Abstract
The mitochondrial uniporter is a selective Ca2+ channel regulated by MICU1, an EF hand-containing protein in the organelle's intermembrane space. MICU1 physically associates with and is co-expressed with a paralog, MICU2. To clarify the function of MICU1 and its relationship to MICU2, we used gene knockout (KO) technology. We report that HEK-293T cells lacking MICU1 or MICU2 lose a normal threshold for Ca2+ intake, extending the known gating function of MICU1 to MICU2. Expression of MICU1 or MICU2 mutants lacking functional Ca2+-binding sites leads to a striking loss of Ca2+ uptake, suggesting that MICU1/2 disinhibit the channel in response to a threshold rise in [Ca2+]. MICU2's activity and physical association with the pore require the presence of MICU1, though the converse is not true. We conclude that MICU1 and MICU2 are nonredundant and together set the [Ca2+] threshold for uniporter activity.
Keywords: calcium, MICU1, MICU2, mitochondria, uniporter
Introduction
Dynamic changes in cytosolic [Ca2+] are important signals by which the cytosol and mitochondria communicate [1–4], with well-documented roles in energetic coupling [5–7] and cell death pathways [8]. Mitochondrial Ca2+ influx is highly energetically favorable due to the electrochemical potential created by the combination of the Ca2+ concentration gradient and the electrical potential across the inner mitochondrial membrane (ΔΨm) [4]. However, Ca2+ uptake into mitochondria must be tightly regulated to ensure that mitochondria can ignore baseline cytosolic fluctuations yet respond effectively to Ca2+ spikes.
Recent identification of the genes encoding the mitochondrial Ca2+ uniporter is now enabling us to probe its physiology at the molecular level. Established uniporter components include the transmembrane pore-forming subunit MCU [9, 10], its paralog MCUb [11], the recently identified transmembrane protein EMRE [12], and the paralogous, EF hand-containing proteins MICU1 [13] and MICU2 [14]. MICU1 was the first component to be identified and, on the basis of its EF hands, was initially proposed to be a [Ca2+]-dependent regulator of uniporter activity, though the mechanism was unclear. Subsequent studies converged on the notion that MICU1 serves as a gatekeeper of the uniporter [15, 16], inhibiting the channel at baseline [Ca2+] levels.
MICU1 is one of three paralogous genes (MICU1, MICU2, and MICU3) that likely arose by gene duplication in vertebrates [14]. All three proteins are of comparable size, have a mitochondrial targeting sequence (MTS) at the amino terminus, and have similar domain architecture with two canonical Ca2+-binding EF hands. MICU1 and MICU2 have been shown to localize to the mitochondrial intermembrane space (IMS) [12, 15]. Several lines of evidence indicate that MICU1 and MICU2 operate together with MCU. For example, immunoprecipitation of the uniporter complex using affinity-tagged MCU pulls down MICU1 and MICU2, but not MICU3, in HEK-293T cells [12]. The RNA expression of MICU1, MICU2, and MCU is strongly correlated across a variety of tissues, whereas MICU3 tends to be highly expressed in the CNS [14]. Given that MICU1 and MICU2 are physically associated within the uniporter complex and are co-expressed across all tissues, it is likely they operate together to regulate the channel.
Previous studies of MICU1, including our own, have been obfuscated by problems associated with incomplete knockdown and measurements of matrix Ca2+ [13–16]. Specifically, limitations of incomplete uniporter protein knockdown can be exacerbated by overexpressing another subunit due to protein cross-stabilization [14]. This shortcoming of RNAi has made it challenging to evaluate the function of EF hand mutant alleles, possibly contributing to discrepancy between previous studies [13, 15, 16]. Additionally, matrix Ca2+ measurements can be confounded by alterations in baseline matrix Ca2+ or matrix Ca2+ buffering. Moreover, previous studies exploring MICU1's physiological role did not consider contributions from MICU2. All of these factors may have contributed to conflicting models. Here, we take advantage of gene knockout (KO) technology, extramitochondrial Ca2+ clearance measurements, and biochemistry to clarify the role of MICU1 as a gatekeeper of the uniporter and to understand the contribution of MICU2. The results indicate that MICU1 and MICU2 are nonredundant and operate together to prevent Ca2+ uptake when outside [Ca2+] is low, but then permit uptake in response to a stimulus above threshold.
Results and Discussion
MICU1 KO and MICU2 KO cells exhibit altered threshold for mitochondrial Ca2+ uptake
To investigate whether MICU1 and MICU2 play unique roles in mitochondrial Ca2+ handling, we used TALE nuclease technology [17] to disrupt MICU1 (MICU1 KO) or MICU2 (MICU2 KO) in HEK-293T cells [12]. We further confirmed elimination of MICU1 and MICU2 by immunoblotting (Supplementary Fig S1A). Disrupting MICU1 leads to decreased protein levels of MICU2 compared to wild-type HEK-293T (WT) cells (to 18 ± 4% of WT levels), similarly to MICU1 knockdown (KD) cells [14]. MICU2 KO cells, on the other hand, have normal MICU1 levels (Supplementary Fig S1A). Overexpression of MICU1-FLAG in MICU1 KO cells rescues MICU2 expression to levels similar to WT. MCU levels appear unaffected in both KO cell lines, which may be distinct from other cell types [14].
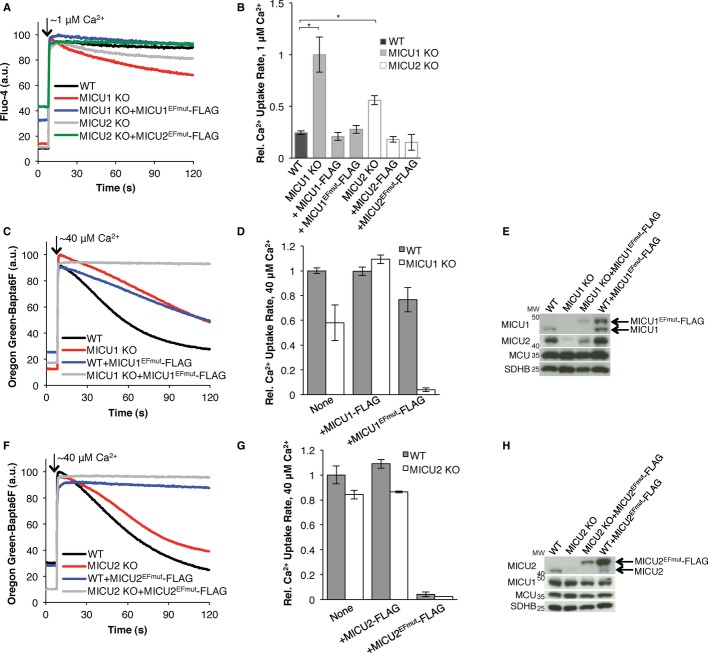
Next, to assess mitochondrial Ca2+ uptake, we measured extramitochondrial Ca2+ clearance using cell-impermeable Ca2+ indicators. This uptake is sensitive to Ru360 (a classical uniporter inhibitor) and CCCP (an uncoupler), but not thapsigargin (a SERCA inhibitor), as expected for mitochondrial Ca2+ uptake (Supplementary Fig S1B and C). Using this assay, MICU1 KO mitochondria display uptake when given a small pulse of Ca2+, which is ignored by WT mitochondria (Fig 1A and B), consistent with previous studies showing that MICU1 sets the threshold for Ca2+ uptake [15, 16]. Interestingly, MICU2 KO cells also exhibit uptake of a small pulse of Ca2+ despite the presence of MICU1, albeit at a slower rate than MICU1 KO cells (Fig 1A and B). Importantly, in both cases, this impaired Ca2+ handling can be rescued by re-expression of the ablated protein, indicating that the phenotype is not a trivial consequence of off-target genome editing (Fig 1B, Supplementary Fig S1D). Thus, the threshold for Ca2+ uptake is affected by loss of either MICU1 or MICU2, as indicated by increased uptake rate when given [Ca2+] below the apparent WT threshold. Although this “gatekeeping” phenotype has been reported for MICU1 [15, 16], this is the first time it is reported for MICU2.
Figure 1.
- Representative Ca2+ uptake traces of WT, MICU1 KO, or MICU2 KO with or without MICU1EFmut or MICU2EFmut exposed to ˜1 μM Ca2+.
- Quantification of the data in (A) and Supplementary Fig S1D.
- Representative Ca2+ uptake traces of WT or MICU1 KO cells with or without MICU1EFmut given ˜40 μM Ca2+.
- Quantification of the data in (C) and Supplementary Fig S1E.
- Immunoblot of whole-cell lysates of cells in (C) using the antibodies indicated on the left.
- Representative Ca2+ uptake traces of WT or MICU2 KO cells with or without MICU2EFmut given ˜40 μM Ca2+.
- Quantification of the data in (F) and Supplementary Fig S1E.
- Immunoblot of whole-cell lysates of cells in (F) using the antibodies indicated on the left.
Data information: Bar graphs represent the rate of Ca2+ uptake from linear fits (50–60 s, n ≥ 3).
Both MICU1 KO and MICU2 KO cell lines exhibit mitochondrial Ca2+ uptake when stimulated with a larger pulse of Ca2+ (Supplementary Fig S1E), consistent with recent reports for MICU1 KD [15, 16]. Interestingly, MICU1 KO mitochondria have moderately reduced uptake kinetics that can be rescued with the WT allele (Fig1C, D; Supplementary Fig S1E). However, MICU1 KD using RNAi does not lead to reduced uptake kinetics with a large Ca2+ pulse [15, 16]. The source for this mild defect in uptake of a large pulse of Ca2+ is unknown. It does not appear to be due to an alteration in mitochondrial membrane potential (ΔΨm), as assessed by TMRM, which is indistinguishable from WT in both MICU1 KO and MICU2 KO cells (Supplementary Fig S2A and B). In addition, the mitochondria do not appear to be significantly loaded with Ca2+ under the conditions of these experiments (Supplementary Fig S2C). In other cell types, loss of MICU1 can lead to a loss of MCU [14], though we cannot detect such differences in MCU levels here.
Overall, genome-editing technology has allowed us to fully disrupt MICU1 or MICU2, and though these cells are viable, each lacks a normal threshold for mitochondrial Ca2+ uptake (Fig 1A and B).
Mutation of MICU1 or MICU2 EF hands abolishes mitochondrial Ca2+ uptake
To further evaluate the functions of MICU1 and MICU2, we introduced mutations in their EF hand domains. We mutated the first and last residues of the predicted 12-residue Ca2+-binding loop of each EF hand to alanine and to lysine, respectively, to disable Ca2+ binding [13]. Since an EF hand mutant should mimic the apo (Ca2+-free) state, MICU1EFmut and MICU2EFmut are expected to simulate the action of WT MICU1 and MICU2 in low, subthreshold [Ca2+]. As expected, the EF hand mutants behave like WT when presented with a low Ca2+ pulse: stable expression of MICU1EFmut or MICU2EFmut in the corresponding KO cells restores the ability of mitochondria to ignore low [Ca2+] similar to WT (Fig 1A and B). These results for MICU1 are consistent with a previous report with MICU1 KD cells [15] yet distinct from another study [16].
Since MICU1EFmut and MICU2EFmut are effectively locked in the apo-like “off” state due to their disrupted Ca2+-binding loops, we predict they should not have the ability to respond to large [Ca2+] pulses. Indeed, mitochondria expressing MICU1EFmut or MICU2EFmut on their respective KO backgrounds exhibit a striking phenotype and cannot uptake even large pulses of Ca2+ (Fig 1C–H). These cells, however, have an intact and depolarizable ΔΨm indistinguishable from WT and do not appear to be loaded with more Ca2+ than WT under the conditions of these experiments (Supplementary Fig S2A and C). Thus, in spite of apparently intact driving force for Ca2+ uptake and full expression of the MCU channel, MICU1EFmut (Fig 1C–E) and MICU2EFmut (Fig 1F–H) are each capable of inhibiting uptake.
We next tested whether this striking result would be recapitulated by expressing MICU1EFmut on a WT background. WT mitochondria stably expressing MICU1EFmut have milder reduction in Ca2+ uptake rate (Fig 1C and D). We hypothesized that this milder phenotype is due to the presence of endogenous MICU1. To test this hypothesis, we decreased endogenous MICU1 expression using RNAi (MICU1 KD) while introducing a hairpin-resistant MICU1EFmut. Consistent with our hypothesis, MICU1 KD cells expressing MICU1EFmut show an intermediate Ca2+ uptake inhibition compared to WT and MICU1 KO cells (Supplementary Fig S3A and B). Thus, Ca2+ uptake inhibition by MICU1EFmut appears to be inversely correlated with endogenous MICU1 levels. Interestingly, expression of MICU1EFmut also increases endogenous MICU1 levels, which is evident in both WT and MICU1 KD cells (Fig 1E, Supplementary Fig S3C). These findings underscore the value of utilizing a KO background and serve as a reminder that endogenous MICU1 levels should be considered when MICU1 is knocked down and a mutant is expressed since endogenous MICU1 levels may increase with expression of the mutant.
In contrast, stable expression of MICU2EFmut in WT cells almost entirely abrogates Ca2+ uptake, comparable to the phenotype observed when it is expressed in MICU2 KO cells (Fig 1F and G). Endogenous MICU2 levels in WT cells are comparable with or without MICU2EFmut expression (Fig 1H). Thus, unlike MICU1, protein-level MICU2-MICU2 stabilization is not obvious and does not alleviate the inhibition of mitochondrial Ca2+ uptake in WT cells expressing MICU2EFmut.
The role of the individual EF hands in MICU1 and MICU2 does not appear to be equivalent in these experiments (Supplementary Fig S3A–F). Mutating each EF hand in MICU1 leads to partial inhibition of mitochondrial Ca2+ uptake, with mutation of EF2 having a stronger effect. Similarly, mutation in individual EF hands in MICU2 leads to partial inhibition of Ca2+ uptake, with EF1 having a stronger impact. Exactly how each EF hand contributes to the physiology of MICU1 and MICU2 is currently not known and will likely require biophysical studies as we move forward.
These experiments help to provide clarity on the function of MICU1 and the role of its EF hands. Initial studies identifying MICU1 as the founding member of the uniporter complex suggested that it was a [Ca2+]-dependent regulator required for Ca2+ uptake. This model was proposed based on the observation that, in response to histamine, the matrix Ca2+ signal in MICU1 KD cells showed abrogated increase compared to WT, and while this phenotype could be rescued with WT MICU1, it could not be rescued with MICU1EFmut. However, variation in baseline [Ca2+], matrix buffering, and matrix pH was not considered in making this model inference. Two subsequent studies, however, further explored the function of MICU1 and show that in the absence of MICU1, Ca2+ uptake is still present, and that MICU1 actually serves as a “gatekeeper,” setting the uniporter's [Ca2+] threshold for Ca2+ uptake. While both converge on the notion that MICU1 is a gatekeeper, each proposes a distinct model for the function of Ca2+ binding to MICU1 [15, 16]. One model [16] suggests that Ca2+ binding to MICU1 is required for channel inhibition, while a second model [15] suggests that Ca2+ binding to MICU1 relieves channel inhibition. The current experiments are clearly more compatible with the latter model (Fig 1). In contrast to the latter study, however, we find that on a MICU1 KO background, MICU1EFmut inhibits Ca2+ uptake of both small and large Ca2+ pulses (Fig 1A–D). Thus, our results clearly support a model for MICU1 and MICU2 inhibition of Ca2+ uptake when [Ca2+] is low and likely MICU1 and MICU2 are in the apo (Ca2+-free) state and subsequent release of this inhibition when MICU1 and MICU2 bind Ca2+.
MICU1-mediated inhibition of Ca2+ uptake does not require MICU2, but MICU2-mediated inhibition of Ca2+ uptake requires MICU1
To further evaluate whether MICU1 and MICU2 play redundant or complementary roles, we next overexpressed either MICU1 or MICU2 to rescue KO of MICU2 or MICU1, respectively. However, we find that neither overexpression of MICU1 on a MICU2 KO background nor MICU2 on a MICU1 KO background rescues the impaired Ca2+ handling (Fig 2A and B). Additionally, to determine whether one protein is sufficient to inhibit Ca2+ uptake without the other, independent of [Ca2+] threshold, we expressed MICU2EFmut in MICU1 KO cells and MICU1EFmut in MICU2 KO cells. In response to a small or large Ca2+ pulse, we find that MICU1EFmut is sufficient to abrogate Ca2+ uptake in the absence of MICU2 (Fig 2A and B). However, MICU2EFmut does not provide further inhibition for MICU1 KO cells at either pulse size (Fig 2A and B) and leads to strikingly more uptake than MICU2EFmut expressed in cells which have MICU1 (Fig 1A,B,F, and G). Thus, MICU1 is capable of inhibiting Ca2+ uptake in the absence of MICU2, but MICU2 appears to require MICU1 as an effector.
Figure 2.
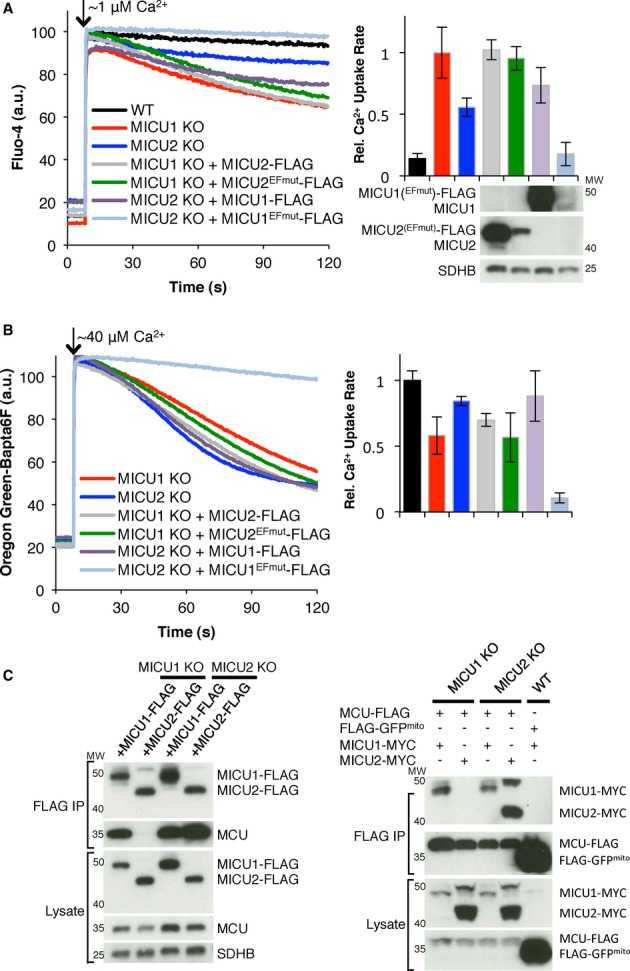
- A, B Mitochondrial Ca2+ uptake in permeabilized HEK-293T cells after CaCl2 pulse was measured by monitoring extramitochondrial Ca2+. MICU1 or MICU2 KO cells stably expressing MICU1, MICU2, MICU1EFmut, or MICU2EFmut were given a (A) ˜1 μM Ca2+ pulse or (B)˜40 μM Ca2+ pulse. Representative traces are shown on the left and quantification of the data is shown in the bar graphs on the right. Immunoblots of whole cell lysates for the corresponding cells are shown under the respective bar graph lane in (A). Note that the WT black bar in (B) is shown for comparison but not present on the trace to the left.
- C FLAG co-immunoprecipitation of MICU1-FLAG, MICU2-FLAG, MCU-FLAG or FLAG-GFPmito expressed in MICU1 KO, MICU2 KO or WT HEK-293T cells with or without transient expression of MICU1-MYC or MICU2-MYC. Lysates and eluates are immunoblotted with anti-FLAG, MCU or SDHB (left side), or FLAG or MYC (right side). Note that the upper band for (FLAG-or MYC-tagged) MICU2 (˜50 kDa) is likely unprocessed MICU2.
Data information: Bar graphs represent the rate of Ca2+ uptake from linear fits (50–60 s, n ≥ 3).
The lack of MICU2EFmut impact on Ca2+ uptake in the absence of MICU1 prompted us to test interaction of MCU and MICU2 in the absence of MICU1. To this end, we tested the ability of MICU1-FLAG and MICU2-FLAG to co-immunoprecipitate MCU on either a MICU1 or MICU2 KO background. MICU1 and MICU2 both are able to pull down MCU in the presence of their paralog (Fig 2C). However, consistent with Ca2+ uptake results, MICU1 is able to co-immunoprecipitate MCU in the absence of MICU2, but MICU2 does not co-immunoprecipitate MCU under the same conditions in the absence of MICU1. Additionally, MCU-FLAG is able to co-immunoprecipitate MICU1 in MICU2 KO cells, but not MICU2 in MICU1 KO cells (Fig 2C). Despite the inability of MICU2 to co-immunoprecipitate MCU, both MICU2 and MICU2EFmut localize properly to the IMS in the absence of MICU1 (Supplementary Fig S4). We speculate that, in the absence of MICU1, MICU2 cannot associate with the MCU complex, including MCU, MCUb, and EMRE, and thus gets targeted for degradation, which would explain its lower abundance in MICU1 KO cells. Collectively, these results suggest that the functional and physical interaction of MICU2 with the MCU complex requires MICU1.
MICU1 C-terminal domain is required for its action and association with the uniporter pore complex
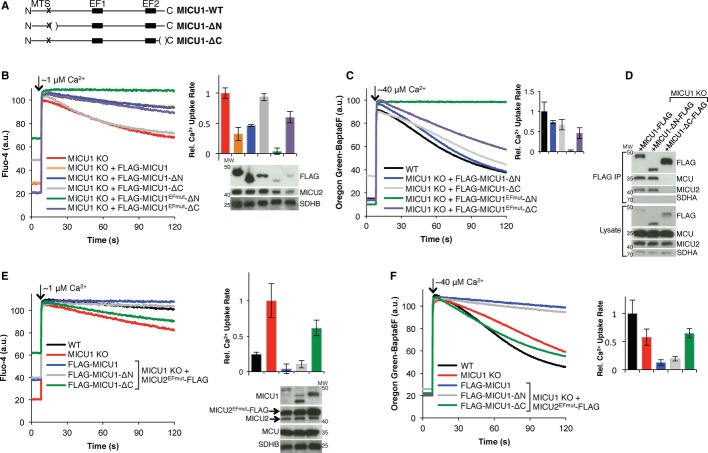
Because our genetic and biochemical studies place MICU1 more proximal to MCU, we explored MICU1 domains that might be important for its function and interaction with the MCU complex. We generated MICU1 truncation mutants and examined their ability to function. The region immediately after the predicted MTS shows little evolutionary conservation. On the other hand, the C-terminus, which is predicted to be predominantly alpha-helical [18, 19], is highly conserved, suggesting that it may be more critical for function. We generated an N-terminal deletion (MICU1-ΔN) by removing 36 amino acids (58–93) after the predicted MTS and a C-terminal deletion (MICU1-ΔC) distal to the second EF hand lacking the final 31 amino acids (445–476) (Fig 3A). While larger N-terminal deletions did not express well, these N-and C-terminal truncated proteins expressed to a comparable level as WT MICU1 (Fig 3B, Supplementary Fig S5A).
Figure 3.
- A Schematic showing the MICU1 truncation mutants used. N and C represent the N-and C-termini, respectively; MTS labels the predicted mitochondrial targeting signal; EF1 and EF2 label the two canonical EF hand domains; parentheses show deleted residues.
- B, C Mitochondrial Ca2+ uptake in digitonin-permeabilized HEK-293T MICU1 KO cells stably expressing MICU1, MICU1-ΔN, MICU1EFmut-ΔN, MICU1-ΔC, or MICU1EFmut-ΔC given a (B) ˜1 μM or (C) ˜40 μM Ca2+ pulse, monitoring extramitochondrial [Ca2+]. Representative traces are shown on the left, and the quantification of uptake rate from linear fits (50–60 s, n ≥ 3) is on the right. Immunoblots of whole-cell lysates for the corresponding cells are shown under the respective bar graph lane.
- D FLAG immunoprecipitation for FLAG-tagged MICU1, MICU1-ΔC, or MICU1-ΔN. Eluates and lysates are immunoblotted with the indicated antibodies.
- E, F Response of digitonin-permeabilized HEK-293T MICU1 KO cells stably expressing MICU2EFmut in addition to MICU1, MICU1-ΔN, or MICU1-ΔC to a (E) ˜1 μM or (F) ˜40 μM Ca2+ pulse, monitoring extramitochondrial [Ca2+]. Representative Ca2+ uptake traces are shown on the left, and quantification of uptake rate from linear fits (50–60 s, n ≥ 3) is on the right. Immunoblots of whole-cell lysates for the corresponding cells are shown under the respective bar graph lane.
Next, we performed Ca2+ uptake assays in MICU1 KO cells stably expressing MICU1-ΔN, MICU1-ΔC, and their EF hand mutants. In response to a low Ca2+ pulse, MICU1-ΔC or MICU1EFmut-ΔC do not fully rescue the increased Ca2+ uptake of MICU1 KO cells, while MICU1-ΔN or MICU1EFmut-ΔN do rescue, resulting in little uptake as WT (Fig 3B). When given a high Ca2+ pulse, only MICU1EFmut-ΔN expression results in abrogated Ca2+ uptake, while MICU1-ΔN, MICU1-ΔC, and MICU1EFmut-ΔC do not (Fig 3C). Thus, deletion from the C-terminus appears to be more deleterious for MICU1 function than deletion from the N-terminus. Because MICU1-ΔC fails to inhibit uniporter Ca2+ uptake, we postulated that it might not interact with the MCU complex appropriately without the C-terminus. Indeed, MICU1-ΔN, but not MICU1-ΔC, co-immunoprecipitates MCU and MICU2 (Fig 3D). Furthermore, despite differences in expression levels, MICU1-ΔN, but not MICU1-ΔC, appears to be co-immunoprecipitated by MCU (Supplementary Fig S5B). However, it is likely that MICU1-ΔC and MICU2 can interact in the cell since MICU2 protein levels are rescued by MICU1-ΔC (Fig 3D), suggesting that the protein cross-stabilization effect of MICU1 on MICU2 is intact, but this interaction is not strong enough to persist under the conditions of the immunoprecipitate. MICU1-ΔC, however, localizes properly to the IMS (Supplementary Fig S4). Mapping the interactions within the MCU complex will be important next steps, and the C-terminus of MICU1 may prove to be important for interaction with the pore complex (MCU, MCUb, and/or EMRE).
Combining the observation that MICU2 does not co-immunoprecipitate MCU in the absence of MICU1 and the C-terminal domain of MICU1 is necessary for MICU1 to co-immunoprecipitate MCU, we postulated that MICU2 would be able to inhibit Ca2+ uptake in MICU1 KO cells in the presence of MICU1-ΔN, but not MICU1-ΔC. To this end, we co-expressed MICU2EFmut-FLAG with either FLAG-MICU1, FLAG-MICU1-ΔN, or FLAG-MICU1-ΔC in MICU1 KO cells and examined their extramitochondrial Ca2+ clearance. As expected, at both pulse sizes, Ca2+ uptake was abrogated in cells expressing FLAG-MICU1 and FLAG-MICU1-ΔN, but not FLAG-MICU1-ΔC in addition to the MICU2EFmut-FLAG, despite each having similar levels of exogenous protein expression (Fig 3E and F). These results provide further in vivo functional support for the co-immunoprecipitation experiments (Fig 2C, 3D, Supplementary Fig S5B).
Collectively, these results demonstrate the functional importance of the highly conserved C-terminal domain of MICU1 (Fig 3). This region may be a candidate for interaction with the rest of the uniporter complex or may be required for proper oligomerization of MICU1. Indeed, the MICU1-MICU1 and MICU1-MICU2 protein cross-stabilizations in HEK-293T cells (Fig 1E, Supplementary Fig S1A, S3C) suggest that MICU1 might form homo-and/or hetero-oligomers. The stoichiometry and composition of the complex has yet to be established and will be important future work.
Proposed model for MICU1 and MICU2 in uniporter physiology
Our results allow us to formulate a model of regulation of the uniporter by MICU1 and MICU2 (Fig 4). MICU1 and MICU2 are located in the IMS [12, 15]. Our biochemical studies indicate that while MICU1 and MCU can associate in the absence of MICU2, MICU2 and MCU association requires the presence of MICU1. Hence, these biochemical studies place MICU1 between MCU and MICU2, though direct interaction studies have not been done. MICU1 and MICU2 serve as negative regulators of Ca2+ transport through the pore complex (Fig 4A), consistent with two recent studies on MICU1 [15, 16]. When the cytosolic [Ca2+] exceeds a threshold sensed by MICU1 and MICU2, the pore is disinhibited and Ca2+ passes through. In the absence of MICU1, this negative regulation is lost and unregulated Ca2+ uptake occurs at [Ca2+] below WT threshold (Fig 4B). In the absence of MICU2, MICU1 is still able to inhibit Ca2+ uptake but likely with a different [Ca2+] threshold (Fig 4B). Perhaps the most striking phenotype reported in the current work is summarized in Fig 4C. When MICU1 or MICU2 cannot respond to [Ca2+] changes, due to nonfunctional EF hands, the uniporter cannot be disinhibited, regardless of [Ca2+], and no Ca2+ uptake is apparent (Fig 4C).
Figure 4.
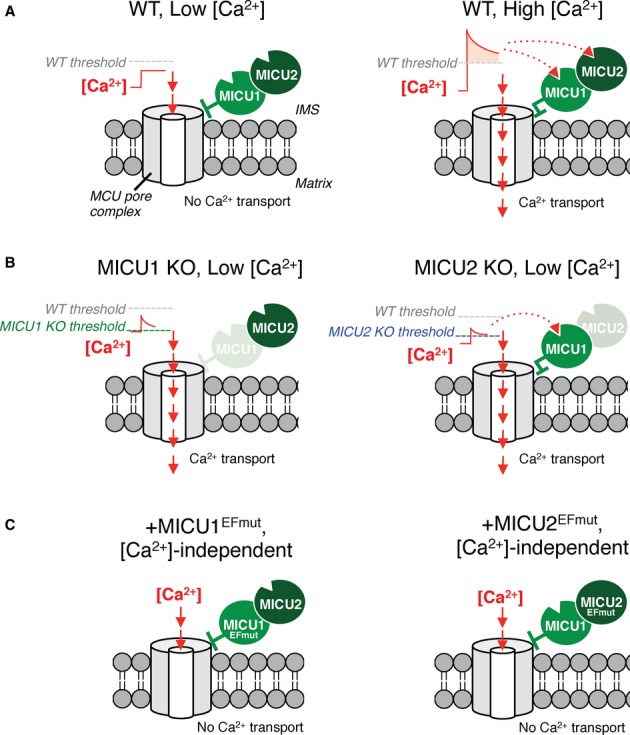
- In WT cells at low external [Ca2+] below threshold, MICU1 and MICU2 inhibit Ca2+ transport (left). When [Ca2+] exceeds threshold, uniporter inhibition is removed and Ca2+ transport proceeds (right).
- MICU1 KO cells have unregulated Ca2+ transport, resulting in uptake even at low [Ca2+] (left); MICU2 KO cells exhibit Ca2+ transport at the low [Ca2+] depicted (right).
- Cells expressing either MICU1EFmut or MICU2EFmut lack Ca2+ transport because Ca2+ is unable to relieve MICU1/2 inhibition of the uniporter, regardless of concentration.
While the current work clearly establishes nonredundant roles for MICU1 and MICU2, much work lies ahead to clarify their physiology. We have observed that expression of MICU1EFmut or MICU2EFmut leads to a near complete loss of Ca2+ uptake. While the simplest explanation is that MICU1EFmut and MICU2EFmut function to “close” the uniporter pore complex, electrophysiological measurements will be required to formally demonstrate this biophysical mechanism. We have shown that MICU1 lacking functional EF hands can inhibit Ca2+ uptake in the absence of MICU2, but the converse is not true. Moreover, we have shown that the physical association of MICU2 with MCU requires MICU1, but not vice versa. MICU1 and MICU2 both appear to be operating as negative regulators of the pore, but the physiological contexts in which each exerts control on the uniporter remain to be determined. Additionally, since only assays of permeabilized cells given pulses of Ca2+ were performed in this study, we cannot rule out the possibility that the mode of Ca2+ delivery may affect the mechanism of Ca2+ uptake through the uniporter, including regulation by MICU1 and MICU2.
There is good reason to believe that the activity and regulation of the uniporter will vary across tissues [3, 14]. The current study has only investigated HEK-293T cells, and it will be important to determine whether the MICU1/MICU2 regulatory complex plays similar roles in other cellular contexts and developmental states. It has recently been reported that loss of MCU and mitochondrial Ca2+ uptake can be tolerated in mouse [20]. In the current study, we have shown that MICU1 and MICU2 play nonredundant roles in inhibiting Ca2+ uptake when outside [Ca2+] is low and alleviate this inhibition at higher [Ca2+]. In vivo loss of MICU1 or MICU2 may therefore have very different consequences than loss of MCU. We anticipate that future in vivo, organismal studies may reveal the precise regulatory logic afforded by MICU1 and MICU2.
Materials and Methods
Cell culture
HEK-293T cells lacking MICU1 or MICU2 were made as described [12]. All other expression of constructs used herein was accomplished by stable transfection followed by antibiotic selection or transient transfection.
Ca2+ uptake assays
Cells were permeabilized in buffer containing 125 mM KCl, 2 mM K2HPO4, 10 μM EGTA, 1 mM MgCl2, 20 mM HEPES at pH 7.2, 0.005% digitonin, 5 mM glutamate and malate, and 1 μM cell-impermeable Fluo-4 or Oregon Green-Bapta6F. 10 μM or 50 μM CaCl2 was injected, resulting in about 1 μM or 40 μM free [Ca2+], respectively. Relative rate of Ca2+ uptake is reported as a linear fit from 50 to 60 s (n ≥ 3). For CCCP mitochondrial Ca2+ release, cells were prepared in buffer containing Oregon Green-Bapta6F and without EGTA.
Membrane potential measurements
TMRM was used to assess ΔΨm in permeabilized cells [21]. Cells were prepared as in Ca2+ uptake assays. For kinetic assays, buffer contained 1.3 μM TMRM. 30 μM CaCl2 was injected. For static measurements, 20 nM TMRM was included in the buffer with or without 2 μM CCCP and TMRM fluorescence is reported as a ratio of excitations at 573 and 546 nm and emission at 590 nm.
Statistical analysis
P-values were computed using Student's t-tests. One-way ANOVA was used when comparing multiple means. P < 0.05 was considered significant.
Biochemistry
Co-immunoprecipitation was performed with FLAG or c-MYC affinity beads using lysis buffer containing 50 mM HEPES (pH 7.4), 150 mM NaCl, 5 mM EGTA, 0.2% DDM, and protease inhibitors. For localization analysis, mitochondria from HEK-293T cells were concentrated and subjected to Proteinase K digestion with a digitonin series, as reported previously [15].
Acknowledgments
We thank members of the Mootha laboratory for constructive feedback, especially Z. Grabarek, T. Kitami, E. Kovács-Bogdán, A. Li, and Y. Sancak. K.J.K. was supported by a graduate research fellowship from the National Science Foundation. This work was supported by National Institutes of Health Grant DK080261. V.K.M. is an Investigator of the Howard Hughes Medical Institute.
Author contributions
KJK and VKM conceived of and designed the project and wrote the manuscript. KJK designed and performed experiments and analyzed data.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://embor.embopress.org
References
- 1.Deluca HF, Engstrom GW. Calcium uptake by rat kidney mitochondria. Proc Natl Acad Sci USA. 1961;47:1744–1750. doi: 10.1073/pnas.47.11.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasington FD, Murphy JV. Ca ion uptake by rat kidney mitochondria and its dependence on respiration and phosphorylation. J Biol Chem. 1962;237:2670–2677. [PubMed] [Google Scholar]
- 3.Kirichok Y, Krapivinsky G, Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature. 2004;427:360–364. doi: 10.1038/nature02246. [DOI] [PubMed] [Google Scholar]
- 4.Gunter TE, Pfeiffer DR. Mechanisms by which mitochondria transport calcium. Am J Physiol. 1990;258:C755–C786. doi: 10.1152/ajpcell.1990.258.5.C755. [DOI] [PubMed] [Google Scholar]
- 5.Denton RM, McCormack JG. The role of calcium in the regulation of mitochondrial metabolism. Biochem Soc Trans. 1980;8:266–270. doi: 10.1042/bst0080266. [DOI] [PubMed] [Google Scholar]
- 6.Hajnoczky G, Robb-Gaspers LD, Seitz MB, Thomas AP. Decoding of cytosolic calcium oscillations in the mitochondria. Cell. 1995;82:415–424. doi: 10.1016/0092-8674(95)90430-1. [DOI] [PubMed] [Google Scholar]
- 7.McCormack JG, Denton RM. The activation of isocitrate dehydrogenase (NAD+) by Ca2+ within intact uncoupled rat brown adipose tissue mitochondria incubated in the presence and absence of albumin [proceedings] Biochem Soc Trans. 1980;8:339. doi: 10.1042/bst0080339. [DOI] [PubMed] [Google Scholar]
- 8.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 9.Stefani De D, Raffaello A, Teardo E, Szabo I, Rizzuto R. A forty-kilodalton protein of the inner membrane is the mitochondrial calcium uniporter. Nature. 2011;476:336–340. doi: 10.1038/nature10230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baughman JM, et al. Integrative genomics identifies MCU as an essential component of the mitochondrial calcium uniporter. Nature. 2011;476:341–345. doi: 10.1038/nature10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffaello A, et al. The mitochondrial calcium uniporter is a multimer that can include a dominant-negative pore-forming subunit. EMBO J. 2013;32:2362–2376. doi: 10.1038/emboj.2013.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sancak Y, et al. EMRE is an essential component of the mitochondrial calcium uniporter complex. Science. 2013;342:1379–1382. doi: 10.1126/science.1242993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perocchi F, Gohil VM, Girgis HS, Bao XR, McCombs JE, Palmer AE, Mootha VK. MICU1 encodes a mitochondrial EF hand protein required for Ca(2+) uptake. Nature. 2010;467:291–296. doi: 10.1038/nature09358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plovanich M, et al. MICU2, a paralog of MICU1, resides within the mitochondrial uniporter complex to regulate calcium handling. PLoS ONE. 2013;8:e55785. doi: 10.1371/journal.pone.0055785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Csordas G, et al. MICU1 controls both the threshold and cooperative activation of the mitochondrial Cap2+ uniporter. Cell Metab. 2013;17:976–987. doi: 10.1016/j.cmet.2013.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallilankaraman K, et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca(2+) uptake that regulates cell survival. Cell. 2012;151:630–644. doi: 10.1016/j.cell.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanjana NE, Cong L, Zhou Y, Cunniff MM, Feng G, Zhang F. A transcription activator-like effector toolbox for genome engineering. Nat Protoc. 2012;7:171–192. doi: 10.1038/nprot.2011.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–W568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan X, et al. The physiological role of mitochondrial calcium revealed by mice lacking the mitochondrial calcium uniporter. Nat Cell Biol. 2013;15:1464–1472. doi: 10.1038/ncb2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Floryk D, Houstek J. Tetramethyl rhodamine methyl ester (TMRM) is suitable for cytofluorometric measurements of mitochondrial membrane potential in cells treated with digitonin. Biosci Rep. 1999;19:27–34. doi: 10.1023/a:1020193906974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.