly, we found that c-MYC-induced neurogenesis was efficiently blocked by NICD1 where enhanced Notch signaling rescued stemness at stage E4 (Fig 4T). Furthermore, we showed that the block of neurogenesis imposed by MYCΔC could be rescued by dnCSL, indicating that c-MYCΔC was unable to prevent neuronal differentiation under these conditions (Fig 4U). Combined, these results suggest that repression of the Notch pathway may participate in neurogenesis driven by MYC overexpression.
Interestingly, real-time PCR showed a difference in downregulation of notch1 by MYC in vivo and in vitro. Failure to efficiently repress notch1 might be responsible for the inability of MYC to induce differentiation and to prevent proliferation in dissociated cells. Despite that MYC overexpression proved to erode notch1 in vivo, this might be only a part of MYC-dependent molecular mechanism responsible for control of stemness versus neuronal differentiation in developing nervous system.
MYC promotes neurogenic cell divisions of RGPs
Previous data proposing a link between Notch and the balance between symmetric proliferative and asymmetric neurogenic division in RGPs 30 led us to hypothesize that MYC affects the cleavage plane orientation during mitosis of RGPs and that MYCN might be differentially expressed in cells undergoing neurogenic versus non-neurogenic divisions. To test this hypothesis, we analyzed the expression pattern of mycn using ISH combined with staining for Phospho-Histone H3 (PH3), Sox2, and NeuroM to outline dividing cells, RGPs, and differentiating neurons, respectively. We found that dividing RGPs were almost devoid of mycn in the dorsal, less neurogenic region of the neural tube, while they were strongly positive for mycn expression when located ventrally at E3-4 in the most neurogenic region as confirmed by NeuroM, ngn2, neurod1, and neurod2 expression (Fig 5A–G; Supplementary Fig S3Q, T, Y). Thus, high mycn expression in dividing RGP cells correlates with zones of increased neurogenesis in the developing neural tube.
Figure 5.
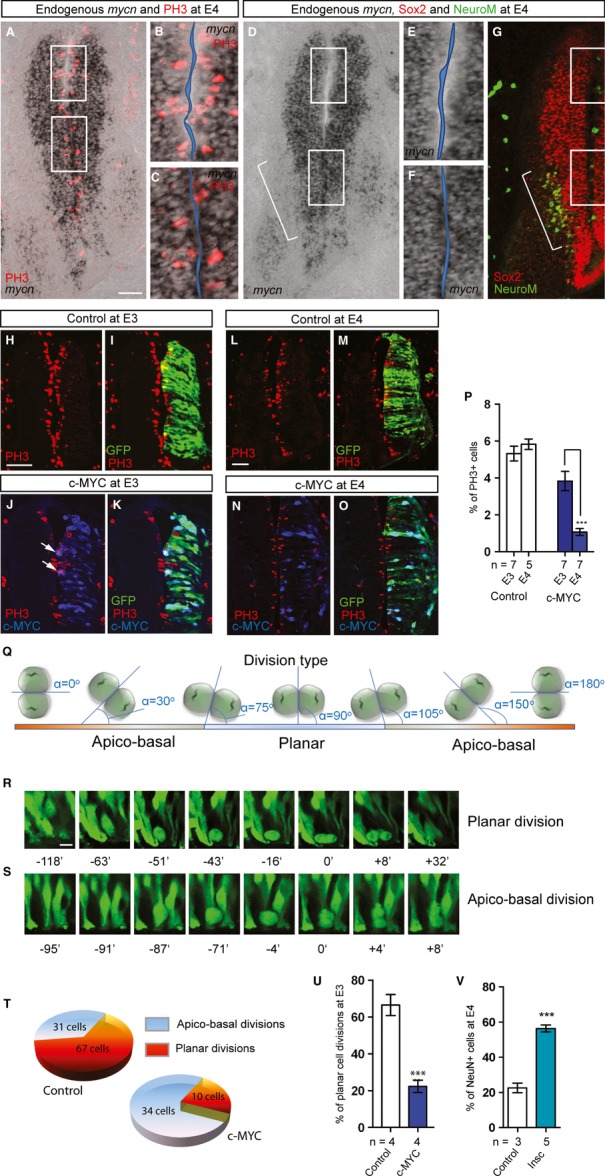
MYC Proteins Influence the Balance of Planar and Apico-Basal Cell Divisions in the Developing Neural Tube.
A-G Endogenous mycn levels by ISH. Dividing cells are identified using staining with an Phospho-Histone H3 (PH3) antibody (A-C). (B) and (C) show magnified dorsal and ventral regions from (A) outlined by white frames, respectively. Progenitors and intermediate neurons are identified by immunostaining against Sox2 and NeuroM on parallel sections (D-G). (E) and (F) show magnified dorsal and ventral regions from (D) outlined by white frames, respectively. Bracket marks the most neurogenic regions as defined by NeuroM expression.
H-P Overexpression of MYC eventually promotes cell cycle exit and neuronal differentiation of RGPs. Electroporation of vector control (H, I, L, M) or c-MYC (J, K, N, O) at E3 and E4. Dividing cells are identified by PH3 immunostaining; transfected cells were detected by GFP and by immunostaining for c-MYC. Scale bars, 50 μm. Quantification of the proportion of dividing PH3+;GFP+ cells in the population of targeted (GFP+) cells (% of PH3+ cells) at E3 and E4 per section is shown in (P). Data are represented as mean ± s.e.m. of n = 5–7 embryos, ***P < 0.001, t-test.
Q The threshold for defining planar versus apico-basal divisions depends on the angle α between the cleavage plane and the lumen wall of the neural tube. The ranges for angle α are set as: 75–105o for planar and 0–74o/106–180o for apico-basal divisions.
R, S Examples of planar (R) and apico-basal (S) divisions following live imaging analysis of neural tube slices electroporated with vector control or MYC constructs. The pictures are taken at different time points before and after cell division at 0 min.
T Raw data representing the total number of planar and apico-basal divisions analyzed in control (88 cells in total) and in conditions with c-MYC overexpression (44 cells in total).
U Quantification of the proportion of planar and apico-basal divisions in chick neural tube slices at E3. Data are represented as mean ± s.e.m. of n = 4 embryos, ***P < 0.001, t-test.
V Overexpression of Inscuteable results in neurogeneisis. The proportion of NeuN+;GFP+ cells in the targeted (GFP+) cells were quantified (NeuN+ cells) at E4. Data are represented as mean ± s.e.m. of n = 3–5 embryos ***P < 0.001, t-test.
Asymmetric inheritance of key molecular features of the neural progenitor during cell division controls differentiation of one of the daughter cells 31, 32. The progenitor's apical footprint and basal process are two morphological domains regulating cell fates if asymmetrically inherited 33, 34. Consistently, by the use of direct clonal analysis and live imaging, the cleavage plane of progenitor cell division has been observed to regulate neurogenesis in the developing chick neural tube, and Notch is involved in a fate choice during this process 27, 35. Increased neurogenic cell divisions are expected to lead to a stable state or reduction in the buildup of cycling neural progenitor cells during development. We therefore addressed changes in numbers of dividing RGPs at 24 and 48 h after electroporation. No major differences in the amount of dividing PH3-positive cells were observed upon electroporation with control vector between E3 and E4 (Fig 5H-P). In contrast, there was a reduction in the numbers of PH3-positive cells at E3 upon c-MYC electroporation, and additionally, a more than a twofold decrease in the number of mitotic cells occurred during the transition from E3 to E4 (Fig 5P). The robust drop in the amount of progenitors corresponded to the increased proportion of differentiated neurons in transfected cells as shown in Fig 1J-M, P-Q, T.
Next, the cleavage plane angles of the electroporated RGPs were examined using live 2-photon imaging. A division was set to be planar when the angle of the cleavage plane was 90° ± 15, and all other angles were considered as apico-basal divisions 36 (Fig 5Q-S). Progenitor cells electroporated with the vector control divided mainly through planar divisions at E3 (66.4% ± 5.7). Intriguingly, planar divisions decreased dramatically upon c-MYC overexpression (22.3% ± 3.4) (Fig 5T-U). These results show that overexpression of c-MYC markedly elevates the proportion of RGPs undergoing apico-basal cell divisions, which in turn could be responsible for stimulated neurogenesis.
To show that an increase in apico-basal cell divisions at E3 and E4 stimulates neurogenesis, we overexpressed Inscuteable (Insc), a key adapter component of the NuMA complex 27. This complex includes the polarity proteins LGN, Par3, Par6, and aPKS and localizes to the cortical layer of the lumen membrane. NuMA is responsible for cell polarization during cell division and is involved in spindle rotation and cleavage plane orientation 37. We found that ectopic introduction of Inscuteable enhanced neurogenesis in a similar manner as MYC overexpression (Fig 5V, Supplementary Fig S5A-F), i.e. without ectopic or premature neuronal differentiation (Supplementary Fig S5G-J). Thus, by in vivo overexpression of one of the molecules involved in polarity and cleavage plane orientation, we could replicate the phenotype caused by MYC. Importantly, expression of inscuteable was slightly elevated 24 h after MYC electroporation (Supplementary Fig S5K-N), which might account for the increase in apico-basal neurogenic divisions following MYC overexpression. Thus, MYC positively regulates the expression of inscuteable in the chick neural tube and thus may convey its neurogenic function through regulation of the machinery responsible for mitotic spindle rotation and cleavage plane orientation.
To conclude, within the organized tissue, MYC proteins regulate neurogenesis in progenitor cells before onset of the neuronal differentiation by controlling Notch signaling. This has a direct impact on the proportion of cells undergoing neurogenic and non-neurogenic cell divisions. Within RGPs, MYC inhibits the Notch signaling pathway, leads to increased apico-basal cell divisions and eventually also to depletion of the stem cells. This role of MYC is important during development of the nervous system where increased MYC expression results in elevated production of neurons and decreased levels of MYC lead to a failure of neurogenesis.
Materials and Methods
Detailed materials and methods are provided in Supplementary Materials and Methods. In ovo electroporation, immunohistochemistry, and in situ hybridizations were performed as described 38. For time-lapse imaging, spinal cord transversal slices (400 μm) were dissected from E3 chick embryos using a Leica vibratome. Video imaging of the in situ dividing cells was performed with a Zeiss LSM510 META NLO (Carl Zeiss, Germany) 2-photon laser scanning microscope. Stacks of ∼50 μm were acquired every 3–5 min for 4–6 h. Ethical permission N200/11 was approved by the Stockholm regional ethics committee for animal research.
Acknowledgments
We are indebted to Drs. K.G. Storey, M. Frank, A. Tikhonenko, J. Holmberg, J. Muhr, D. Hagey, F. Lallemend, J. Lovén, and U. Lendahl for constructs, antibodies, and constructive discussions; M. Klarqvist, Dr R. Erlandsson, and Dr E. Fredlund for bioinformatic assistance; R. van Eijk and Dr U. Westermark for technical assistance and all members of our laboratories for helpful discussions. This work was supported by grants from the Swedish Research Council, the Swedish Cancer Society, the Swedish Childhood Cancer Foundation, the Knut and Alice Wallenberg Foundation (CLICK) and ERC (232675). N. Z. was the recipient of a stipend financed by the Cancer Concern, Los Angeles, and the Cancer Research Institute, New York, USA. I.A. and M.W. were supported by Assistant Professorships from the Swedish Research Council. M.A.H. was recipient of a Senior Investigator Award from the Swedish Cancer Society.
Author contributions
N.Z., I.A, N.F., and M.W. performed the research; P.E. and P.U. supervised and designed parts of the study; N.Z., I.A., M.W., and M.A.H. analyzed the data, designed the study, and wrote the manuscript; M.A.H. supervised the research.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://embor.embopress.org
References
- 1.Eilers M, Eisenman RN. Myc's broad reach. Genes Dev. 2008;22:2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmid P, Schulz WA, Hameister H. Dynamic expression pattern of the myc protooncogene in midgestation mouse embryos. Science. 1989;243:226–229. doi: 10.1126/science.2911736. [DOI] [PubMed] [Google Scholar]
- 3.Kato K, Kanamori A, Wakamatsu Y, Sawai S, Kondoh H. Tissue distribution of N-myc expression in the early organogenesis period of the mouse embryo. Dev Growth Differ. 1991;33:29–36. doi: 10.1111/j.1440-169X.1991.00029.x. [DOI] [PubMed] [Google Scholar]
- 4.Pirity M, Blanck JK, Schreiber-Agus N. Lessons learned from Myc/Max/Mad knockout mice. Curr Top Microbiol Immunol. 2006;302:205–234. doi: 10.1007/3-540-32952-8_8. [DOI] [PubMed] [Google Scholar]
- 5.Charron J, Malynn BA, Fisher P, Stewart V, Jeannotte L, Goff SP, Robertson EJ, Alt FW. Embryonic lethality in mice homozygous for a targeted disruption of the N-myc gene. Genes Dev. 1992;6:2248–2257. doi: 10.1101/gad.6.12a.2248. [DOI] [PubMed] [Google Scholar]
- 6.Stanton BR, Perkins AS, Tessarollo L, Sassoon DA, Parada LF. Loss of N-myc function results in embryonic lethality and failure of the epithelial component of the embryo to develop. Genes Dev. 1992;6:2235–2247. doi: 10.1101/gad.6.12a.2235. [DOI] [PubMed] [Google Scholar]
- 7.Baudino TA, McKay C, Pendeville-Samain H, Nilsson JA, Maclean KH, White EL, Davis AC, Ihle JN, Cleveland JL. c-Myc is essential for vasculogenesis and angiogenesis during development and tumor progression. Genes Dev. 2002;16:2530–2543. doi: 10.1101/gad.1024602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- 9.Nagao M, Campbell K, Burns K, Kuan CY, Trumpp A, Nakafuku M. Coordinated control of self-renewal and differentiation of neural stem cells by Myc and the p19ARF-p53 pathway. J Cell Biol. 2008;183:1243–1257. doi: 10.1083/jcb.200807130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wey A, Martinez Cerdeno V, Pleasure D, Knoepfler PS. c- and N-myc regulate neural precursor cell fate, cell cycle, and metabolism to direct cerebellar development. Cerebellum. 2010;9:537–547. doi: 10.1007/s12311-010-0190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zindy F, Knoepfler PS, Xie S, Sherr CJ, Eisenman RN, Roussel MF. N-Myc and the cyclin-dependent kinase inhibitors p18Ink4c and p27Kip1 coordinately regulate cerebellar development. Proc Natl Acad Sci USA. 2006;103:11579–11583. doi: 10.1073/pnas.0604727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Varlakhanova NV, Cotterman RF, deVries WN, Morgan J, Donahue LR, Murray S, Knowles BB, Knoepfler PS. myc maintains embryonic stem cell pluripotency and self-renewal. Differentiation. 2010;80:9–19. doi: 10.1016/j.diff.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, et al. The N-Myc-DLL3 cascade is suppressed by the ubiquitin ligase Huwe1 to inhibit proliferation and promote neurogenesis in the developing brain. Dev Cell. 2009;17:210–221. doi: 10.1016/j.devcel.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirabayashi Y, Itoh Y, Tabata H, Nakajima K, Akiyama T, Masuyama N, Gotoh Y. The Wnt/beta-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development. 2004;131:2791–2801. doi: 10.1242/dev.01165. [DOI] [PubMed] [Google Scholar]
- 16.Kuwahara A, Hirabayashi Y, Knoepfler PS, Taketo MM, Sakai J, Kodama T, Gotoh Y. Wnt signaling and its downstream target N-myc regulate basal progenitors in the developing neocortex. Development. 2010;137:1035–1044. doi: 10.1242/dev.046417. [DOI] [PubMed] [Google Scholar]
- 17.Claveria C, Giovinazzo G, Sierra R, Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500:39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- 18.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 19.Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19:282–294. doi: 10.1101/gad.1257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mizuguchi R, Sugimori M, Takebayashi H, Kosako H, Nagao M, Yoshida S, Nabeshima Y, Shimamura K, Nakafuku M. Combinatorial roles of olig2 and neurogenin2 in the coordinated induction of pan-neuronal and subtype-specific properties of motoneurons. Neuron. 2001;31:757–771. doi: 10.1016/s0896-6273(01)00413-5. [DOI] [PubMed] [Google Scholar]
- 21.Roztocil T, Matter-Sadzinski L, Alliod C, Ballivet M, Matter JM. NeuroM, a neural helix-loop-helix transcription factor, defines a new transition stage in neurogenesis. Development. 1997;124:3263–3272. doi: 10.1242/dev.124.17.3263. [DOI] [PubMed] [Google Scholar]
- 22.Partanen JI, Nieminen AI, Makela TP, Klefstrom J. Suppression of oncogenic properties of c-Myc by LKB1-controlled epithelial organization. Proc Natl Acad Sci USA. 2007;104:14694–14699. doi: 10.1073/pnas.0704677104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Royer C, Lu X. Epithelial cell polarity: a major gatekeeper against cancer? Cell Death Differ. 2011;18:1470–1477. doi: 10.1038/cdd.2011.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F, et al. Transcription factor RBP-J-mediated signaling represses the differentiation of neural stem cells into intermediate neural progenitors. Mol Cell Neurosci. 2009;40:442–450. doi: 10.1016/j.mcn.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Zhong W, Chia W. Neurogenesis and asymmetric cell division. Curr Opin Neurobiol. 2008;18:4–11. doi: 10.1016/j.conb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 27.Das RM, Storey KG. Mitotic spindle orientation can direct cell fate and bias Notch activity in chick neural tube. EMBO Rep. 2012;13:448–454. doi: 10.1038/embor.2012.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung CN, Hamaguchi Y, Honjo T, Kawaichi M. Site-directed mutagenesis study on DNA binding regions of the mouse homologue of Suppressor of Hairless, RBP-J kappa. Nucleic Acids Res. 1994;22:2938–2944. doi: 10.1093/nar/22.15.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmberg J, Hansson E, Malewicz M, Sandberg M, Perlmann T, Lendahl U, Muhr J. SoxB1 transcription factors and Notch signaling use distinct mechanisms to regulate proneural gene function and neural progenitor differentiation. Development. 2008;135:1843–1851. doi: 10.1242/dev.020180. [DOI] [PubMed] [Google Scholar]
- 30.Dave RK, et al. Sonic hedgehog and notch signaling can cooperate to regulate neurogenic divisions of neocortical progenitors. PLoS One. 2011;6:e14680. doi: 10.1371/journal.pone.0014680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Knoblich JA. Mechanisms of asymmetric stem cell division. Cell. 2008;132:583–597. doi: 10.1016/j.cell.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 32.Izumi H, Kaneko Y. Evidence of asymmetric cell division and centrosome inheritance in human neuroblastoma cells. Proc Natl Acad Sci USA. 2012;109:18048–18053. doi: 10.1073/pnas.1205525109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alexandre P, Reugels AM, Barker D, Blanc E, Clarke JD. Neurons derive from the more apical daughter in asymmetric divisions in the zebrafish neural tube. Nat Neurosci. 2010;13:673–679. doi: 10.1038/nn.2547. [DOI] [PubMed] [Google Scholar]
- 34.Shitamukai A, Konno D, Matsuzaki F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J Neurosci. 2011;31:3683–3695. doi: 10.1523/JNEUROSCI.4773-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vilas-Boas F, Fior R, Swedlow JR, Storey KG, Henrique D. A novel reporter of notch signalling indicates regulated and random Notch activation during vertebrate neurogenesis. BMC Biol. 2011;9:58. doi: 10.1186/1741-7007-9-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morin X, Jaouen F, Durbec P. Control of planar divisions by the G-protein regulator LGN maintains progenitors in the chick neuroepithelium. Nat Neurosci. 2007;10:1440–1448. doi: 10.1038/nn1984. [DOI] [PubMed] [Google Scholar]
- 37.Kiyomitsu T, Cheeseman IM. Cortical dynein and asymmetric membrane elongation coordinately position the spindle in anaphase. Cell. 2013;154:391–402. doi: 10.1016/j.cell.2013.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adameyko I, et al. Sox2 and Mitf cross-regulatory interactions consolidate progenitor and melanocyte lineages in the cranial neural crest. Development. 2012;139:397–410. doi: 10.1242/dev.065581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


