Abstract
The spindle assembly checkpoint (SAC) delays progression into anaphase until all chromosomes have aligned on the metaphase plate by inhibiting Cdc20, the mitotic co-activator of the APC/C. Mad2 and BubR1 bind and inhibit Cdc20, thereby forming the mitotic checkpoint complex (MCC), which can bind stably to the APC/C. Whether MCC formation per se is sufficient for a functional SAC or MCC association with the APC/C is required remains unclear. Here, we analyze the role of two conserved motifs in Cdc20, IR and C-Box, in binding of the MCC to the APC/C. Mutants in both motifs assemble the MCC normally, but IR motif integrity is particularly important for stable binding to the APC/C. Cells expressing Cdc20 with a mutated IR motif have a compromised SAC, as uninhibited Cdc20 can compete with the MCC for APC/C binding and activate it. We thus show that stable MCC association with the APC/C is critical for a functional SAC.
Keywords: APC/C, SAC, Cdc20, MCC
Introduction
Correct segregation of sister chromatids during mitosis depends on a surveillance mechanism referred to as the spindle assembly checkpoint (SAC). The SAC is composed of a set of conserved proteins that all accumulate at improperly attached kinetochores during prometaphase and this generates an inhibitory ‘anaphase wait’ signal [1, 2]. The SAC prevents further mitotic progression by inhibiting Cdc20, the mitotic co-activator of the Anaphase Promoting Complex/Cyclosome (APC/C). The APC/C is a large E3 ubiquitin ligase responsible for marking multiple substrates for degradation and requires one of two co-activators, Cdc20 or Cdh1, for its activity [3]. Binding and activation of the APC/C complex by co-activators depend on a conserved IR motif at the extreme C-terminus of the co-activators as well as a C-Box motif in the N-terminus [4, 5]. The IR motif binds to TPR containing subunits of the APC/C while the C-Box is likely to be closer to the catalytic subunits in agreement with a direct role of this motif in activating the APC/C [4, 6–9]. The APC/C complex recognizes destruction motifs in substrates via a combined interphase between the co-activator and the APC10 subunit [10–12]. Two critical mitotic substrates of APC/C-Cdc20 are Cyclin B1 and Securin as the degradation of these results in mitotic exit and sister chromatid separation respectively [13].
During an active checkpoint Cdc20 is inhibited by the binding of the checkpoint proteins Mad2 and BubR1-Bub3 (we will refer to the BubR1-Bub3 complex as BubR1 throughout) to form the mitotic checkpoint complex (MCC) [14–19]. Although Mad2 and BubR1 alone can inhibit Cdc20 directly, they strongly synergize explaining the need for both proteins for a functional SAC [20–23]. Mad2 binds to a short consensus sequence in the N-terminus of Cdc20 while BubR1 interacts with the Cdc20 propeller domain and additional contacts between Mad2 and BubR1 help to stabilize the entire MCC [24, 25]. The binding of Mad2 and BubR1 interferes with important functional motifs in Cdc20 as the Mad2 binding motif is also critical for APC/C binding and BubR1 appears to act as a pseudo-substrate by competing with substrate binding through a conserved KEN box motif [21, 26–29]. In addition the MCC complex can bind stably to the APC/C and the cryo-EM structure of the APC/C-MCC complex suggests that this repositions Cdc20 away from APC10 thus preventing functional substrate interactions [24, 30]. Whether the stable association of the MCC with the APC/C is required for a functional SAC is not clear as apo-APC/C is present during an active SAC and the association of the MCC with the APC/C has also been proposed to regulate both Cdc20 and MCC stability [31–34].
Here we reveal a critical role of the IR motif of Cdc20 in stable binding of the MCC to the APC/C and show that this is important for a functional SAC. Since mutation of the IR motif per se does not affect the formation of MCC, this reveals that stable MCC association with the APC/C is critical for a functional SAC.
Results and Discussion
The C-Box and IR motifs of Cdc20 are required for mitotic progression
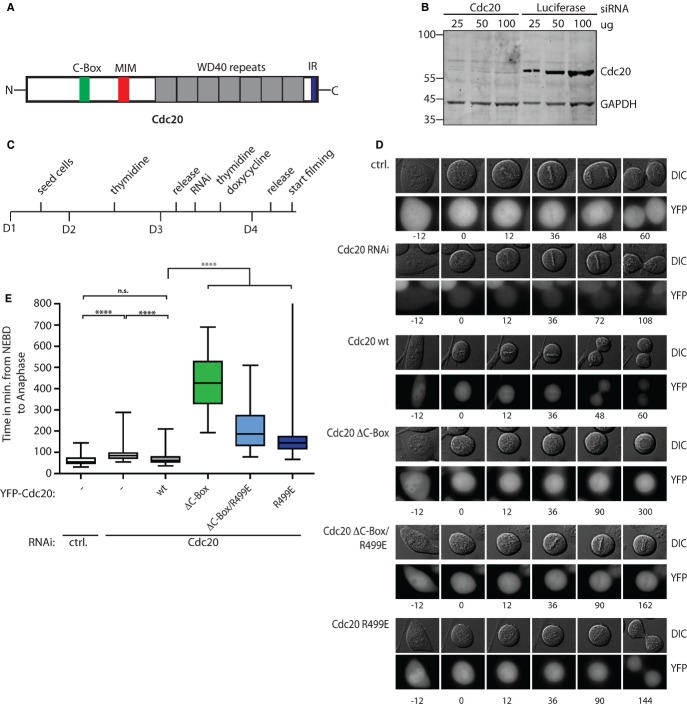
As Cdc20 is required for binding of the MCC to the APC/C we wanted to investigate the role of functional important motifs in Cdc20 for this interaction. At the extreme C-terminus, Cdc20 harbors an IR dipeptide motif that has previously been shown to bind to TPR subunits of the APC/C and in the N-terminal unstructured tail, Cdc20 contains a conserved C-Box that also contributes to the binding of co-activators to the APC/C (Fig 1A). The role of these motifs in Cdc20 regulation during an active SAC has however not been carefully investigated in human cells. To analyze these motifs we generated stable inducible isogenic HeLa cell lines using the Flp-in system expressing siRNA-resistant forms of YFP-tagged Cdc20. We generated cell lines expressing Cdc20 wild-type (WT), Cdc20 R499E were the arginine of the IR motif was mutated to glutamic acid, Cdc20 ΔC-Box where amino acids 78–80 were deleted removing three conserved residues of the C-Box and finally a form encompassing both mutations (referred to as Cdc20 ΔC-Box/R499E). Due to the fact that the different Cdc20 proteins displayed different stabilities we optimized the induction time so that almost equal levels of the different exogenous Cdc20 proteins were present which was approximately 5 times the endogenous level (Supplementary Fig 1A). All cell lines were homogenous and at least 70% of the cells expressed Cdc20 upon induction as determined by microscopy.
Figure 1.
- Schematic representation of Cdc20 with important functional motifs investigated in this study. Indicated are the seven WD40 repeats (grey), the C-Box (green) and MIM (Mad2 interacting motif) (red) in the unstructured N-terminal part and the IR motif (blue) in the C-terminus.
- Blot of Cdc20 RNAi-treated cells and control-treated cells is shown with the indicated amount of total extract loaded.
- Experimental setup of RNAi rescue time-lapse experiments.
- Representative DIC and fluorescence images of RNAi rescue time-lapse experiments. Ctrl RNAi-treated cells progress through mitosis normally with a median time of 54 min. Cdc20 RNAi-treated cells arrest at metaphase with a median time of 84 min, which is a significant increase compared to control (P < 0.0001). This phenotype is only rescued in Cdc20 WT-expressing cells, not in Cdc20 ΔC-Box-, Cdc20 ΔC-Box/R499E-or Cdc20 R499E-expressing cells.
- Quantification of mitotic timing (NEBD to anaphase, arrest or apoptosis). N > 47 per conditions shown as a box-and-whisker plot with the median indicated by a line in the box representing the lower and upper quartile. Mann-Whitney test was performed for statistical analysis (P-value: ****P < 0.0001, n.s., non significant). Only cells recorded for at least 2 h were included in the analysis.
Source data are available online for this figure.
We first analyzed the ability of the different Cdc20 proteins to rescue the mitotic arrest induced by depleting the endogenous Cdc20 by RNAi using live-cell imaging and scoring the time from nuclear envelope breakdown (NEBD) to anaphase (Fig 1C–E). Using the outlined protocol Cdc20 was depleted efficiently such that approximately 15% endogenous protein remained (Fig 1B). Although the removal of endogenous Cdc20 induced a 30-min metaphase delay, this was not an extensive delay likely reflecting that very little Cdc20 is required for mitotic progression as also observed by others [35]. The mitotic delay observed upon Cdc20 RNAi was fully rescued by expressing Cdc20 WT but none of the Cdc20 mutants restored mitotic timing confirming that the mutation of the C-box or IR motif results in inactive Cdc20 molecules as expected. Surprisingly, the expression of the inactive Cdc20 molecules resulted in a much longer metaphase delay than the Cdc20 RNAi condition suggesting that the Cdc20 mutants interfere with the activity of the remaining endogenous Cdc20. This increase in the metaphase delay did not appear to be due to an effect of the mutant Cdc20 molecules on the stability of the remaining endogenous Cdc20 as far as we can judge by Western blot (Supplementary Fig S1B). Although Cdc20 ΔC-Box and Cdc20 R499E might maintain some capacity to compete with endogenous Cdc20 for APC/C binding, this is unlikely the mechanism by which Cdc20 ΔC-Box/R499E induces a prolonged metaphase delay. Potentially the inactive Cdc20 molecules interfere with activating mechanisms of the remaining endogenous Cdc20 either spatially or by sequestering factors required for this or through dimerization.
The C-Box and IR motifs of Cdc20 are required for APC/C binding during an active checkpoint
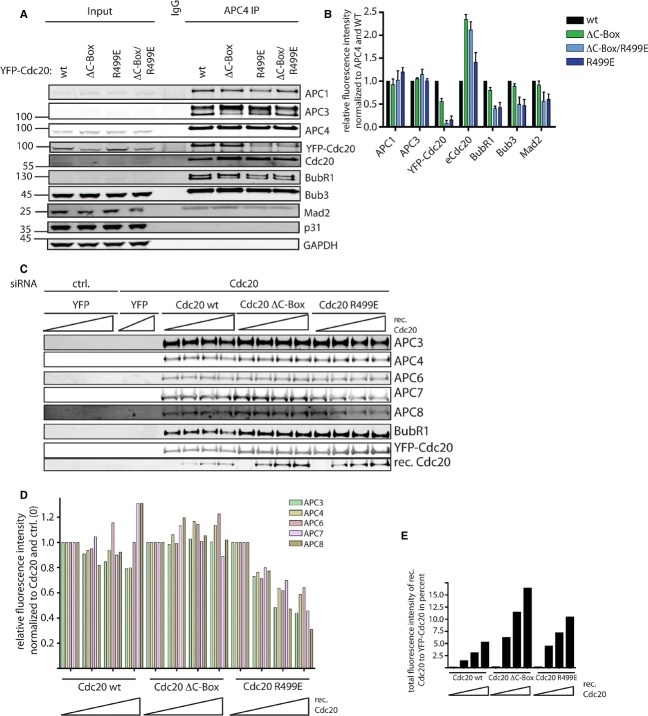
The above experiments validated the cell lines and Cdc20 mutants and we next analyzed the interaction with the APC/C and MCC components during a SAC arrest. In the stable cell lines we removed endogenous Cdc20 by RNAi and arrested cells in mitosis with nocodazole and collected cells by mitotic shake-off and then performed Cdc20 purifications using a GFP-binder affinity resin or APC/C purifications using an APC4 monoclonal antibody (Fig 2). All Western blots were analyzed using Li-cor quantitative Western blot technology allowing quantification of experiments. In Cdc20 purifications we observed no changes in the interaction with MCC components with either Cdc20 R499E or Cdc20 ΔC-Box while Cdc20 ΔC-Box/R499E displayed slightly elevated levels of all MCC components as well as a 4-fold increase in the SAC antagonist p31comet (Fig 2A and B). The increase in p31comet levels associated with Cdc20 ΔC-Box/R499E likely reflects that p31comet cannot associate efficiently with the MCC when bound to the APC/C as reported by the Taylor lab [36] and we have also been unable to detect APC/C subunits in p31comet purifications (Supplementary Fig 1C). The elevated levels of MCC components in Cdc20 ΔC-Box/R499E could be due to inefficient disassembly of the MCC complex which constantly occurs during an active SAC and which might involve APC/C activity [31, 32, 34]. However when we forced MCC disassembly by adding an inhibitor of Aurora B we did not observe any difference in the rates of MCC disassembly compared to Cdc20 WT arguing that this is not the reason (Supplementary Fig 1D–E). Therefore we suspect that the fact that Cdc20 ΔC-Box/R499E cannot bind the APC/C makes MCC formation more efficient.
Figure 2.
- Endogenous Cdc20 was depleted by RNAi, cells were synchronized for 24 h in thymidine, released into nocodazole for 11 h and harvested by mitotic shake-off. 24 h prior to shake-off, expression of the indicated constructs was induced by doxycycline (10 ng/ml) and YFP-Cdc20 purified. The binding of MCC and APC/C components was analyzed by Western blot.
- Quantification of MCC and APC/C components normalized to YFP-Cdc20 signal and with Cdc20 WT set to 1. Mean of two independent experiments shown with the black squares showing the values obtained in the two experiments.
- Purification of the APC/C complex using an APC4 monoclonal antibody. The cells were synchronized as in (A) and either induced with 0.5 ng/ml or 10 ng/ml doxycycline for 24 h. The binding of MCC components and Cdc20 proteins was analyzed by Western blot.
- Quantification of three independent experiments as in (C) with the mean and standard deviation shown.
Source data are available online for this figure.
In Cdc20 R499E and Cdc20 ΔC-Box purifications all of the APC/C subunits we analyzed were reduced and combining these two mutations almost completely abolished APC/C binding (Fig 2A and B). A similar result was obtained when we purified the APC/C complex and as expected the MCC levels on the APC/C mirrored that of Cdc20 (Fig 2C and D). To analyze this further we purified Cdc20 WT, Cdc20 ΔC-Box and Cdc20 R499E from nocodazole-arrested cells and washed the complexes with buffers containing increasing concentrations of NaCl (0, 150, 300 mM) (Supplementary Fig 2A–B). This experiment confirmed the importance of the C-box and IR motif in binding Cdc20 to the APC/C as the mutants were more sensitive to an increase in salt concentration compared to the WT protein. In all the assays we have conducted the R499E mutation affects APC/C binding more than the C-box which mirrors observations with the co-activator Cdh1 [4]. In line with this we observed that Cdc20 R499E is more stable during a SAC arrest than Cdc20 ΔC-Box (Supplementary Fig 2C). As the degradation of Cdc20 ΔC-Box required the presence of nocodazole this suggests that stable MCC-APC/C interaction is required for Cdc20 degradation.
Our results show that during an active SAC both the C-Box and IR motif of Cdc20 contribute to the binding of the MCC to the APC/C complex. These observations are in disagreement with observations from the Pines lab that observed no effect on APC/C binding when they mutated the C-box or deleted the IR motif in Cdc20 [21]. One possibility for the difference could be that deleting the IR motif, rather than mutating it to IE, did not interfere with MCC formation or alternatively that the buffer conditions used in that study were unable to reveal an effect of deleting the IR motif. To investigate this we obtained the cell lines used in the Pines lab study and purified FLAG-tagged Cdc20 WT or Cdc20 ΔIR from nocodazole-treated cells under different buffer conditions and either in the presence or absence of endogenous Cdc20. Similar to our results using Cdc20 R499E we observed a strong reduction in APC/C binding when the IR motif was deleted and this was particularly clear at high salt concentrations and in the presence of endogenous Cdc20 (Supplementary Fig 3A–C). We suspect that the reason why no effect of deleting the IR motif was observed in that previous study is due to the buffer conditions used. Testing a range of buffer conditions is ideally needed to probe the effect of mutations on protein-protein interactions.
We have recently shown a requirement for the APC3 subunit in stable binding of the MCC to the APC/C [37] and since APC3 is the likely IR receptor this is in agreement with the observations presented here on the role of the IR motif. Although Cdc20 shifts in position on the APC/C when part of the MCC complex [30] the APC3-IR interaction could be maintained due to the flexible C-terminus of Cdc20.
Stable association of the MCC with the APC/C is required for a functional checkpoint
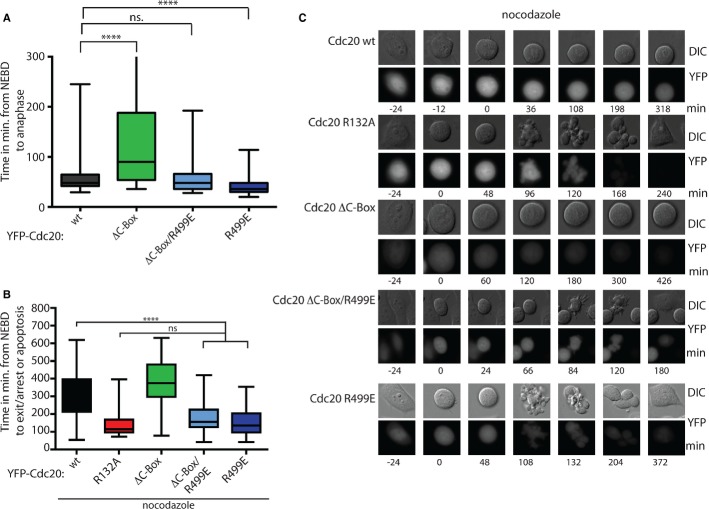
Although our biochemical analysis of Cdc20 R499E and Cdc20 ΔC-Box did not reveal major differences in MCC assembly we were surprised that in the presence of endogenous Cdc20 a marked difference in mitotic duration was observed. Cdc20 ΔC-Box expressing cells were specifically delayed in mitosis and this delay required an intact IR motif as Cdc20 ΔC-Box/R499E did not delay (Fig 3A). In contrast, Cdc20 R499E-expressing cells progressed faster through mitosis (mean 43 ± 1.7 min.) than Cdc20-expressing cells (mean 64 ± 4.4 min.) and these results suggested that there could be an important functional difference between the IR motif and the C-Box in the SAC. To address this, we challenged cells with nocodazole to engage the SAC and monitored cell fate by time-lapse microscopy. While Cdc20 WT and Cdc20 ΔC-Box delayed in response to nocodazole, it was clear that the mutation of the IR motif resulted in a less efficient SAC as many Cdc20 R499E and Cdc20 ΔC-Box/R499E-expressing cells exited mitosis within 2 h (Fig 3B and C). Similar observations were obtained with Cdc20 ΔIR (Supplementary Fig 3D). Although cells expressing Cdc20 R499E or Cdc20 DC-Box/R499E had a compromised SAC, the effect was not as severe as when we directly reduced MCC formation by mutating the Mad2 binding site (Cdc20 R132A).
Figure 3.
- Cells expressing the indicated Cdc20 proteins were monitored by time-lapse microscopy in the presence of endogenous Cdc20 and the time from NEBD to anaphase measured. Cells expressing the indicated Cdc20 proteins were synchronized by a double thymidine block. 24 h prior to imaging doxycycline (10 ng/ml) was added to induce protein expression and cells were filmed every 6 min by time-lapse microscopy for 18 h. At least 146 cells were analyzed per condition shown as a box-and-whisker plot with the median indicated by a line in the box representing the lower and upper quartile. Mann–Whitney test was performed for statistical analysis (P-value: ****P < 0.0001, ns, non significant).
- Similar experiment as in (A) but in the presence of 30 ng/ml nocodazole. Time from anaphase to mitotic exit, arrest or apoptosis is measured. N > 35 shown as a box-and-whisker plot with the median indicated by a line in the box representing the lower and upper quartile. Mann–Whitney test was performed for statistical analysis (P-value: ****P < 0.0001, ns, non significant).
- Representative DIC and fluorescent images for the experiments quantified in (B).
Source data are available online for this figure.
Given that mutation of the IR motif affected the SAC in the presence of the endogenous Cdc20, we investigated binding of the MCC and endogenous Cdc20 in the different cell lines. We purified the APC/C complex from nocodazole-arrested cells and quantified the MCC components and exogenous and endogenous Cdc20. In these experiments it was evident that binding to the APC/C of Cdc20 R499E and Cdc20 ΔC-Box/R499E was strongly impaired in the presence of endogenous Cdc20 while Cdc20 WT and Cdc20 ΔC-Box still could bind (Fig 4A and B). Although mutation of either the C-Box or IR motif resulted in higher levels of endogenous Cdc20, the levels of MCC components were specifically reduced in the R499E mutants. This suggests that when the IR motif is mutated, but not the C-Box, uninhibited Cdc20 could compete with the MCC for binding to the APC/C explaining the compromised SAC. To confirm this we added increasing concentrations of recombinant Cdc20 to nocodazole extracts from the different YFP-Cdc20-expressing cell lines and measured the ability of the recombinant Cdc20 to compete for APC/C binding. Similar to the in vivo purifications, the mutation of either the C-box or the IR motif allowed for more recombinant Cdc20 to bind but specifically in the R499E mutant we observed a reduction in APC/C subunits with increasing concentrations of recombinant Cdc20 (Fig 4C–E). The reason for increased levels of recombinant Cdc20 in Cdc20 ΔC-box is likely that recombinant Cdc20 can bind to the C-box binding site on APC/C and that this APC/C complex will have two Cdc20 molecules bound to it at the same time.
Figure 4.
- The APC/C complex was purified from nocodazole-arrested cells using an APC4 monoclonal antibody and expressing the indicated exogenous Cdc20 proteins. The binding of endogenous and exogenous Cdc20 as well as MCC components was analyzed by Western blotting.
- Quantification of the experiment described in (A) with the signal normalized to APC4 and to Cdc20 WT. The mean and standard deviation from 3 independent experiments is shown.
- From the indicated YFP-Cdc20-expressing cells a mitotic lysate was prepared and increasing concentrations of recombinant Cdc20 added corresponding to 5-, 25-and 50-fold above endogenous Cdc20 levels respectively. Following incubation for 1 h at 37°C the YFP-tagged Cdc20 was purified and the amount of recombinant Cdc20 as well as APC/C subunits bound analyzed by Western blot.
- Quantification of the indicated APC subunits normalized to YFP-Cdc20 with increasing concentrations of recombinant Cdc20 added.
- Quantification of recombinant Cdc20 bound relative to YFP-Cdc20.
Source data are available online for this figure.
Here we reveal that stable association of the MCC with the APC/C critically depends on the conserved C-box and IR motifs of Cdc20. Although both motifs contribute to the stability of the APC/C-MCC complex, the IR motif is more critical and its mutation allows free Cdc20 to bind and activate the APC/C resulting in a SAC override. The fact that stable MCC association with the APC/C is critical for a functional SAC is puzzling as there is also a pool of apo-APC/C present during an active SAC in human cells [30]. Likely there are important spatial and functional aspects of APC/C and SAC regulation that needs to be dissected before this can be fully understood.
Materials and Methods
Cloning and generation of stable cell lines
Full-length RNAi-resistant Cdc20 was amplified by PCR and cloned into the BamHI and NotI sites of pcDNA5/FRT/TO 3*FLAG-Venus. Cdc20 R499E, Cdc20ΔC-Box, Cdc20ΔC-Box/R499E and Cdc20 ΔIR were obtained by quickchange using the following primers: deletion of C-Box (5′GCAAACCTGGCGGTATCCCCCATCGCAGTG), R499E mutation (5′CCACCAAGGCATCGAGTGAGCGGCCGC), deletion of the IR motif (5′ATCCACCAAGGCTGAGCGGCCGC). The generation of stable HeLa cell lines expressing constructs under the control of a doxycline-inducible promotor was done as previously described [33].
Antibodies
The following antibodies were used at the indicated dilutions for Western blot. Cdc20 mouse monoclonal (sc-13162, 1:1,000; Santa Cruz), Cdc20 rabbit polyclonal (A301-180A-1, 1:1000; Bethyl Laboratories), p31 rabbit polyclonal (1:500; raised against full length protein), GAPDH rabbit polyclonal (sc-25778, 1:500; Santa Cruz), Mad2 rabbit polyclonal (A300-300A, 1:500; Bethyl Laboratories), Mad2 mouse monoclonal (1:500; gift from A. Musacchio), Bub3 mouse monoclonal (611731, 1:500; BD Biosciences), BubR1 rabbit polyclonal (A300-995A, 1:1,000; Bethyl Laboratories), APC1 rabbit polyclonal (A301-653A-1, 1:500; Bethyl Laboratories), APC4 mouse monoclonal (1:500; raised against C-terminal peptide), APC3 mouse monoclonal (610454, 1:500; BD Biosciences), APC8 (611402, 1:200; Biolegend), APC6 goat polyclonal (sc6395, 1:250; Santa Cruz), APC7 rabbit polyclonal (A302-551-1, 1:500; Bethyl Laboratories), FLAG rabbit polyclonal (F7425, 1:500; Sigma).
RNAi
Endogenous Cdc20 was depleted using RNAiMAX from Life Technologies according to manufacturer's instructions. Cdc20 siRNA (5′ CGGAAGACCUGCCGUUACAUU) was obtained from Sigma or Dharmacon and used at a concentration of 50 to 125 nM. In biochemical experiments cells were treated for 36 h prior to harvesting.
Live-cell imaging
Cells grown in 8-well slides (Ibidi) were cultured in complete DMEM and subjected to a double thymidine block. Twenty-four hours prior to imaging the expression of indicated constructs was induced using 10 ng/ml doxycycline. Two hours after release from the second thymidine block the medium was changed to L-15 medium (Life Technologies) supplemented with 10% fetal bovine serum (Hyclone) prior to imaging. 30 ng/ml nocodazole was added when indicated. The slide was mounted onto a Delta Vision Elite microscope (GE Healthcare) and cells were filmed for 16–18 h in 6–7 min intervals using a 40X, 1.35 NA, WD 0.10 objective. All data analysis was performed using the softWoRx software (GE Healthcare).
Statistical analysis was performed with Prism software and a Mann–Whitney test was used.
Purification of MCC and APC/C complexes
MCC and APC/C complexes were purified from cells after 24 h of thymidine treatment, released for 11 h into nocodazole (200 ng/ml) and harvested by mitotic shake-off. 24 h prior to mitotic shake-off the expression of the indicated constructs was induced using 10 ng/ml doxycycline (if not stated differently). Cells were lysed in lysis buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.8, 1 mM EDTA, 1 mM DTT and 0.1 to 1% NP40). MCC complexes were immunoprecipitated using GFP-Trap (ChoromoTek) for 10 min at 4°C or using anti-FLAG beads (Sigma) for 1 h at 4°C. APC/C complexes were immunoprecipitated using a mouse monoclonal APC4 antibody cross-linked to Protein G-Sepharose 4B (Life Technologies) for 1 h at 4°C. Precipitated protein complexes were washed in lysis buffer and eluted in 4X SDS sample buffer (Life Technologies).
Acknowledgments
We thank Stephen Taylor for providing the HeLa FRT TRex cell line and Giuseppe Cazzamali and Michael Williamson for producing recombinant Cdc20. We are grateful to Jon Pines and Daisuke Izawa for proving FLAG tagged Cdc20 cell lines. We thank Tiziana Lischetti and Marie Sofie Yoo Larsen for comments on the manuscript. This work was supported by grants from the Danish Cancer Society, the Lundbeck Foundation and the Novo Nordisk Foundation to JN.
Author contributions
JBH performed and designed all experiments; JN assisted with clonings and wrote the main part of the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://embor.embopress.org
References
- 1.Musacchio A. Spindle assembly checkpoint: the third decade. Philos Trans R Soc Lond B Biol Sci. 2011;366:3595–3604. doi: 10.1098/rstb.2011.0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lara-Gonzalez P, Westhorpe FG, Taylor SS. The spindle assembly checkpoint. Curr Biol. 2012;22:R966–R980. doi: 10.1016/j.cub.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 3.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 4.Vodermaier HC, Gieffers C, Maurer-Stroh S, Eisenhaber F, Peters J-M. TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr Biol. 2003;13:1459–1468. doi: 10.1016/s0960-9822(03)00581-5. [DOI] [PubMed] [Google Scholar]
- 5.Schwab M, Neutzner M, Möcker D, Seufert W. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC. EMBO J. 2001;20:5165–5175. doi: 10.1093/emboj/20.18.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kimata Y, Baxter JE, Fry AM, Yamano H. A role for the Fizzy/Cdc20 family of proteins in activation of the APC/C distinct from substrate recruitment. Mol Cell. 2008;32:576–583. doi: 10.1016/j.molcel.2008.09.023. [DOI] [PubMed] [Google Scholar]
- 7.Schreiber A, Stengel F, Zhang Z, Enchev RI, Kong EH, Morris EP, Robinson CV, Fonseca da PC, Barford D. Structural basis for the subunit assembly of the anaphase-promoting complex. Nature. 2011;470:227–232. doi: 10.1038/nature09756. [DOI] [PubMed] [Google Scholar]
- 8.Wendt KS, Vodermaier HC, Jacob U, Gieffers C, Gmachl M, Peters JM, Huber R, Sondermann P. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8:784–788. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 9.Thornton BR, Ng TM, Matyskiela ME, Carroll CW, Morgan DO, Toczyski DP. An architectural map of the anaphase-promoting complex. Genes Dev. 2006;20:449–460. doi: 10.1101/gad.1396906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Passmore LA, Barford D. Coactivator functions in a stoichiometric complex with anaphase-promoting complex/cyclosome to mediate substrate recognition. EMBO Rep. 2005;6:873–878. doi: 10.1038/sj.embor.7400482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonseca da PCA, Kong EH, Zhang Z, Schreiber A, Williams MA, Morris EP, Barford D. Structures of APC/C(Cdh1) with substrates identify Cdh1 and Apc10 as the D-box co-receptor. Nature. 2011;470:274–278. doi: 10.1038/nature09625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carroll CW, Enquist-Newman M, Morgan DO. The APC subunit Doc1 promotes recognition of the substrate destruction box. Curr Biol. 2005;15:11–18. doi: 10.1016/j.cub.2004.12.066. [DOI] [PubMed] [Google Scholar]
- 13.Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- 14.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 16.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 19.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 20.Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 21.Izawa D, Pines J. Mad2 and the APC/C compete for the same site on Cdc20 to ensure proper chromosome segregation. J Cell Biol. 2012;199:27–37. doi: 10.1083/jcb.201205170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chao WCH, Kulkarni K, Zhang Z, Kong EH, Barford D. Structure of the mitotic checkpoint complex. Nature. 2012;484:208–213. doi: 10.1038/nature10896. [DOI] [PubMed] [Google Scholar]
- 25.Tipton AR, Wang K, Link L, Bellizzi JJ, Huang H, Yen T, Liu ST. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J Biol Chem. 2011;286:21173–21179. doi: 10.1074/jbc.M111.238543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Lees E. Identification of an overlapping binding domain on Cdc20 for Mad2 and anaphase-promoting complex: model for spindle checkpoint regulation. Mol Cell Biol. 2001;21:5190–5199. doi: 10.1128/MCB.21.15.5190-5199.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.King EMJ, Sar van der SJA, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lara-Gonzalez P, Scott MIF, Diez M, Sen O, Taylor SS. BubR1 blocks substrate recruitment to the APC/C in a KEN-box-dependent manner. J Cell Sci. 2011;124:4332–4345. doi: 10.1242/jcs.094763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Herzog F, Primorac I, Dube P, Lénárt P, Sander B, Mechtler K, Stark H, Peters JM. Structure of the anaphase-promoting complex/cyclosome interacting with a mitotic checkpoint complex. Science. 2009;323:1477–1481. doi: 10.1126/science.1163300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mansfeld J, Collin P, Collins MO, Choudhary JS, Pines J. APC15 drives the turnover of MCC-CDC20 to make the spindle assembly checkpoint responsive to kinetochore attachment. Nat Cell Biol. 2011;13:1234–1243. doi: 10.1038/ncb2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- 33.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uzunova K, Dye BT, Schutz H, Ladurner R, Petzold G, Toyoda Y, Jarvis MA, Brown NG, Poser I, Novatchkova M, Mechtler K, Hyman AA, Stark H, Schulman BA, Peters JM. APC15 mediates CDC20 autoubiquitylation by APC/C(MCC) and disassembly of the mitotic checkpoint complex. Nat Struct Mol Biol. 2012;19:1116–1123. doi: 10.1038/nsmb.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolthuis R, Clay-Farrace L, Zon van W, Yekezare M, Koop L, Ogink J, Medema R, Pines J. Cdc20 and Cks direct the spindle checkpoint-independent destruction of cyclin A. Mol Cell. 2008;30:290–302. doi: 10.1016/j.molcel.2008.02.027. [DOI] [PubMed] [Google Scholar]
- 36.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124(Pt 22):3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sedgwick GG, Hayward DG, Di Fiore B, Pardo M, Yu L, Pines J, Nilsson J. Mechanisms controlling the temporal degradation of Nek2A and Kif18A by the APC/C-Cdc20 complex. EMBO J. 2013;32:303–314. doi: 10.1038/emboj.2012.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.