Abstract
In plants, transposable elements (TEs) are kept inactive by transcriptional gene silencing (TGS). TGS is established and perpetuated by RNA-directed DNA methylation (RdDM) and maintenance methylation pathways, respectively. Here, we describe a novel RdDM function specific for shoot apical meristems that reinforces silencing of TEs during early vegetative growth. In meristems, RdDM counteracts drug-induced interference with TGS maintenance and consequently prevents TE activation. Simultaneous disturbance of both TGS pathways leads to transcriptionally active states of repetitive sequences that are inherited by somatic tissues and partially by the progeny. This apical meristem-specific mechanism is mediated by increased levels of TGS factors and provides a checkpoint for correct epigenetic inheritance during the transition from vegetative to reproductive phase and to the next generation.
Keywords: Arabidopsis, chromatin, epigenetics, shoot apical meristem, transcriptional gene silencing
Introduction
Genomes of higher plants contain a high proportion of transposable elements (TEs). Nearly all TE families are represented with some potentially mobile copies. This endangers genome stability, especially if transposition were to occur in cells forming the germline and offspring. Plant evolution has brought about an efficient protection mechanism against extensive TE activity by preventing their expression via transcriptional gene silencing (TGS). This is mediated by epigenetic regulation through DNA methylation and repressive histone modifications 1, 2. In short, TE transcription triggers an RNA-directed DNA methylation (RdDM) mechanism that involves de novo DNA methylation 3, 4. Depending on the sequence context, different pathways ensure correct maintenance and transmission of established DNA methylation patterns 5, 6. Conversely, disturbance of RdDM and maintenance pathways allows the transcription of specific TEs 1–8.
Unlike in mammals, where DNA methylation is largely erased and then re-established during germ cell maturation and zygote formation, plant DNA methylation is considered to be generally stable 1. However, reinforced silencing of TEs has been proposed in gametes and the early embryo via mobile siRNAs produced in companion cells 9–11. This suggests an important role for RdDM in surveying the genome of gametes and early zygotes.
Gamete formation in plants occurs late during development, and cells undergo numerous cell divisions before flowering. In addition, the formation of secondary meristems widens the range of cells that can contribute to progeny. Therefore, any loss of TE silencing during the vegetative phase 12, 13 can lead to the transmission of active TEs to the next generation.
Here, we show that release of TGS control upon treatment with the DNA methylation inhibitor zebularine 14 is observed only in tissues inherited from the embryo, but not in newly developing parts of the plant. Functional analysis identifies RdDM as an important regulator of TGS maintenance in newly formed tissues, and lack thereof leads to an increased inheritance of active states to the next generation. Various meristematic tissues display enhanced expression of genes required for TGS, and we propose that this tissue-specific coordinated expression is required to enforce epigenomic stability and germline protection during vegetative growth.
Results and Discussion
DNA methylation inhibitors cause tissue-specific transcriptional reactivation of repetitive DNA
The cytidine analog zebularine induces transient DNA hypomethylation and transcriptional activation of otherwise silent sequences in wild-type (WT) Arabidopsis 15. To analyze the mechanism governing re-methylation and re-silencing, we applied zebularine to the line TS-GUS (6b5, L5) 16 containing a transcriptionally silent β-glucuronidase transgene that is activated throughout the entire plant in the background of epigenetic mutants like ddm1 (Fig 1A) 5. While mock-treated plants showed no GUS staining (Fig 1B), growth in the presence of 20 or 40 μM zebularine or 400 μM 5-azadeoxycytidine released GUS silencing in cotyledons, but neither in true leaves of all stages nor in floral tissues (Fig 1C and D and Supplementary Fig S1). In addition, no GUS signal was detected in selfed progenies from zebularine-treated WT plants (Supplementary Fig S1), suggesting that the loss of silencing was restricted to embryonic tissues only. This was confirmed after zebularine application to a TS-GFP reporter line containing a repetitive silent GFP marker 17 that showed an even sharper separation between GFP-positive cotyledons, hypocotyl, and root and the GFP-negative true leaves around the SAM (Fig 1E and F, arrowhead).
Figure 1.
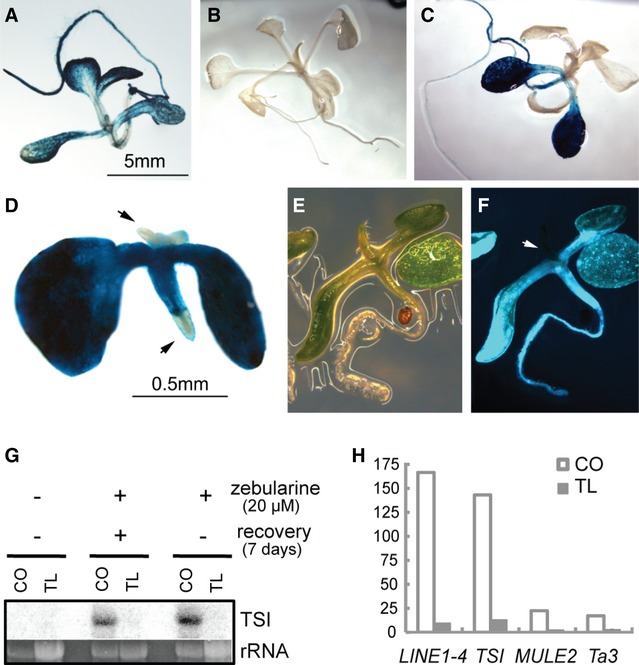
DNA methylation inhibitors induce tissue-specific reactivation of transcriptionally silenced repeats.
A TS-GUS expression in ddm1.
B GUS signal is absent in mock-treated WT TS-GUS plants.
C, D Tissue-specific reactivation of TS-GUS after treatment with (C) 40 μM zebularine for 3 weeks and (D) 400 μM 5-azacytidine for 3 days and subsequent recovery for 1 week.
E, F Tissue-specific reactivation of a transcriptionally silent TS-GFP transgene after the treatment with 40 μM zebularine under visible (E) and GFP fluorescence (F) light.
G Northern blot detection of transcription from TSI repeats in cotyledons (CO) and true leaves (TL) of 14-day-old mock- or 20 μM zebularine-treated seedlings.
H Quantitative reverse transcription PCR measurements of zebularine-induced transcription of TEs in cotyledons and the first pair of true leaves of WT plants. Based on the pool of approximately 20 plants in one biological replicate.
Data information: In (D) and (F): arrows point to meristem tissues lacking GUS and GFP signals.
Zebularine-induced tissue-specific reactivation holds true also for endogenous repeats. We dissected cotyledons and the first pair of true leaves from plantlets grown for 14 days either on drug-free medium, continuously on zebularine, or on 20 μM zebularine for 7 days followed by 7-day recovery on drug-free medium. Northern blot and/or quantitative reverse transcription PCR (qRT–PCR) analysis of Transcriptionally Silent Information (TSI) repeats, LINE1-4, MULE2, and Ta3 revealed no signal in mock-treated plants, while zebularine treatment—independent of recovery—released silencing only in cotyledons (Fig 1G and H).
To exclude that the lack of reactivation in true leaves was due to reduced uptake of zebularine, loss of inhibitor activity, or its dilution via DNA replication, we compared DNA methylation of centromeric repeats between cotyledons and the first pair of true leaves, for mock- and zebularine-treated plants. Methylation-sensitive Southern blots indicate that zebularine treatment reduces methylation in both tissues (Supplementary Fig S1C). Furthermore, fluorescence in situ hybridization (FISH) with centromeric repeat sequences revealed reduced heterochromatin condensation in nuclei from cotyledons and true leaves of inhibitor-treated plants (Supplementary Fig S1D). However, the degree of decondensation was less complete in true leaves (i.e., nuclei with full decondensation of all chromocenters), which may indicate slight differences in zebularine activity or stability in specific tissues.
Taken together, the tissue-specific activation of silent repeats after zebularine treatment argues for a regulatory mechanism that corrects the loss of TGS during early vegetative growth.
RdDM components secure repeat silencing in true leaves in spite of inhibitor treatment
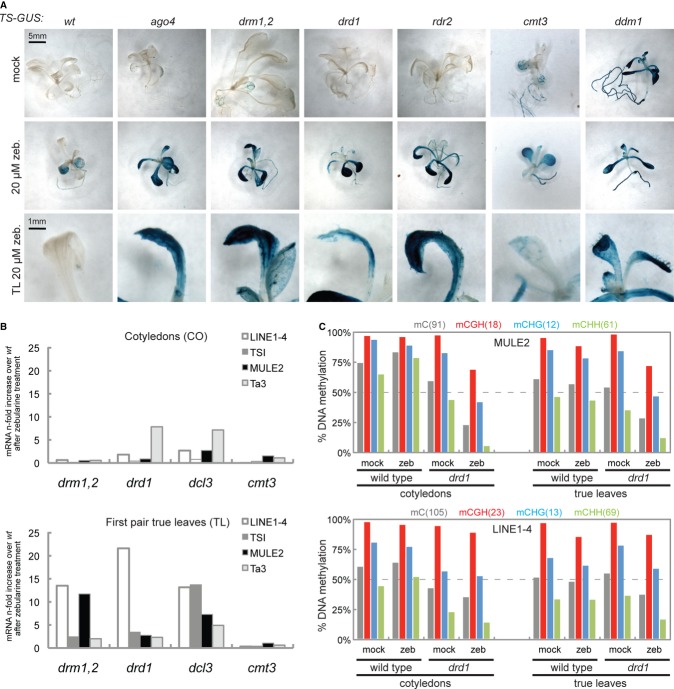
To investigate the molecular basis of the tissue-specific difference in TGS, we introgressed the TS-GUS transgene into mutants associated with TGS and chromatin regulation. In agreement with a previous report 5, during mock treatment, we observed full TS-GUS reactivation only in ddm1 or met1, and none or weak cotyledon-specific activation in cmt3, kyp, lhp1, fas1, fas2, hda6, and RdDM mutants (Fig 2A, upper panel and Supplementary Fig S2). These tissue-specific activation patterns resembled those after zebularine treatment and prompted us to expose the low activating mutants to 20 μM zebularine, scoring for potential combinatorial effects. Remarkably, zebularine treatment led to strong GUS expression in true leaves of ago4, drm1/drm2, drd1, and rdr2, while no true leaf GUS staining was observed in cmt3, fas1, fas2, kyp, lhp1, or hda6, suggesting that RdDM components are involved in mediating re-silencing (Fig 2A and Supplementary Fig S2B, bottom panel).
Figure 2.
- Representative examples of GUS staining of whole seedlings (top and middle row) or their first true leaves (bottom row) after mock or zebularine treatments.
- Quantitative reverse transcription PCR measurements of zebularine-induced reactivation from TEs in cotyledons (CO) and true leaves (TL) in transcriptional gene silencing mutants compared to WT. Based on the pool of approximately 20 plants in one biological replicate.
- DNA methylation analysis by bisulfite sequencing. Shown is percent cytosine methylation in all sequence contexts for MULE2 and LINE1-4 in true leaves and cotyledons of mock- and zebularine-treated WT and drd1. A minimum of 15 unique clones were scored per experiment.
Source data are available online for this figure.
In addition, we observed elevated true leaf-specific transcription for LINE1-4, MULE2, and TSI in zebularine-treated drm1/drm2, drd1, and dcl3, in comparison with treated WT plants (Fig 2B and Supplementary Fig S2). Activation of Ta3, an element regulated mainly by methylation of histone H3 lysine 9 and CHG 18, was not further induced in true leaves of zebularine-treated mutant plants (Fig 2B and Supplementary Fig S2). A similar response of LINE1-4 in WT accessions Col-0 and Ws-2 indicated that this was independent of the different genetic background of the mutants (Supplementary Fig S2D).
In order to measure the combined effect of zebularine and defective RdDM on DNA methylation in true leaves, we performed bisulfite sequencing at defined copies of LINE1-4 and MULE2 in WT and drd1 plants (Fig 2C). Compared to untreated WT plants, lack of DRD1 resulted in a reduction in methylated cytosines by 15% in cotyledons (mostly at CHG and CHH), while in true leaves, only minor changes could be measured (< 10% reduction). Importantly, additional zebularine treatment in drd1 plants resulted in a more pronounced hypomethylation compared to zebularine treatment or lack of drd1 alone (Fig 2C). The additive effect of zebularine—although at low dose—exceeded the 15% reduction at both analyzed targets in true leaves, compared to untreated WT controls, and affected all sequence contexts (Fig 2C). Surprisingly, we observed that asymmetric methylation increased after zebularine treatment in cotyledons (Fig 2C).
To further validate that RdDM antagonizes DNA methylation interference by zebularine, we germinated WT and drd1 TS-GUS plants on zebularine-free medium and transferred the seedlings after 6 days to zebularine-containing medium for additional 6 days (Fig 3A). Owing to the dependence on DNA replication, zebularine-mediated reactivation was observed only in tissues that proliferated during drug treatment such as newly grown parts of the root (Fig 3A). Thus, absence of replication in developed hypocotyl, cotyledons, and adult root regions protected against zebularine-mediated reactivation (Fig 3A). In addition, the lack of drd1 resulted in TS-GUS reactivation in true leaves, validating that RdDM antagonizes the effect of zebularine treatment in true leaves (Fig 3A).
Figure 3.
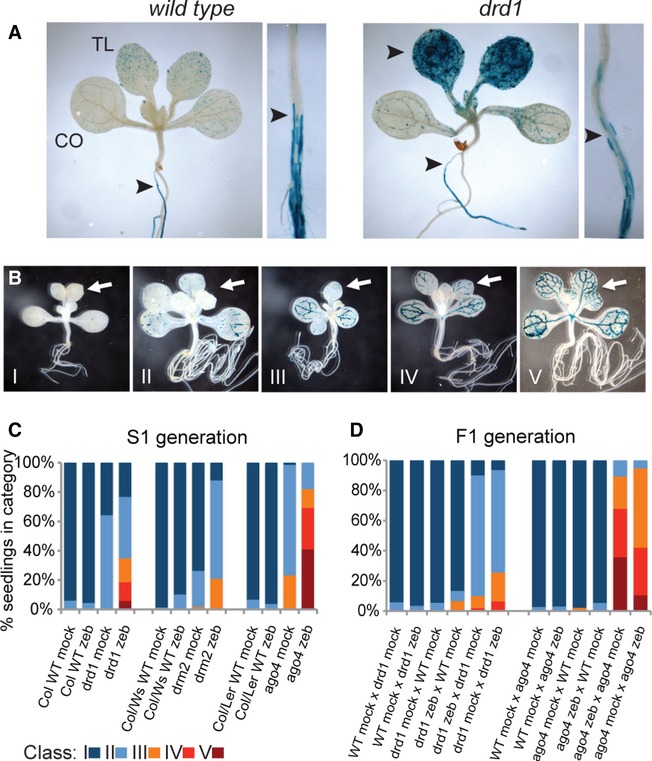
Zebularine treatment of RdDM mutants results in stochastic and transgenerational inheritance of active repetitive elements.
A TS-GUS reactivation in WT and drd1 after 6-day mock treatment followed by 6-day 20 μM zebularine treatment. Arrowheads indicate zebularine-mediated reactivation in tissues grown in the presence of the inhibitor.
B Representative examples of selfed (S1) seedlings from zebularine-treated S0 mutant plants with varying degree of TS-GUS reactivation in cotyledons and true leaves (I–V). Categories show plants with increasing degree of TS-GUS reactivation in true leaves (arrows).
C, D Percentage of S1 seedlings with varying GUS reactivation from S0 mock-treated and 20 μM zebularine-treated plants (C). Percentage of F1 seedlings with varying GUS reactivation from crosses between WT and mutant plants with or without zebularine (D). Approximately 100 seedlings were scored per genotype and treatment. The classification is based on (B).
Taken together, these results indicate that release of repression from a subset of TEs in true leaves requires simultaneous chemical interference with DNA methylation and genetic interference with the RdDM pathway. Hence, we suggest a central role of RdDM in mediating re-silencing of TEs in vegetative tissues by correcting for induced inefficiency in TGS maintenance. In contrast to TE re-silencing after genetic deletions of DDM1 that usually requires several generations 19, partial removal of methylation by zebularine is restored immediately.
Lack of RdDM components allows the inheritance of inhibitor-activated states
To address the inheritance of reactivated states, we screened TS-GUS activity in adult tissues that developed after recovery from zebularine treatment. Except for noticeable TS-GUS activity in the rosette leaves of 3-week-old drd1 and in the vascular system of 5-week-old ago4 plants, GUS expression in the remaining mutant lines was restricted to rare sectors varying in shape, size, and position between individual plants (Supplementary Fig S3B). These apparently stochastic effects were also evident from quantitative mRNA measurements for the expression of endogenous TEs, where independent biological replicates showed drastic differences between mutants or between targets (Supplementary Fig S3C).
The reactivation in adult tissues made us ask whether such stochastic activation can be transmitted to the next generation. Selfed progeny (S1) of mock- and zebularine-treated WT, drd1, ago4, and drm1/drm2 (S0) were grown on zebularine-free media and compared by GUS staining (Fig 3B). WT seedlings showed no staining, irrespective of the treatment, indicating full re-establishment of TS-GUS silencing. S1 plantlets obtained from mock-treated mutants displayed low GUS levels in cotyledons, as observed previously (Fig 2A). However, zebularine treatment of the parental plants during the first 3 weeks of vegetative growth led to an enhanced GUS staining in cotyledons of drm1/drm2 and furthermore in true leaves of drd1 and ago4 S1 progeny (Fig 3B and C). The differential degree of inheritance between individual RdDM mutants stems most likely from variable strength of silencing in the parental plants. Importantly, inheritance of active GUS was found in reciprocal crosses with mock-treated drd1 and ago4 plants, but was abolished in crosses with WT plants (Fig 3D).
The compromised re-silencing of repeats in zebularine-treated RdDM mutants provides evidence for a safeguarding function of the RdDM pathway during vegetative growth and consequently for the next generation. Genetic deletions of AGO4, DRD1, or DRM2 allowed the formation of clonal patches of active transgenes in later developing parts of the plant and increased the frequency of transmission of the active state to progeny. The mosaic-like expression patterns in the progeny likely reflect incomplete demethylation in different cells during zebularine treatment, resulting in epigenetic chimeras and differential representation of the affected cells after subsequent cell divisions. Crosses with WT plants providing functional AGO4 and DRD1 could prevent the transmission of the activated state to the next generation, demonstrating the requirement of the RdDM pathway for restoring silencing at re-activated repetitive elements.
Expression of RdDM and chromatin regulator genes is significantly increased in the SAM
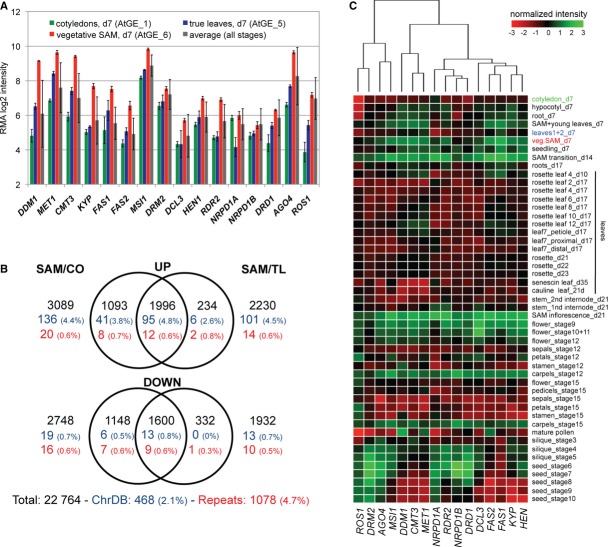
The above observations suggested a qualitative and quantitative difference in the degree of TGS control and its reinforcement in different tissues. Based on the clear exemption of the meristematic region from TS-GFP and TS-GUS activation in zebularine-treated seedlings (Fig 1D and F), we argued that the SAM could play a primary role in mediating this tissue-specific response. We compared gene expression in the vegetative SAM, cotyledons, true leaves, and a set of 49 different tissues in published ATH1 microarray data 20. First, we focused on a set of 16 genes known to be involved in TGS (Fig 4A). All of them had highest expression levels in the SAM sample, compared to cotyledons, true leaves, or average intensities calculated across all tissues (Fig 4A, and validated by qRT–PCR for a subset of genes; Supplementary Fig S4). This indicated that the stringent silencing observed in true leaves might originate from a high abundance of TGS factors in the SAM. This is in agreement with gene expression analysis of cells in the Arabidopsis shoot apical stem cell niche 21. In contrast, a control group of housekeeping genes failed to show similar differences between the analyzed samples (Supplementary Fig S4B).
Figure 4.
- Robust multiarray averaging (RMA)-normalized expression array values from probe sets corresponding to selected chromatin regulator genes in 7-day-old cotyledons (green), true leaves (blue), and vegetative shoot apical meristem (SAM) (red) compared to the average intensity in 49 different tissues or developmental stages (gray, see also C). Standard deviation from three biological replicates (individual tissues) or across all arrays (average) is indicated.
- Venn diagrams representing significant differential gene expression between SAM and cotyledons or true leaves. The total number of differentially expressed probes and the percent (in parentheses) are shown.
- Heatmap and hierarchical clustering visualizing normalized tissue-specific expression of selected TGS genes across 49 different Arabidopsis tissues or developmental stages.
We observed that genes involved in the maintenance of TGS (MET1, DDM1, CMT3, or FAS1) were less expressed in cotyledons compared to true leaves, most likely owing to lower proliferation rates in cotyledons. Nevertheless, a direct comparison between both tissues failed to detect significant differences in the expression of additional genes involved in chromatin regulation between cotyledons and true leaves (Supplementary Table S1 and Fig 4C). We next explored whether the elevated expression of chromatin regulators in the SAM tissue was due to a global increase in transcription by directly comparing expression to cotyledons or true leaves. No significant difference was found in the number of up- and down-regulated protein-coding genes, suggesting that the overall transcriptional activity is not elevated in SAM tissues (Fig 4B and Supplementary Table S2). However, genes encoding chromatin regulators were sixfold enriched in the SAM up-regulated (4.4%) versus the SAM down-regulated probe sets (0.7%) (Fig 4B; Supplementary Fig S4C and E and Supplementary Table S2). Nevertheless, the lower abundance of chromatin regulators in young and adult leaf tissues is sufficient to maintain TGS under standard conditions (Fig 4B; Supplementary Fig S4D and Supplementary Table S2).
Besides the vegetative SAM, we observed coordinated and increased expression of genes involved in establishment and maintenance of TGS in other meristematic tissues with rapidly dividing cells, including all apical meristems at different developmental stages (e.g., vegetative growth and transition to flowering), early stages of flower development, and all stages of carpel development (Fig 4C and Supplementary Fig S4F). In contrast, tissues growing mostly by cell expansion, such as hypocotyl, stem internodes, cotyledons, or differentiated leaves, had generally lower expression from the same set of genes.
Taken together, this suggests that the elevated expression of the RdDM pathway and other chromatin regulators in meristems functions as a relay mechanism that ensures correct propagation of silent states to new tissues and organs, including the germline. Cell-type specific differences in TGS were previously reported for gametophytes and early embryonic phases of plant development where specific components of TGS are coordinately up- or down-regulated in terminally differentiated companion cells 9–11. Although still a matter of debate 22, it has been proposed that this could lead to the generation of small RNAs complementary to TEs that reinforce silencing in the germline 9, 11. This suggests silencing checkpoints throughout gametogenesis and seed development. Preferential reinforcement of silencing in meristematic tissues, as reported here, would present a similar checkpoint during vegetative growth prior to formation of the next generation. Combined action of all three checkpoints could provide a robust surveillance mechanism that ensures silencing of TEs during vegetative growth and sexual propagation.
Materials and Methods
Plant material, growth conditions, and chemical treatments
The A. thaliana Col-0 TS-GUS (L5, 6b5) line 5, 16 was crossed with the mutants: rdr2-1 23; drd1-6 24; kyp (SALK 041474); fas1 (SAIL_662.D10); fas2 (SALK_033228); hda6 allele rts1-1 25; cmt3 in Ws-2 26, drm1/drm2 double mutant in Ws-2 7; ago4-1 in Ler1 27 and ddm1-5 in Zh 28. The segregating F2 plants were genotyped and lines homozygous for the TS-GUS locus and the mutations or the WT alleles were used for analyses. Plant treatments were performed as described 15. In brief, sterilized seeds were grown on agar-solidified germination medium containing 20 or 40 μM zebularine (Sigma) in growth chambers under 16-h light/8-h dark cycles at 21°C. Recovery was allowed after transferring zebularine-treated seedlings to drug-free medium or soil. 5-Azacytidine (Sigma) treatment was performed by germinating seeds for 3 days in water containing 400 μM 5-azacytidine (refreshed every 24 h) and subsequent recovery on drug-free medium for 7 days. To analyze the long-term effects, three plants with or without zebularine treatment were transferred to soil and analyzed 3 weeks later for TS-GUS activation in cauline leaves. Their seeds (S1) were grown on zebularine-free media and analyzed.
GUS and GFP detection and FISH
GUS staining was performed as described 15. Samples were analyzed using a Leica MZ16FA binocular microscope with a Leica DFC300FX CCD camera. GFP was analyzed under UV illumination with a Leica GFP1 filter (425/60–480 nm). Nuclei were isolated, centromeric repeat probes prepared and FISH performed as described 15. Images were acquired using a Zeiss Axioplan 2 microscope.
Tissue dissection, nucleic acid isolation, and gel-blots
Cotyledons, the first pair of true leaves, and tissues enriched for SAM and RAM were dissected from 2-week-old seedlings grown either on 20 μM zebularine or on drug-free medium. DNA was extracted with Phytopure (GE Healthcare) and RNA with RNeasy (Qiagen). Gel blot analyses were performed as described 15.
Bisulfite sequencing
DNA was isolated from cotyledons and true leaves of WT and drd1 grown on mock and 20 μM zebularine-containing media for 14 days. Samples were bisulfite-treated by EpiTect kit (Qiagen), and MULE2 and LINE1-4 (Chr_2:6,881,271-6,881,800; Chr_2:378,248-378,792, respectively) were amplified from the converted DNA using primers listed in the Supplementary Table S4. At least 15 unique reads per sample were analyzed by CyMATE 29.
qRT–PCR
DNase I-treated RNA was reverse-transcribed with random hexamer primers using RevertAid MuLV-RTase, RNaseH- (MBI Fermentas). qRT–PCR was done with SensiMix Plus SYBR and Fluorescein kit (Quantace) in an iQ5 system (Bio-Rad). PCR primers are given in Supplementary Table S4. Relative mRNA abundance was normalized to EIF4A1 or ACTIN2 mRNA.
Microarray data analysis
Affymetrix ATH1 gcRMA-normalized data 20 were downloaded from http://www.weigelworld.org. Heatmaps for selected chromatin regulators were generated according to z-scores across all samples, allowing hierarchical clustering using the heatmap.2 package in R. Changes in gene expression were calculated by contrasting vegetative_SAM_d7 (AtGE_6) to cotyledons_d7 (AtGE_1) or true_leaves_d7 (AtGE_5) in R using the Limma package. Only log2 fold-changes > 1 or < −1 with adjusted P values < 0.05 were considered significant. 475 and 1155 probe sets corresponding to chromatin regulators (Chromatin Data Base, http://www.chromdb.org/) and TEs 11, respectively, were considered (Supplementary Table S3).
Acknowledgments
We thank H. Vaucheret, T. Blevins, and F. Meins for donation of reporter lines; B. Wohlrab, N. Lettner, and B. Eilts for technical assistance; and T. Friese for editing the manuscript. This work was funded by FWF (P18986 to OMS), GEN-AU (GZ 200.140-VI/1/2006 to OMS), EMBO (TB), GMI, FMI, and MPIPZ.
Author contributions
TB, AF, and AP conceived, performed, and analyzed the experiments; AP, TB, and OMS wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://embor.embopress.org
References
- 1.Feng S, Jacobsen SE, Reik W. Epigenetic reprogramming in plant and animal development. Science. 2010;330:622–627. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henderson IR, Jacobsen SE. Epigenetic inheritance in plants. Nature. 2007;447:418–424. doi: 10.1038/nature05917. [DOI] [PubMed] [Google Scholar]
- 3.Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, McCombie RW, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 4.Matzke M, Kanno T, Huettel B, Daxinger L, Matzke AJM. Targets of RNA-directed DNA methylation. Curr Opin Plant Biol. 2007;10:512–519. doi: 10.1016/j.pbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Elmayan T, Proux F, Vaucheret H. Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr Biol. 2005;15:1919–1925. doi: 10.1016/j.cub.2005.09.044. [DOI] [PubMed] [Google Scholar]
- 6.Huettel B, Kanno T, Daxinger L, Aufsatz W, Matzke AJM, Matzke M. Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 2006;25:2828–2836. doi: 10.1038/sj.emboj.7601150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao X, Jacobsen SE. Role of the Arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol. 2002;12:1138–1144. doi: 10.1016/s0960-9822(02)00925-9. [DOI] [PubMed] [Google Scholar]
- 8.Lippman Z, May B, Yordan C, Singer T, Martienssen R. Distinct mechanisms determine transposon inheritance and methylation via small interfering RNA and histone modification. PLoS Biol. 2003;1:e67. doi: 10.1371/journal.pbio.0000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ibarra CA, Feng X, Schoft VK, Hsieh T-F, Uzawa R, Rodrigues JA, Zemach A, Chumak N, Machlicova A, Nishimura T, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337:1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 11.Slotkin RK, Vaughn M, Borges F, Tanurdžić M, Becker JD, Feijó JA, Martienssen RA. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136:461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pecinka A, Dinh HQ, Baubec T, Rosa M, Lettner N, Scheid OM. Epigenetic regulation of repetitive elements is attenuated by prolonged heat stress in Arabidopsis. Plant Cell. 2010;22:3118–3129. doi: 10.1105/tpc.110.078493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tittel–Elmer M, Bucher E, Broger L, Mathieu O, Paszkowski J, Vaillant I. Stress-induced activation of heterochromatic transcription. PLoS Genet. 2010;6:e1001175. doi: 10.1371/journal.pgen.1001175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou L, Cheng X, Connolly BA, Dickman MJ, Hurd PJ, Hornby DP. Zebularine: a novel DNA methylation inhibitor that forms a covalent complex with DNA methyltransferases. J Mol Biol. 2002;321:591–599. doi: 10.1016/S0022-2836(02)00676-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baubec T, Pecinka A, Rozhon W, Mittelsten Scheid O. Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 2009;57:542–554. doi: 10.1111/j.1365-313X.2008.03699.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morel J-B, Mourrain P, Béclin C, Vaucheret H. DNA methylation and chromatin structure affect transcriptional and post-transcriptional transgene silencing in Arabidopsis. Curr Biol. 2000;10:1591–1594. doi: 10.1016/s0960-9822(00)00862-9. [DOI] [PubMed] [Google Scholar]
- 17.Blevins T. Molecular Genetic Analysis of siRNA Biogenesis and Function in Arabidopsis thaliana. Switzerland: University of Basel; 2009. PhD Thesis, http://oatd.org/oatd/record?record=oai\:edoc.unibas.ch\:1029. [Google Scholar]
- 18.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 19.Teixeira FK, Heredia F, Sarazin A, Roudier F, Boccara M, Ciaudo C, Cruaud C, Poulain J, Berdasco M, Fraga MF, et al. A role for RNAi in the selective correction of DNA methylation defects. Science. 2009;323:1600–1604. doi: 10.1126/science.1165313. [DOI] [PubMed] [Google Scholar]
- 20.Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nat Genet. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- 21.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grant–Downton R, Kourmpetli S, Hafidh S, Khatab H, Le Trionnaire G, Dickinson H, Twell D. Artificial microRNAs reveal cell-specific differences in small RNA activity in pollen. Curr Biol. 2013;23:R599–R601. doi: 10.1016/j.cub.2013.05.055. [DOI] [PubMed] [Google Scholar]
- 23.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:e104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kanno T, Mette MF, Kreil DP, Aufsatz W, Matzke M, Matzke AJM. Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr Biol. 2004;14:801–805. doi: 10.1016/j.cub.2004.04.037. [DOI] [PubMed] [Google Scholar]
- 25.Aufsatz W, Mette MF, van der Winden J, Matzke M, Matzke AJM. HDA6 a putative histone deacetylase needed to enhance DNA methylation induced by double-stranded RNA. EMBO J. 2002;21:6832–6841. doi: 10.1093/emboj/cdf663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartee L, Malagnac F, Bender J. Arabidopsis cmt3 chromomethylase mutations block non-CG methylation and silencing of an endogenous gene. Genes Dev. 2001;15:1753–1758. doi: 10.1101/gad.905701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zilberman D, Cao X, Jacobsen SE. ARGONAUTE4 Control of locus-specific siRNA accumulation and DNA and histone methylation. Science. 2003;299:716–719. doi: 10.1126/science.1079695. [DOI] [PubMed] [Google Scholar]
- 28.Mittelsten Scheid O, Afsar K, Paszkowski J. Release of epigenetic gene silencing by trans-acting mutations in Arabidopsis. Proc Natl Acad Sci USA. 1998;95:632–637. doi: 10.1073/pnas.95.2.632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hetzl J, Foerster AM, Raidl G, Mittelsten Scheid O. CyMATE: a new tool for methylation analysis of plant genomic DNA after bisulphite sequencing. Plant J. 2007;51:526–536. doi: 10.1111/j.1365-313X.2007.03152.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.