Abstract
The spindle assembly checkpoint (SAC) ensures accurate chromosome segregation by delaying entry into anaphase until all sister chromatids have become bi-oriented. A key component of the SAC is the Mad2 protein, which can adopt either an inactive open (O-Mad2) or active closed (C-Mad2) conformation. The conversion of O-Mad2 into C-Mad2 at unattached kinetochores is thought to be a key step in activating the SAC. The “template model” proposes that this is achieved by the recruitment of soluble O-Mad2 to C-Mad2 bound at kinetochores through its interaction with Mad1. Whether Mad1 has additional roles in the SAC beyond recruitment of C-Mad2 to kinetochores has not yet been addressed. Here, we show that Mad1 is required for mitotic arrest even when C-Mad2 is artificially recruited to kinetochores, indicating that it has indeed an additional function in promoting the checkpoint. The C-terminal globular domain of Mad1 and conserved residues in this region are required for this unexpected function of Mad1.
Keywords: Mad1, Mad2, mitosis, SAC
Introduction
The SAC ensures accurate chromosome segregation by delaying anaphase entry by inhibiting Cdc20, the mitotic co-activator of the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase essential for targeting cyclin B1 and securin for degradation [1]. Cdc20 is inhibited by the direct binding of Mad2 and the BubR1-Bub3 checkpoint proteins forming the mitotic checkpoint complex (MCC) [2–6]. Current models propose that the Mad2-Cdc20 complex represents the initial inhibitory complex formed that is then converted into the MCC by binding of BubR1-Bub3. Following this Mad2 is removed by a p31-dependent mechanism to generate the Cdc20-BubR1-Bub3 complex potentially representing the final inhibitor [7–10].
Given the importance of the Mad2-Cdc20 complex, it is critical to understand how unattached kinetochores catalytically generate this complex. An important feature of Mad2 is that it exists in at least two conformations, namely an active closed conformation (C-Mad2) that is the conformation observed when Mad2 is bound to its ligands Mad1 and Cdc20, and an inactive open conformation (O-Mad2), which is the predominant conformation of soluble Mad2 [11–15]. The Mad1-Mad2 complex is an extremely stable complex displaying little exchange of bound C-Mad2, and Mad1 makes contacts with the kinetochore to position C-Mad2 at this structure [16–18]. Based on the observation that C-Mad2 can catalyze the conversion of O-Mad2 into C-Mad2-Cdc20 in vitro and that C-Mad2 and O-Mad2 can dimerize the “template model” proposes that unattached kinetochores act to generate C-Mad2 by recruitment of O-Mad2 to the C-Mad2-Mad1 complex localized at unattached kinetochores [19–21]. This model explains the need for both soluble Mad2 and the Mad1-Mad2 complex, the observed FRAP kinetics of Mad2, and the requirement for the Mad2 dimerization interface for a functional SAC [20, 22–25].
In the template model, the active molecule at the kinetochore is C-Mad2, while Mad1 merely acts to bring this molecule to the kinetochore. In agreement with this, no differences in the ability to promote O-Mad2 conversion have been observed when C-Mad2 and C-Mad2-Mad1 were compared in in vitro assays [19].
Surprisingly, we show here that Mad1 is absolutely essential for generating an active SAC even when C-Mad2 is constitutively recruited to kinetochores. We find that the C-terminal globular domain of Mad1 and conserved residues in this region are critical for this role of Mad1 in the SAC. Our work reveals an unexpected direct role of Mad1 in the SAC.
Results and Discussion
Constitutive recruitment of Mad2 to kinetochores results in a mitotic arrest
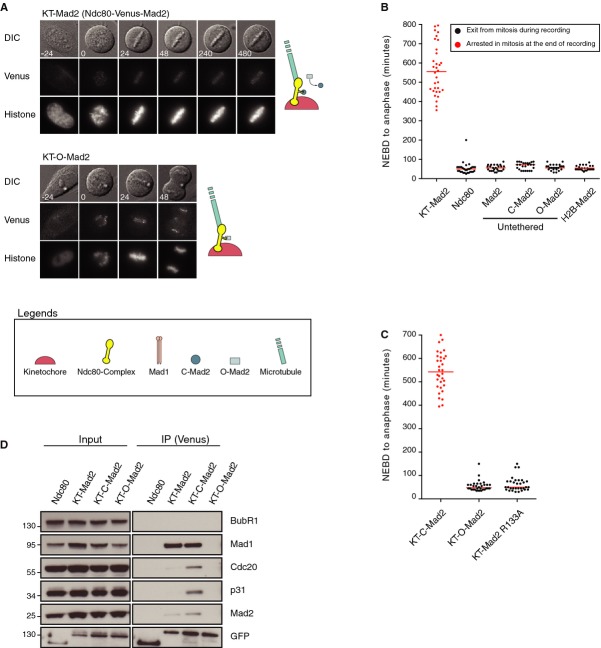
To investigate whether the only function of Mad1 in the SAC is to recruit Mad2 to kinetochores, we needed to bypass the requirement of Mad1 for Mad2 kinetochore targeting. To this end, we targeted Mad2 to kinetochores by fusing it to the C-terminus of the outer kinetochore protein Ndc80 (referred to as KT-Mad2). The KT-Mad2 fusion protein localized strongly to kinetochores, and even at low levels, a strong mitotic arrest was observed with all chromosomes aligned on the metaphase plate (Fig 1A and B). The expression level of KT-Mad2 in the stable cell line was very low compared to endogenous Mad2 (Supplementary Fig S1A). This metaphase arrest persisted for hours until the metaphase plate collapsed likely due to cohesion fatigue [26, 27]. When we expressed soluble Mad2, C-Mad2 (Mad2 L13A), or O-Mad2 (Mad2 V193N), only negligible effects on mitotic progression were observed and similarly targeting Mad2 to chromosomes via fusion to H2B did not arrest cells in mitosis (Fig 1B). Analysis of recombinant Mad2 L13A and Mad2 V193N on a Resource Q column confirmed that they were largely in the closed or open confirmation, respectively, similar to what has been reported (Supplementary Fig S1B) [21]. The failure of soluble Mad2 proteins to induce a metaphase arrest was not due to low expression levels as they were expressed at much higher levels than KT-Mad2 (Supplementary Fig S1C). These results show that Mad2 needs to be specifically targeted to kinetochores to induce a mitotic arrest similar to what has been described for Mad1 and Mps1 [28, 29].
Figure 1.
- A Still images from a time-lapse movie of a stable HeLa cell line expressing the KT-Mad2 (Ndc80-Venus-Mad2) fusion protein or the KT-O-Mad2 protein, and a histone marker. Time is in minutes and t = 0 at NEBD.
- B, C NEBD-Anaphase times in stable HeLa cell lines expressing the indicated Mad2 constructs as measured by time-lapse microscopy. Each dot represents a single cell and red dots are cells that were still arrested when the recording ended. The red line indicates the median.
- D The indicated Ndc80 fusion proteins were purified using GFP binder resin from nocodazole-arrested cells and analyzed for their ability to bind the indicated proteins by Western blot.
To investigate the conformational requirements of the kinetochore-targeted Mad2, we used the same approach to target C-Mad2, O-Mad2, and Mad2 R133A that has a mutation in the dimerization interface, hereby preventing Mad2 dimerization [16, 21]. While targeting of C-Mad2 to kinetochores produced a strong metaphase arrest, cells expressing similar levels of targeted O-Mad2 or Mad2 R133A did not arrest (Fig 1C). Purification of the different Ndc80 fusions from stable cell lines arrested with nocodazole revealed that the tethered Mad2 molecules behaved as expected in that only KT-Mad2 and KT-C-Mad2 bound to Mad1 (Fig 1D). These observations are in agreement with the template model and provide further in vivo evidence for this model.
Mad1 is required for the KT-Mad2-induced mitotic arrest
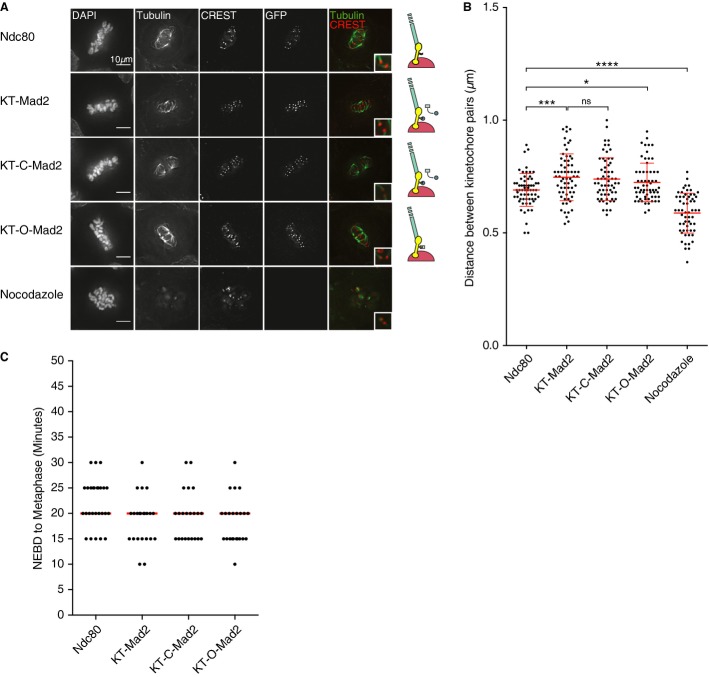
Since the Ndc80 complex is essential for microtubule binding and end-on attachment, we wanted to exclude that the observed arrest upon kinetochore targeting of active Mad2 was due to a secondary defect in kinetochore–microtubule interactions. To exclude this, we analyzed the metaphase-arrested cells by immunofluorescence after cold treatment, as this will depolymerize non-kinetochore microtubules. Cells transfected with the different Mad2 tethering constructs all exhibited robust K-fibers and end-on attachment to kinetochores, and in addition, measurement of the distance between sister kinetochore pairs revealed that tension was applied (Fig 2A and B). Measuring the time from nuclear envelope breakdown to alignment of all chromosomes at the metaphase plate revealed no major difference between cells expressing Ndc80-Venus and KT-Mad2 (Fig 2C). These results and the observation that KT-O-Mad2 does not affect chromosome segregation argue that the arrest observed in KT-Mad2 and KT-C-Mad2 is due to the persistent presence of these proteins rather than subtle defects in kinetochore–microtubule interactions.
Figure 2.
- Cells were transfected with the indicated Ndc80 fusion proteins or treated with nocodazole and treated on ice prior to fixation and stained with the indicated antibodies to monitor microtubule–kinetochore interactions.
- The distance between kinetochore pairs on metaphase plates was determined by measuring the distance between the two CREST pairs from the images in (A). At least 60 pairs were measured from at least 9 different cells for each condition, and a red line indicates the mean and standard deviation is indicated. Each dot represents one kinetochore pair. A t-test was used to compare the different conditions *P ≤ 0.05, ***P ≤ 0.001, ****P ≤ 0.0001, ns: non-significant P > 0.05.
- The time from NEBD to the alignment of all chromosomes on the metaphase plate was measured from the time-lapse movies for the indicated cell lines. The median is indicated by the red line and was 20 minutes for all conditions.
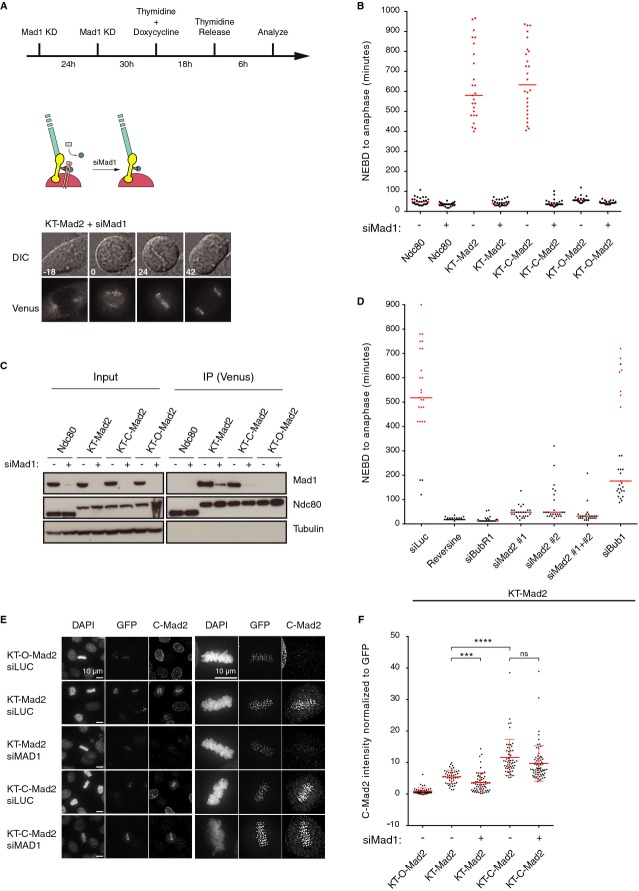
Given that KT-Mad2 and KT-C-Mad2 bind Mad1, we could now test whether Mad1 was still required to obtain a strong metaphase arrest. When we depleted Mad1 in cells by approximately 90% using RNAi, the cell lines expressing KT-Mad2 and KT-C-Mad2 no longer arrested for a prolonged time and the levels of Mad1 bound by these molecules were strongly reduced (Fig 3A–C). A similar result was obtained using KT-Mad2 L13Q, a mutation that also maintains Mad2 in the closed conformation (Supplementary Fig S1D) [21]. Thus, Mad1 binding to the kinetochore-targeted Mad2 molecules is essential for inducing a prolonged mitotic arrest. In addition to a requirement for Mad1, all other checkpoint components we tested: Mps1, Bub1, BubR1, and soluble Mad2, were required for a strong mitotic arrest (Fig 3D). This shows that KT-Mad2 induces a SAC arrest requiring all components of the pathway. In the metaphase-arrested cells, we could still detect Bub1 and BubR1 at kinetochores although at lower levels than in prometaphase cells (Supplementary Fig S2A and B). Their localization is likely also critical for the observed metaphase arrest.
Figure 3.
- Still images from time-lapse movies of a stable HeLa cell line expressing KT-Mad2 where Mad1 has been depleted by RNAi. The protocol for depletion of Mad1 is depicted above.
- NEBD-Anaphase times were measured from time-lapse movies for the indicated Ndc80 fusions either depleted of Mad1 or control-depleted. Each dot represents a single cell and red dots represent cells that were still arrested when the recording ended. The median is indicated by a red line.
- Purification of the indicated Ndc80 fusions from stable HeLa cell lines treated with nocodazole using GFP binder. Cells were either depleted of Mad1 or treated with a control oligo as indicated and probed for Ndc80 and Mad1.
- A stable HeLa cell line expressing KT-Mad2 was depleted of soluble Mad2 using two different oligos targeting the 3′ UTR of Mad2 or depleted of BubR1 or Bub1 using RNAi. Mps1 was inhibited using reversine. For each condition, the NEBD-Anaphase time was measured from time-lapse movies and each dot represents a single cell analyzed.
- The indicated Ndc80 fusions were transfected into HeLa cells and either control-depleted or depleted of Mad1 using RNAi. The cells were fixed and stained for expression of the fusion protein (GFP) and closed Mad2 (C-Mad2).
- The level of C-Mad2 and GFP signal was quantified from deconvoluted images using 3 z-stacks 200 nm apart encompassing the bulk of kinetochore signal, and then the C-Mad2 signal was normalized to GFP. Each dot represents a single kinetochore, and at least 47 kinetochores from at least seven different cells were analyzed. The mean is indicated by a red line and standard deviation as well. A t-test was used to compare the different conditions ***P ≤ 0.001, ****P ≤ 0.0001, ns: non-significant P > 0.05.
That Mad1 was still required for a SAC arrest despite the continuous kinetochore targeting of C-Mad2 suggested additional critical roles of Mad1 in the SAC beyond Mad2 kinetochore recruitment. To make this conclusion, we needed to rule out that the requirement of Mad1 did not reflect that KT-Mad2 and KT-C-Mad2 depended on Mad1 binding for maintaining the closed conformation. To address this, we used a mouse monoclonal antibody generated in the laboratory specific for the closed conformation of Mad2 (see Supplementary Fig 2C–E for characterization of this antibody). We stained metaphase plates from cells expressing the different KT-Mad2 molecules in the presence or absence of Mad1 depletion. The antibody did not stain cells expressing KT-O-Mad2 as expected, while clear kinetochore staining coinciding with the GFP signal was observed in cells expressing KT-Mad2 and KT-C-Mad2 (Fig 3E and F). The levels of closed Mad2 at kinetochores were higher in KT-C-Mad2-expressing cells than in KT-Mad2-expressing cells, showing that the L13A mutation maintains Mad2 in a closed conformation in vivo similar to what was observed in our biochemical analysis of Mad2 L13A (Supplementary Fig S1B). Upon Mad1 depletion, the staining with the closed specific Mad2 antibody decreased in KT-Mad2 cells, but not in KT-C-Mad2 (Fig 3E and F). Furthermore, the level of p31, a C-Mad2-specific ligand, at metaphase kinetochores in KT-C-Mad2-expressing cells was not affected by Mad1 depletion (Supplementary Fig S3A). Comparison of the level of C-Mad2 at kinetochores in KT-C-Mad2 and KT-Mad1 revealed no major differences (Supplementary Fig S3B). Combined, these results reveal that in KT-C-Mad2 the closed conformation of Mad2 is maintained when Mad1 is depleted and can provide levels of C-Mad2 at kinetochores to the same degree as KT-Mad1. The dependency on Mad1 for an arrest in KT-C-Mad2-expressing cells therefore argues for additional roles of Mad1 in the SAC beyond C-Mad2 kinetochore recruitment.
The C-terminal domain of Mad1 is critical for a functional SAC
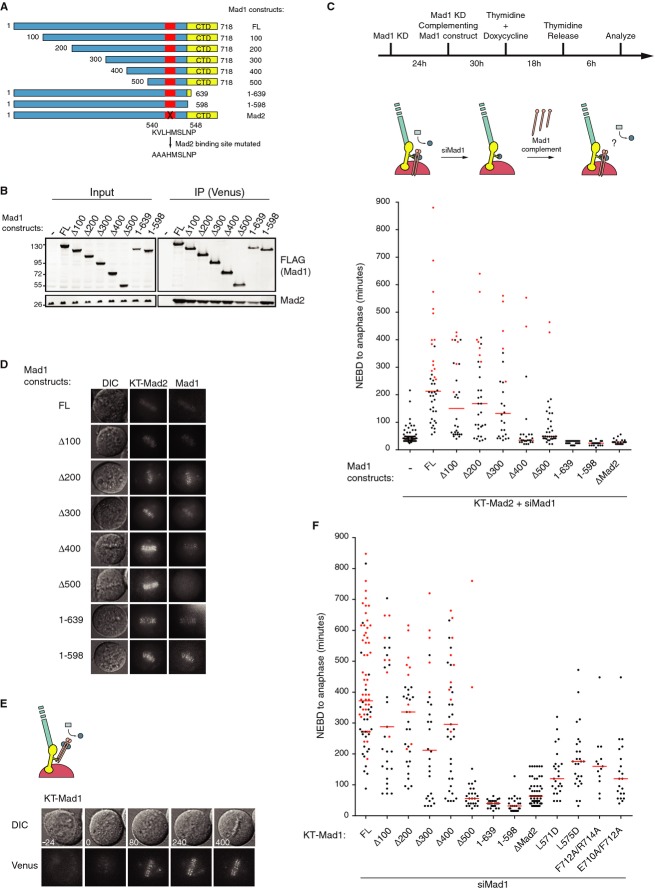
To gain further mechanistic insight into how Mad1 acts in the SAC, we first investigated which domains of Mad1 are required for its function in the SAC when its role in kinetochore recruitment of C-Mad2 is bypassed. Mad1 is a 718 amino acid long protein consisting of a long coiled-coil region preceding the Mad2-binding site followed by two alpha helixes that pair and end in a small globular domain (Fig 4A) [12, 30]. We generated a panel of N-terminal and C-terminal truncations of Mad1 all containing the Mad2-binding site. As a control, we also generated the full-length Mad1 protein with a mutated Mad2-binding site (Fig 4A). The Mad1 constructs were made resistant to the Mad1 RNAi oligo and tagged with mTurquoise (TFP) at the N-terminus. All the constructs were able to bind Mad2 as expected (Fig 4B).
Figure 4.
- Schematic of the different Mad1 constructs with the Mad2-binding site and globular domain (CTD) indicated.
- Stable cell lines expressing the indicated FLAG-Venus-Mad1 constructs were purified from nocodazole-arrested cells, and their ability to bind Mad2 was determined by Western blotting.
- The stable KT-Mad2 cell line was depleted for Mad1 and complemented with the indicated Mad1 constructs. NEBD-Anaphase times were determined for the different Mad1 constructs expressing similar levels. The red line indicates the median, and each dot represents a single cell analyzed.
- Still images from time-lapse analysis of stable Venus-Mad1 cell lines transfected with TFP-tagged KT-Mad2.
- Still images from time-lapse movies of a cell expressing Ndc80-Venus-Mad1 (KT-Mad1) with time given in minutes and time at NEBD set to zero.
- NEBD-Anaphase times determined from time-lapse movies of cells expressing the indicated Ndc80 fusion proteins. Each dot represents a single cell analyzed and red dots represent cells still arrested in mitosis when the recording ended. The red lines indicate the median.
We then depleted Mad1 from the cell line expressing KT-Mad2 and complemented the cells with the different Mad1 constructs (Fig 4C). As overexpression of Mad1 abrogates the SAC due to sequestering of soluble Mad2, only cells expressing low levels of Mad1 were able to restore the metaphase arrest and all constructs were analyzed in this range of expression. All Mad1 constructs were recruited to the kinetochores by KT-Mad2 except Mad1 Δ1-500 (Fig 4D). As expected, full-length Mad1 restored the KT-Mad2-induced arrest and this depended on its Mad2-binding site. More importantly, the C-terminal globular domain was absolutely required for restoring the arrest as cells complemented with Mad1 1-639 divided with almost normal mitotic timing (Fig 4C). A similar requirement for the C-terminal globular domain of Mad1 was observed in KT-C-Mad2-expressing cells (Supplementary Fig S3C).
In a complementary approach, we fused the different Mad1 truncations to the C-terminus of Ndc80-Venus to monitor their ability to induce a mitotic arrest (referred to as KT-Mad1). Similar to what has been reported for a Mis12-Mad1 fusion protein [28], the constitutive targeting of Mad1 to Ndc80 resulted in a strong metaphase arrest (Fig 4E–F). Again in this assay, we observed a clear dependency on the C-terminal globular domain of Mad1 to obtain a metaphase arrest. The targeted Mad1 constructs, except for Mad1 ΔMad2, recruited C-Mad2 to kinetochores as evidenced by quantitative immunofluorescence with the closed specific Mad2 antibody (Supplementary Fig S4A and B), reaffirming that failure in inducing an arrest is not due to the lack of C-Mad2 at the kinetochore. Also, we observed that mutation of conserved surface-exposed residues in the C-terminal globular domain (residues 710, 712, 714) strongly reduced the ability of tethered Mad1 to induce a metaphase arrest (Fig 4F). These residues were also required for soluble Mad1 function (Supplementary Fig S4C). In addition, two mutations, Mad1 L571D and Mad1 L575D, described to affect the pairing of the two terminal alpha helices [12], which positions the C-terminal domain, had reduced activity in this assay (Fig 4F).
The results presented in this study reveal an essential role of Mad1 in the SAC unrelated to its role in recruiting C-Mad2 to kinetochores. We show a critical role for the globular C-terminal domain of Mad1 and conserved residues within this domain that have been shown not to affect kinetochore recruitment or dimerization of Mad1 [30]. Similar conclusions have been obtained by the Hauf laboratory in fission yeast [31]. Potentially, this function of the Mad1 C-terminal domain is conserved.
Previous work from the Hardwick laboratory has revealed a checkpoint-dependent complex between Mad1 and Bub1 in budding yeast that depends on the conserved RLK motif of Mad1 (residues 617-619 in human Mad1) [32]. Using in vitro translated proteins, an interaction has also been reported for the human proteins [33]. In budding yeast, the Mad1-Bub1 interaction is required for a functional SAC in part due to the fact that the interaction is required for Mad1 kinetochore localization. Since we observe a dependency for Bub1 even when the Mad1-Mad2 complex is tethered, it could be that the Mad1-Bub1 interaction is required for a functional SAC in addition to playing a role in Mad1-Mad2 recruitment. However, analysis of endogenous and exogenous Mad1 by exhaustive mass spectrometry as well as extensive yeast two-hybrid screens has failed to detect Bub1 as a binding partner for Mad1. Potentially, this interaction in human cells is very weak compared to budding yeast or the role of the Mad1 C-terminal domain observed here is unrelated to Bub1 binding. Defining this function of Mad1 will be an important future goal.
Materials and Methods
Cloning
Using a forward primer for Venus and a reverse primer for either Mad2 or Mad1, the appropriate Venus-Mad fragments were amplified and inserted into pcDNA5/FRT/TO Ndc80-Venus [34] by using ApaI and NotI restriction enzymes that removed Venus and allowed in-frame insertion of the various Venus-Mad PCR products. All constructs were fully sequenced.
Antibodies
All antibodies used are specified in Supplementary Data 1.
RNAi depletion and rescue
For efficient Mad1 depletion, cells were subjected to a double knock-down protocol using 10 nM of Mad1 siRNA oligo (Ambion Silencer Select, s15905) with transfection on days one and two. As a control, 50 nM of Luciferase siRNA oligo (SIGMA, VC300B2) was used. Cells were analyzed on day three or four as indicated. In RNAi rescue experiments, cells were co-transfected with the Mad1 siRNA oligo and the complementing plasmid constructs on day two.
For depletion of Mad2, BubR1, and Bub1, the following oligos were used from Sigma: 5′ GAUGGUGAAUUGUGGAAUA (BubR1), 5′ GAGUGAUCACGAUUUCUAA (Bub1), 5′ CCUGAAAUCAAGUCAUCUA (MAD2 #1), 5′ ACUGAACUGUGUUAAUUG (MAD2 #2).
Purification of complexes/immunoprecipitation analysis
Cells were lysed in lysis buffer (50 mM Tris–HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM DDT, and 0.1% NP40). Complexes were immunoprecipitated in lysis buffer with antibodies coupled to Protein G-Sepharose 4B (Invitrogen) or GFP-Trap (ChromoTek) beads as indicated and incubated at 4°C for 2 h or 30 min, respectively. Precipitated protein complexes were washed three times in lysis buffer and eluted in 2× SDS sample buffer.
Immunofluorescence
After thymidine treatment, cells were released and given MG132 for 2 hours prior to fixation to keep cells in mitosis. The cells were pre-fixed for 20 s in 4% formaldehyde, permeabilized in 0.5% Triton X-100, and then fixed for 20 min in 4% formaldehyde. For cold treatment, cells were put on ice 10 min prior to fixation. The fixed cells were quenched with 25 mM glycine for 20 min, incubated with primary antibodies for 2 h at room temperature or overnight at 4°C, followed by 1 h of incubation with appropriate secondary antibodies and DAPI (1:1,000). For detection of the transfected constructs, GFP antibody or GFP-booster (1:200, ChromoTek) was used.
Acknowledgments
We thank Stephen Taylor for providing the HeLa/FRT/TRex cell line and Silke Hauf for sharing unpublished results. Mia F. Nielsen and Tine K. Nielsen kindly prepared recombinant Mad2 protein. This work was supported by grants to JN from the Novo Nordisk Foundation and the Lundbeck Foundation.
Author contribution
TK performed biochemical analysis and live cell analysis of tethered proteins. MSYL performed immunofluorescence analysis and helped with live cell analysis. GGS and WS performed the characterization of the C-Mad2 antibody. JOS and JVS assisted with MS analysis of Mad1 complexes. JN assisted with clonings and designing of experiments and wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://embor.embopress.org
References
- 1.Pines J. Cubism and the cell cycle: the many faces of the APC/C. Nat Rev Mol Cell Biol. 2011;12:427–438. doi: 10.1038/nrm3132. [DOI] [PubMed] [Google Scholar]
- 2.Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- 3.Kim SH, Lin DP, Matsumoto S, Kitazono A, Matsumoto T. Fission yeast Slp1: an effector of the Mad2-dependent spindle checkpoint. Science. 1998;279:1045–1047. doi: 10.1126/science.279.5353.1045. [DOI] [PubMed] [Google Scholar]
- 4.Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardwick KG, Johnston RC, Smith DL, Murray AW. MAD3 encodes a novel component of the spindle checkpoint which interacts with Bub3p, Cdc20p, and Mad2p. J Cell Biol. 2000;148:871–882. doi: 10.1083/jcb.148.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson J, Yekezare M, Minshull J, Pines J. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat Cell Biol. 2008;10:1411–1420. doi: 10.1038/ncb1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westhorpe FG, Tighe A, Lara-Gonzalez P, Taylor SS. p31comet-mediated extraction of Mad2 from the MCC promotes efficient mitotic exit. J Cell Sci. 2011;124:3905–3916. doi: 10.1242/jcs.093286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulukian A, Han JS, Cleveland DW. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev Cell. 2009;16:105–117. doi: 10.1016/j.devcel.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Han JS, Holland AJ, Fachinetti D, Kulukian A, Cetin B, Cleveland DW. Catalytic Assembly of the Mitotic Checkpoint Inhibitor BubR1-Cdc20 by a Mad2-Induced Functional Switch in Cdc20. Mol Cell. 2013;51:92–104. doi: 10.1016/j.molcel.2013.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- 12.Sironi L, Mapelli M, Knapp S, Antoni De A, Jeang K-T, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- 14.Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- 15.Fava LL, Kaulich M, Nigg EA, Santamaria A. Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 2011;30:3322–3336. doi: 10.1038/emboj.2011.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. EMBO J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen RH, Brady DM, Smith D, Murray AW, Hardwick KG. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol Biol Cell. 1999;10:2607–2618. doi: 10.1091/mbc.10.8.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simonetta M, Manzoni R, Mosca R, Mapelli M, Massimiliano L, Vink M, Novak B, Musacchio A, Ciliberto A. The influence of catalysis on mad2 activation dynamics. PLoS Biol. 2009;7:e10. doi: 10.1371/journal.pbio.1000010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Antoni De A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, Musacchio A. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- 21.Mapelli M, Massimiliano L, Santaguida S, Musacchio A. The Mad2 conformational dimer: structure and implications for the spindle assembly checkpoint. Cell. 2007;131:730–743. doi: 10.1016/j.cell.2007.08.049. [DOI] [PubMed] [Google Scholar]
- 22.Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 23.Chung E, Chen R-H. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nezi L, Rancati G, Antoni De A, Pasqualato S, Piatti S, Musacchio A. Accumulation of Mad2-Cdc20 complex during spindle checkpoint activation requires binding of open and closed conformers of Mad2 in Saccharomyces cerevisiae. J Cell Biol. 2006;174:39–51. doi: 10.1083/jcb.200602109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, Musacchio A. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. EMBO J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daum JR, Potapova TA, Sivakumar S, Daniel JJ, Flynn JN, Rankin S, Gorbsky GJ. Cohesion fatigue induces chromatid separation in cells delayed at metaphase. Curr Biol. 2011;21:1018–1024. doi: 10.1016/j.cub.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stevens D, Gassmann R, Oegema K, Desai A. Uncoordinated loss of chromatid cohesion is a common outcome of extended metaphase arrest. PLoS ONE. 2011;6:e22969. doi: 10.1371/journal.pone.0022969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maldonado M, Kapoor TM. Constitutive Mad1 targeting to kinetochores uncouples checkpoint signalling from chromosome biorientation. Nat Cell Biol. 2011;13:475–482. doi: 10.1038/ncb2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jelluma N, Dansen TB, Sliedrecht T, Kwiatkowski NP, Kops GJPL. Release of Mps1 from kinetochores is crucial for timely anaphase onset. J Cell Biol. 2010;191:281–290. doi: 10.1083/jcb.201003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim S, Sun H, Tomchick DR, Yu H, Luo X. Structure of human Mad1 C-terminal domain reveals its involvement in kinetochore targeting. Proc Natl Acad Sci USA. 2012;109:6549–6554. doi: 10.1073/pnas.1118210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinrich S, Sewart K, Windecker H, Langegger M, Schimdt N, Hustedt N, Hauf S. Mad1 contribution to spindle assembly checkpoint signalling goes beyond presenting Mad2 at kinetochores. EMBO Rep. 2014 doi: 10.1002/embr.201338114. DOI 10.1002/embr.201338114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brady DM, Hardwick KG. Complex formation between Mad1p, Bub1p and Bub3p is crucial for spindle checkpoint function. Curr Biol. 2000;10:675–678. doi: 10.1016/s0960-9822(00)00515-7. [DOI] [PubMed] [Google Scholar]
- 33.Seeley TW, Wang L, Zhen JY. Phosphorylation of human MAD1 by the BUB1 kinase in vitro. Biochem Biophys Res Commun. 1999;257:589–595. doi: 10.1006/bbrc.1999.0514. [DOI] [PubMed] [Google Scholar]
- 34.Zhang G, Kelstrup CD, Hu X-W, Hansen MJK, Singleton MR, Olsen JV, Nilsson J. The Ndc80 internal loop is required for recruitment of the Ska complex to establish end-on microtubule attachment to kinetochores. J Cell Sci. 2012;125:3243–3253. doi: 10.1242/jcs.104208. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.