Abstract
The cytochrome P450 (P450) superfamily of enzymes plays a prominent role in drug metabolism. Although mice are a widely used preclinical model in pharmacology, the expression of murine P450 enzymes at the protein level has yet to be fully defined. Twenty-seven proteins belonging to P450 subfamilies 1A, 2A, 2B, 2C, 2D, 2E, 2F, 2J, 2U, 3A, 4A, 4B, 4F, and 4V were readily detectable in Balb/c mouse tissue using a global mass spectrometry–based proteomics approach. Subsequently, a targeted mass spectrometry–based assay was developed to simultaneously quantify these enzymes in ranges of femtomoles of P450 per microgram of total protein concentration range. This screen was applied to mouse liver microsomes and tissue lysates of kidney, lung, intestine, heart, and brain isolated from mixed-sex fetuses; male and female mice that were 3–4 weeks, 9–10 weeks, and 8–10 months of age; and pregnant mice. CYP1A2 was consistently more abundant in male mouse liver microsomes compared with age-matched females. Hepatic expression of CYP2B9 was more abundant in 3- to 4-week-old male and female mice than in mice of other ages; in addition, CYP2B9 was the only enzyme that was detectable at higher levels in pregnant mouse liver microsomes compared with age-matched females. Interestingly, sexually dimorphic expression of CYP2B9, 2D26, 2E1, and 4B1 was observed in kidney only. The targeted proteomics assay described here can be broadly used as a tool for investigating the expression patterns of P450 enzymes in mice.
Introduction
The cytochrome P450 (P450) enzymes play major roles in the metabolism of a wide variety of small molecules, including drugs, steroids, and carcinogens. P450-dependent catalysis dominates drug metabolism, accounting for an approximate 75% of biotransformation of clinically relevant drugs (Guengerich, 2008). Metabolism of a substrate by P450 enzymes generally results in the formation of a more hydrophilic and readily excretable product. As such, P450 activity is a critical contributor to the clearance of many drugs. P450 enzymes are divided into families and subfamilies based on amino acid–sequence identity. P450s belonging to the 1A, 2A, 2B, 2C, 2D and 3A subfamilies have been demonstrated to participate primarily in drug metabolism (Guengerich and Cheng, 2011). The P450 family contains 57 human and 102 murine putatively functional genes (Nelson et al., 2004). Although mice are widely used in preclinical pharmacological studies, current understanding of the expression patterns of murine P450 family members at the protein level is limited, largely because of the lack of availability of antibodies that react with specific mouse P450 enzymes. As a result, much of what is known regarding the expression of murine P450s has relied solely on the analysis of mRNA levels.
The application of mass spectrometry–based global proteomics facilitates the high-throughput analysis of complex protein samples; however, a limitation of this technique is that the stochastic nature of its scan-dependent acquisition can result in bias toward the detection of higher abundance proteins. As a result, lower abundance proteins can be difficult to quantitate when global profiling is performed. The use of targeted proteomics can overcome this limitation since data acquisition is aimed at the detection of specific proteins of interest. One such targeted method is selected reaction monitoring (SRM), which is generally performed using a triple-quadrupole mass spectrometer. In this approach, sensitivity and selectivity are increased through the monitoring of a particular fragment ion of a selected precursor ion (Lange et al., 2008). The specific pair of mass-to-charge ratios associated with the precursor ion and the fragment ion is referred to as a “transition.” The use of these transitions allows for a selected number of peptides to be monitored instead of all peptides present in the mixture being measured simultaneously, as is the case in global proteomics. This feature of SRM-targeted proteomics allows for the reproducible detection and quantification of both low- and high-abundance proteins in complex mixtures over a wide dynamic range (Picotti et al., 2009).
In the present study, we describe a targeted SRM-based proteomics approach that can be used to simultaneously quantitate 27 murine P450 enzymes, including those belonging to subfamilies involved in drug metabolism. In developing the assay, we first applied global proteomics to mouse tissue to compile a list of P450 proteins that were detectable via mass spectrometry. We then identified transitions unique to each P450 that would facilitate detection of these enzymes in a multiplexed fashion in a complex protein mixture. Through the incorporation of heavy isotope–labeled peptides, quantitation of the targeted P450 proteins was achieved over several orders of magnitude. To demonstrate the utility of the assay, we characterized P450-enzyme expression in the liver, kidney, lung, intestine, heart, and brain of male and female Balb/c mice at 3–4 weeks, 9–10 weeks, and 8–10 months of age. In addition, P450 protein levels were measured in each of these organs in pregnant mice while fetal P450 enzyme abundance was analyzed in liver and brain. This assay can be employed broadly and leveraged to gain insight into expression patterns of murine P450s, including in response to drug treatments, physiologic states, and genetic modification.
Materials and Methods
Chemicals and Reagents.
Efavirenz [(4S)-6-chloro-4-(2-cyclopropylethynyl)-4-(trifluoromethyl)-2,4-dihydro-1H-3,1-benzoxazin-2-one] was purchased from Toronto Research Chemicals (Toronto, ON, Canada). NADPH was purchased from BD Biosciences (San Jose, CA). Iodoacetamide, phenylmethylsulfonyl fluoride, and urea were obtained from Sigma-Aldrich (St. Louis, MO). Hexanes and ethyl acetate were purchased from Thermo Fisher Scientific (Waltham, MA). All other chemicals were of the highest grade commercially available.
Mouse Tissue Preparation.
Liver microsomes were prepared from the livers of male and female mice 3–4 weeks, 9–10 weeks, and 8–10 months of age in addition to pregnant mice (10–12 weeks of age) and fetuses (gestation day 16) as previously described (Raucy and Lasker, 1991). Mice were killed as approved by the Johns Hopkins University Animal Care and Use Committee. Tissue total protein lysates were prepared from kidney, lung, intestine, heart, and brain of male and female mice in each age-group. Brain was the only organ besides liver that was collected from fetuses. All fetal brains from an individual pregnant mother were pooled and lysed. Tissue was homogenized in lysis buffer containing 20 mM Tris HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM ethylene glycerol tetraacetic acid, 1% Triton X-100, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 μg/ml leupeptin, and 1 mM phenylmethylsulfonyl fluoride (Cell Signaling, Danvers, MA). Samples were centrifuged at 10,000g for 30 minutes at 4°C, and the supernatants were saved. The microsomes and lysates that were isolated from four individual mice within the same age, organ, and sex group were pooled. Fetal liver microsomes and brain lysates from all pregnant mice were pooled. Protein concentration was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific), and protein was frozen at –80°C until use.
In-Gel Peptide Isolation for Global Proteomics.
Protein (25 μg) reduction and alkylation was performed in NuPAGE LDS Sample Buffer (Life Technologies, Grand Island, NY). NuPAGE Sample Reducing Agent (Life Technologies) was added (50 mM dithiothreitol) and heated at 70°C for 10 minutes. Samples were cooled to room temperature before incubation with 1.2 μl of 1 M iodoacetamide (100 mM) for 20 minutes in the dark. NuPAGE Sample Reducing Agent (80 mM) was added for continuation of the reaction for 20 minutes in the dark. A 10% Tris-glycine gel (Bio-Rad, Hercules, CA) was prepared with 25 μg of protein loaded in each well. NuPAGE antioxidant (500 μl; Life Technologies) was mixed in the running buffer. The gel was stained with SimplyBlue SafeStain (Life Technologies) according to manufacturer’s instructions. The 45- to 62-kDa band region of the gel was removed, corresponding to the range of molecular masses of murine P450s. The stain was removed from the gel by incubation for 2 hours with 200 mM ammonium bicarbonate and 40% acetonitrile. Gels were dried under vacuum and trypsinized at 1:20 trypsin/protein ratio for 18 hours at 37°C. The resulting peptide solution was removed and the remaining peptides were extracted from the gel using 0.1% trifluoroacetic acid, 50 mM ammonium bicarbonate, and 50% acetonitrile by incubation at 37°C for 1 hour. All of the peptides of each sample were combined before drying under vacuum. Samples were reconstituted in 10 μl of mobile phase A, 2 μl of which were injected for liquid chromatography–mass spectrometry (LC-MS) analysis.
Global Proteomics LC-MS Assay.
Protein discovery experiments were performed by the Mass Spectrometry and Proteomics Facility at Johns Hopkins School of Medicine. Total protein lysates (heart, kidney, brain, lung, and intestine) and liver microsomes isolated from a 9- to 10-week-old male and a 9- to 10-week-old female Balb/c mouse were analyzed using global proteomics to generate a list of P450 enzymes for incorporation into a targeted proteomics assay. An Eksigent NanoLC-2D Plus HPLC system (Eksigent/AB Sciex, Framingham, CT) was used in tandem with the high resolution Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Peptides were separated on an in-house 10-cm column packed with 5 μm of Magic C18 beads (Michrom Bioresource/Bruker, Auburn, CA). Gradient solvents were composed of 2% acetonitrile 0.1% formic acid in LC-MS grade water (solvent A) and 90% acetonitrile 0.1% formic acid (solvent B). Flow rate was set to 300 nl/min for the 90-minute gradient. A 40-minute blank was run between each sample. The sample gradient increased from 2 to 8% to 5 minutes, 8 to 32% to 70 minutes, 32 to 50% to 75 minutes, 50 to 100% to 80 minutes, 100% to 84 minutes, and then 2% to 90 minutes. Eluting peptides were sprayed into the mass spectrometer through a 10-μm emitter tip (New Objective, Woburn, MA). Peptides were analyzed at resolution 30,000 within a mass range of m/z 300–2000. The eight most abundant peptides were individually selected at isolation width 1.9 Da, and fragmented using collision energy 35 eV. Fragments were scanned at resolution 7500 within m/z 350–1800. Data-dependent acquisition was used for confident identification of as many peptides as possible. Dynamic exclusion of 30 seconds with a repeat count of 1 maximized the number of possible identifications. Monoisotopic ion precursor selection (MIPS) was “on.” Lock mass was “on” (siloxane 371 Da). Tandem mass spectra were extracted, and charge state was deconvoluted and deisotoped by Proteome Discoverer (version 1.3; Thermo Fisher Scientific). All tandem mass spectrometry (MS/MS) spectra were analyzed with Mascot (version 2.2; Matrix Science, London, UK) using the National Center for Biotechnology Information (NCBI) 167nr Database, Musculus species with acquired raw MS/MS data, trypsin as the enzyme, one missed cleavage allowed, precursor mass tolerance of 2 ppm, fragment mass tolerance of 0.02 Da, and formation of y- and b-ions. Oxidation of methionine and carbamidomethyl of cysteine were specified in Mascot as variable modifications. For each sample, spectra were analyzed by database searching using the Mascot search algorithm. Scaffold is a proteomic data viewing software provided by Proteome Software (Portland, OR).
In-Solution Peptide Isolation for Targeted Proteomics.
A proteomic SRM assay was developed to quantify the P450 proteins identified in the data-dependent mass spectrometry experiment. Samples for the targeted assay were prepared using the filter-aided sample preparation method developed by Matthias Mann (Wisniewski et al., 2009). The targeted assay was applied to the previously described microsomes, including male and female Balb/c mice of three ages plus pregnant mice and fetuses. In addition, pooled tissue lysates (n = 4) from these mice were analyzed, including kidney, lung, intestine, heart, and brain. Tissue lysates or microsomes (100 μg) were diluted in 50 μl of 50 mM ammonium bicarbonate. Proteins were reduced with NuPAGE Sample Reducing Agent for 60 minutes at 60°C. Free cysteines were alkylated in 100 mM iodoacetamide at room temperature for 15 minutes. Protein was washed with urea (9 M) three times with 300 μl on a 30-kDa filter (Sigma-Aldrich), followed by centrifugation at 14,000g for 10 minutes. Urea was removed using five washes of 300 μl of 25 mM ammonium bicarbonate. Isotope-labeled peptides were added (6000 fmol of CYP1A2 peptide and 600 fmol of CYP2B10, CYP2D22, CYP3A11, and CYP4V2 peptides in a 10:1:1:1:1 peptide ratio mix). Proteins were digested with 10 μg of trypsin for 18 hours. An additional 1 μg of fresh trypsin was added after 15 hours. The peptides were then collected by combining the filtrate and one 200-μl 25 mM ammonium bicarbonate wash. Sample was dried under vacuum at 60°C. Each sample was resuspended in 40 μl of mobile phase A directly before analysis. Each injection into the mass spectrometer contained 5 μg of sample protein in addition to 30 fmol of CYP2B10, CYP2D22, CYP3A11, and CYP4V2 peptides, and 300 fmol of CYP1A2 peptide in a volume of 2 μl.
Targeted Proteomics LC-MS Assay.
To write the mass spectrometric method and view the targeted method data, the open source program Skyline (MacCoss Laboratory, University of Washington School of Medicine) was used. P450 protein sequences obtained from the NCBI Reference Sequence Database (NCBI RefSeq Database) were imported into Skyline in the FASTA format. Skyline performed an in-silico digestion of the proteins at lysines and arginines, allowing 0 missed cleavages, +2 and +3 charges, and lengths of 5–25 residues. Basic Local Alignment Search Tool (NCBI BLAST) was used to determine uniqueness of a peptide to the protein. Multiple rounds of transition selection were performed, prioritizing peptides that do not contain ragged ends, methionines, cysteines, glutamines, and asparagines. One peptide for quantification and one peptide for protein confirmation were chosen for the final assay. The four transitions with the highest signal-to-noise ratio were retained for the peptide intended for quantification; replicate analysis of the sample allowed for confident retention time identification. Five representative peptides were chosen for absolute quantitation, as indicated in Table 1. A synthetic peptide standard, either isotope-labeled or -unlabeled, was used for each peptide monitored in the assay. Isotopically labeled peptides were synthesized for a representative group of P450 enzymes to facilitate absolute quantitation of these proteins. Heavy labeled peptides (C-terminal [13C]/[15N) were synthesized by New England Peptide (Gardner, MA), and unlabeled peptides were synthesized by Sigma Biosciences (Rockville, MD).
TABLE 1.
SRM assay design for tryptic peptides and isotope-labeled peptides
The P450 enzymes targeted for quantitation as well as the protein accession numbers, peptide sequences, and transitions used for quantitation of each P450 in the targeted mass spectrometric assay are outlined. The protein list was generated from a global proteomics screen of liver microsomes and tissue protein lysates isolated from 9- to 10-week-old male and female Balb/c mice. All peptides selected are specific to the protein, with the exception of CYP4A10 peptide, which is also predicted to be formed from CYP4A32. The multiple reaction monitoring (MRM) assay refers to the single peptide and transition pair, which form the complete SRM assay method. The heavy labeled (13C and 15N) synthetic peptides included in the assay for quantitation are represented in boldface with an asterisk next to the heavy labeled residue.
| Enzyme | Accession No. | Peptide Sequence | MRM (Parent/Transition) |
|---|---|---|---|
| 1A2 | NP_034123.1 | T254FNDNFVLFLQK265 | 743.390/747.476 (y6) |
| TFNDNFVLFLQK* | 747.398/755.491 (y6) | ||
| 2A12 | NP_598418.1 | E88ALVDHAEEFSGR100 | 730.344/1047.449 (y9) |
| 2B9 | NP_034130.1 | D263YIDTYLLR271 | 586.303/780.425 (y6) |
| 2B10 | NP_034129.1 | G99TVAVVEPTFK109 | 574.322/819.461 (y7) |
| GTVAVVEPTFK* | 578.329/827.475 (y7) | ||
| 2C29 | NP_031841.3 | N49ISQSFTNFSK59 | 636.815/830.404 (y7) |
| 2C37 | NP_034131.2 | Y308AILLLLK315 | 473.820/712.533 (y6) |
| 2C39 | NP_034133.2 | F126TLTTLR132 | 426.253/603.382 (y5) |
| 2C50 | NP_598905.2 | G384TNVITSLSSVLR396 | 673.886/862.499 (y8) |
| 2C54 | NP_996260.1 | E85ALVDHGDVFAGR97 | 693.344/973.449 (y9) |
| 2C67 | NP_001019890.1 | V125FTINTLR132 | 482.285/503.294 (y4) |
| 2C70 | NP_663474.2 | E85ALIDQGDEFSDK97 | 733.836/1040.417 (y9) |
| 2D9 | NP_034136.2 | D408ESVWEKPLR417 | 629.825/828.473 (y6) |
| 2D10 | NP_034135.2 | F369GDIAPLNLPR379 | 606.840/709.436 (y6) |
| 2D22 | NP_062797.3 | T263TWDPTQPPR272 | 599.796/695.383 (y6) |
| TTWDPTQPPR* | 604.800/705.392 (y6) | ||
| 2D26 | NP_083838.1 | G121VILAPYGPEWR132 | 679.367/454.302 (b5) |
| 2E1 | NP_067257.1 | G113IIFNNGPTWK123 | 623.833/963.468 (y8) |
| 2F2 | NP_031843.2 | S48QDLLTSLTK57 | 553.309/662.408 (y6) |
| 2J5 | NP_034137.1 | L50PFVGNFFQIDTK62 | 763.406/1012.510 (y8) |
| 2U1 | NP_082092.2 | E464TFIPFGIGK473 | 554.805/618.361 (y6) |
| 3A11 | NP_031844.1 | L332QDEIDEALPNK343 | 692.851/358.208 (y3) |
| LQDEIDEALPNK* | 696.858/366.223 (y3) | ||
| 3A13 | NP_031845.1 | D244VISFFTTSVER255 | 700.856/839.426 (y7) |
| 3A25 | NP_062766.2 | F447ALISIK453 | 396.255/573.397 (y5) |
| 4A10 | NP_034141.3 | T215YLQAIGDLNNLFHSR230 | 621.323/887.448 (y7) |
| 4A12A | NP_803125.2 | S214YIQAVEDLNDLVFSR229 | 934.973/1207.595 (y10) |
| 4B1 | NP_031849.1 | G125LLVLEGPK133 | 463.289/642.382 (y6) |
| 4F13 | NP_570952.1 | S453PLAFIPFSAGTR465 | 682.372/735.378 (y7) |
| 4V2 | NP_598730.1 | V391FPSVPLFAR400 | 566.829/886.515 (y8) |
| VFPSVPLFAR* | 571.833/896.523 (y8) |
designating the last letter only, which is intended to indicate the only heavy labeled residue of the entire boldface peptide sequence.
Mass spectrometry and liquid chromatography parameters for the quantitative proteomics assay were determined separately from the peptide and transition selection in Skyline (MacLean et al., 2010). Liquid chromatography was performed by a nanoAcquity ultra-performance liquid chromatograph (Waters, Wexford, Ireland) in tandem with a triple quadrupole TSQ Vantage mass spectrometer (Thermo Fisher Scientific). Peptides were separated using a Halo reverse phase C18 column with 2.7-μm bead diameter and dimensions 2.1 × 100 mm (MAC-MOD Analytical, Chadds Ford, PA). The column was heated to 40°C, and the samples were maintained at 6°C. The flow rate was 100 μl/min. Mobile phase A was 0.1% formic acid in water, with mobile phase B as 0.1% formic acid in 90% acetonitrile. The 30-minute chromatography sequence started at 5% B for 2 minutes, increased to 60% B to 26 minutes, 98% to 27 minutes, 5% to 28 minutes and remained at 5% to 30 minutes. Mass-to-charge ratio of all quantitative and confirmatory peptides and transitions are indicated in Table 1 and Supplementary Table 1. Cycle time was 5 seconds, peak widths for Q1 and Q3 were 0.70 full width at half-maximum (FWHM), and chromatographic filter peak width was set to 50 seconds. All ions were monitored in positive mode and analyzed using Xcalibur software (version 2.1; Thermo Fisher Scientific).
Proteomics Quantification.
The isotope-labeled peptide standards were prepared in triplicate by spiking Balb/c mouse liver microsomes, following the sample preparation procedure described above, with dilutions of synthetic isotope-labeled peptide mix. Isotope-labeled peptide mix was serially diluted, resulting in a final calibration curve spanning five orders of magnitude (1.5, 3, 15, 30, 75, 150, and 300 fmol and 1.5, 3, and 15 pmol). Quantification was performed using the peptide transition with highest signal-to-noise ratio in the extracted chromatogram of each peptide. Calibrant peak areas were normalized to the 30-fmol peak area and fit to a 1/y2-weighted linear regression in GraphPad Prism version 6.02 for Windows (GraphPad Software Inc., San Diego, CA). Analyte concentration was calculated from the analyte/isotope-labeled peptide area-under-the-curve ratio using the linear regression equation from the normalized isotope calibrants. The calibration curves for the synthetic unlabeled peptides were prepared by serial dilution of the equimolar peptide mix from a 500 μM stock in triplicate. Direct injection (2 μl) of the dilutions produced calibration points spanning six orders of magnitude (1.5, 3, 15, 30, 75, 150, and 300 fmol and 1.5 and 3 pmol). At least six of the dilutions were used for each peptide calibration curve according to linear range, except the peptides representing proteins 2C37 and 4A10, which contained five points. Unlabeled peptide standards were averaged (n = 3) and fit to a 1/y2-weighted linear regression in GraphPad Prism. Relative quantification of peptides was performed using the analyte peptide peak area and the regression equation to calculate the approximate concentration of peptide in the tissue.
Efavirenz Metabolite LC-MS Assay.
The efavirenz (EFV) metabolism assay was performed as previously described (Avery et al., 2013) using a final concentration of 7 μM EFV. Determination of EFV metabolite formation was performed using a Dionex UltiMate 3000 ultra high-performance liquid chromatograph (Thermo Fisher Scientific) in tandem with a triple quadrupole TSQ Vantage mass spectrometer (Thermo Fisher Scientific). Samples were injected onto a XTerra reverse phase column, with 2.5-μm C18 beads and 2.1 × 50-mm dimensions (Waters, Milford, MA). Column and samples remained at room temperature during analysis. Sample was injected by the Dionex autosampler at a draw speed of 5 μl/s, a dispense speed of 20 μl/s, followed by injection wash of 100 μl and loop wash by a factor of 2. Flow rate was 0.4 ml/min, with mobile phase A as 0.1% formic acid in water and mobile phase B as 0.1% formic acid in acetonitrile. The column was conditioned at 35% B. From 0.2 to 5 minutes, mobile phase B increased linearly to 60%. Remaining sample was eluted at 95% B for 0.8 minutes, and the column equilibrated at 5% B for 0.2 minutes. EFV and fluorinated EFV were monitored using SRM at m/z of precursor/product ion pairs from 313.85/243.98 to 298.00/227.90 as was reported previously (Avery et al., 2013). Scan width was 0.002, scan time was 0.001, peak widths for Q1 and Q3 were 0.70 FWHM, and collision energies were 18 and 20, respectively. Mono- and di- hydroxylated EFV metabolites were monitored in product-scanning mode. The expected m/z of the parent ions were 329.821 and 345.789, respectively. Scan time was 0.1, peak widths for Q1 and Q3 were 0.70 FWHM, and collision energies were 14, 17, and 14, respectively. Signal intensities of EFV metabolites were normalized using the intensity of the fluorinated EFV peak intensity. Product ions were scanned from m/z 50 to 370. All ions were monitored in negative ionization mode.
Statistical Analysis.
Graphs and one-way analysis of variance (ANOVA) F-statistics with Bonferroni corrections were performed using GraphPad Prism version 6.02 for Windows.
Results
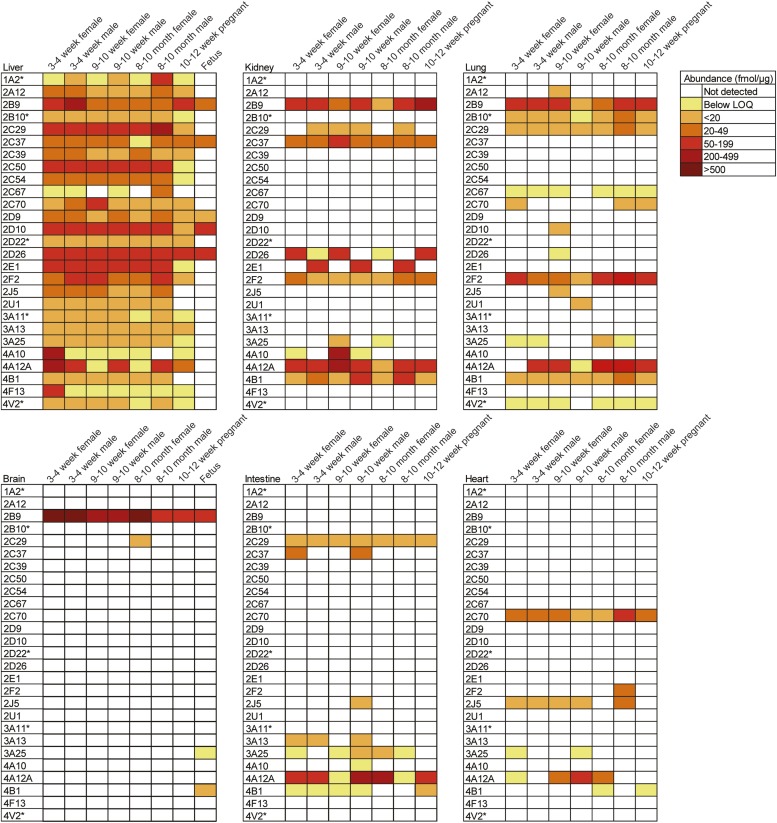
A targeted proteomics method using mass spectrometry in selected reaction monitoring mode was developed to simultaneously measure protein levels of murine CYPs 1A2, 2A12, 2B9, 2B10, 2C29, 2C37, 2C39, 2C50, 2C54, 2C67, 2C70, 2D9, 2D10, 2D22, 2D26, 2E1, 2F2, 2J5, 2U1, 3A11, 3A13, 3A25, 4A10, 4A12A, 4B1, 4F13, and 4V2. These P450 enzymes were identified for incorporation into this multiplexed assay based on an initial global unbiased proteomics screen that detected each of these proteins in tissue lysates and liver microsomes isolated from 9–10-week-old male or female Balb/c mice. The multiplexed screen was refined using the mouse liver microsomes. The workflow for the development of the assay and subsequent sample analysis is depicted in Fig. 1. Liver was chosen for these initial studies because this organ is the predominant site of drug metabolism and as such contains an abundance of P450 enzymes. In addition, because P450s are microsomal proteins, we isolated microsomes from mouse livers to increase the ability to detect P450 enzymes and also to allow for parallel analysis of protein levels and metabolic activity toward a substrate. The peptides and SRM transitions optimized to detect each protein are listed in Table 1 (SRM transitions used for quantitation) and Supplemental Table 1 (all SRM transitions used for detection of each P450-specific peptide). The limit of quantitation and linear range of each peptide quantitative assay is shown in Supplemental Table 2. Differential expression of several P450 enzymes was observed in liver microsomes isolated from male and female Balb/c mice 3–4 weeks, 9–10 weeks, and 8–10 months of age (Fig. 2). In addition, liver microsomes from 10- to 12-week-old pregnant mice and mixed-sex fetuses (gestation day 16) were screened (Fig. 2). The most consistently abundant proteins across age in both males and nonpregnant females were CYPs 2C29, 2D26, 2D10, and 2E1. Expression of CYP1A2 appeared to be sexually dimorphic as it was more abundant in male mice compared with females across all age-groups investigated. CYP4A12A protein levels were found to be lower in liver microsomes isolated from adult (9- to 10-week-old and 8- to 10-month-old) female mice compared with age-matched males. CYP2E1 appeared to be less abundant in livers from pregnant mice than in all other groups, whereas the levels of CYP2B9 detected were higher in pregnant mice than in nonpregnant adult females and adult males. Of note, the levels of hepatic CYP2B9 in 3–4-week-old males and females were greater than adult males and nonpregnant adult females. CYPs 4A10 and 4F13 were detected in high abundance in 3–4 weeks of age female mouse liver microsomes, whereas the levels of these enzymes were below the limit of quantitation at all other ages. Fetal liver expressed CYPs 2B9, 2C37, 2D9, 2D10, and 2D26.
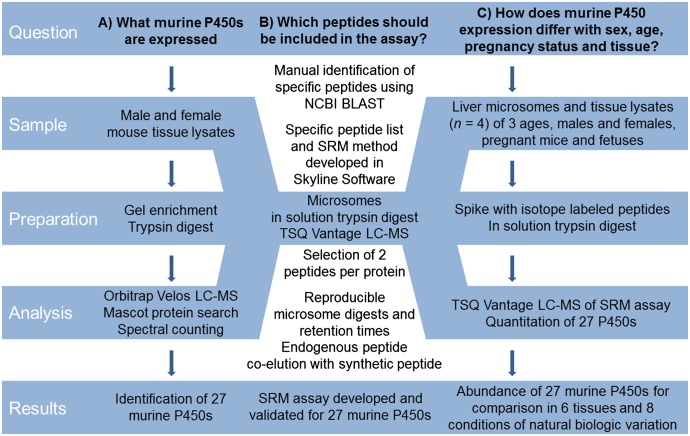
Fig. 1.
Mass spectrometry workflows for identification and quantitation of murine P450 enzymes. A global proteomics approach was used to compile a list of P450s expressed in mouse liver, kidney, lung, heart, intestine, and brain (A); develop an SRM assay for the proteins identified (B); and use the SRM assay developed to apply a targeted approach to quantitate P450 expression in mouse liver microsomes and tissue lysates (C).
Fig. 2.
Cytochrome P450 quantitation in murine liver microsomes and tissue lysates. Liver microsomes, kidney, lung, brain, intestine, and heart were prepared from pregnant mice, fetuses, and male and female Balb/c mice 3–4 weeks, 9–10 weeks, and 8–10 months of age. The proteins were harvested from four individual age- and sex-matched mice and pooled. The resulting pooled samples were then assayed in triplicate. Proteins were analyzed using the targeted SRM assay on a Thermo TSQ Vantage triple quadrupole mass spectrometer. Color coding denotes the abundance of each P450 enzyme measured using the peptides listed in Table 1. The asterisks indicate P450 enzymes subjected to absolute quantitation using isotopically labeled peptides. Unlabeled-peptide synthetic standards were used to perform relative quantitation of all other P450 proteins. Quantitation was performed using Skyline software, and peak signal-to-noise ratio was manually validated.
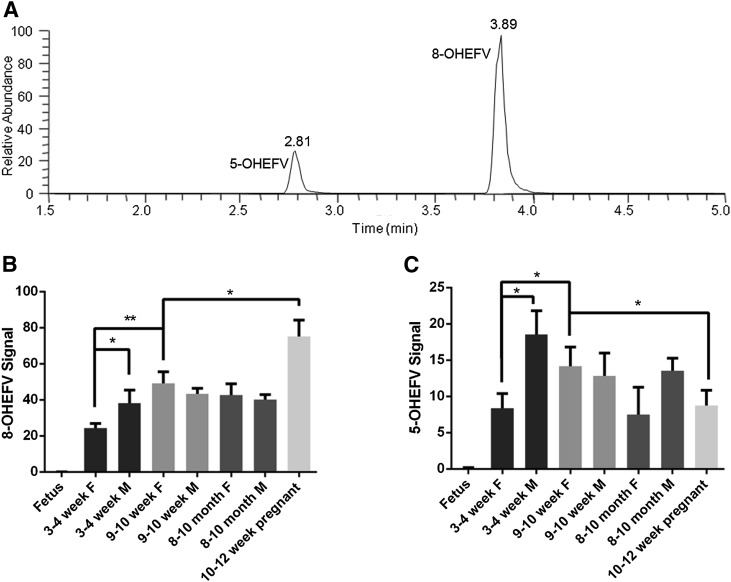
Activity of the liver microsomes that were analyzed using proteomics was probed in parallel using the anti-HIV drug efavirenz as a substrate. EFV was chosen for the activity assays because we and others have demonstrated that multiple human P450 subfamilies can contribute to the oxidative metabolism of this drug (Ward et al., 2003; Ogburn et al., 2010; Avery et al., 2013). Metabolism of EFV in mice has yet to be reported. Mouse microsomal metabolism of EFV resulted in the formation of two mono-oxygenated metabolites (Fig. 3A) as measured using ultra high-performance liquid chromatography–tandem mass spectrometry (uHPLC-MS/MS). The metabolite with retention time 3.89 minutes (Fig. 3A) was identified as 8-hydroxyEFV (8-OHEFV), which is the primary metabolite of EFV formed in humans, based on the comparison of the retention time and fragmentation pattern of this analyte with a synthetic standard. The fragmentation spectrum of the metabolite with retention time of 2.81 minutes (Supplemental Fig. 1) was largely identical to that of 8-OHEFV, indicating that the oxygen insertion was occurring on the same ring that encompasses the 8-position, although the metabolite that eluted at 2.81 appeared to be more hydrophilic based on the retention time. A 7-hydroxyEFV metabolite has also been previously described; however, this synthetic standard eluted later than the 2.81-minute metabolite. As such, we propose that this metabolite is 5-hydroxyEFV (5-OHEFV). No additional metabolites were detectable in these mouse liver microsome assays.
Fig. 3.
Metabolism of EFV across age, sex, and pregnancy statuses using mouse liver microsomes. A representative chromatogram depicting the elution of monohydroxylated EFV metabolites is shown (A). EFV metabolism assays were performed using liver microsomes isolated from pregnant mice, fetuses, and male and female Balb/c mice 3–4 weeks, 9–10 weeks, and 8–10 months of age. Formation of 8-OHEFV (B) and 5-OHEFV (C) was measured and normalized to the fluorinated EFV analog used as an internal standard. *P < 0.05 and **P < 0.01 as determined by one-way analysis of variance.
Formation of 8-OHEFV (Fig. 3B) and 5-OHEFV (Fig. 3C) by liver microsomes isolated from livers of 3- to 4-week-old females was lower than that of 9- to 10-week-old female liver microsomes (P = 0.0045 and P = 0.0326, respectively). In addition, 8-OHEFV and 5-OHEFV were less abundant in the incubations containing the 3- to 4-week-old female liver microsomes compared with males of the same age (P = 0.0412 and P = 0.0150, respectively). Interestingly, sex differences in EFV-metabolite formation were not observed using liver microsomes isolated from the 9- to 10-week- or 8- to 10-month-old mice. Liver microsomes from pregnant mice (10–12 weeks of age) demonstrated significantly higher 8-OHEFV production in comparison with 9- to 10-week-old females (P = 0.0289), whereas 5-OHEFV was significantly lower (P = 0.0355). Fetal liver microsomes did not demonstrate activity toward EFV.
To gain an understanding of murine P450 protein expression in extrahepatic tissues, kidney, lung, brain, intestine, and heart, total protein lysates from the Balb/c mice were analyzed using the targeted proteomics assay. Total protein lysates were analyzed for these tissues as opposed to microsomes because the protein yields for microsomal preparations from these organs were far below that of liver. Ten of the 27 P450 enzymes were detected in kidney lysates (Fig. 2). CYPs 4A12A, 2B9, and 4A10 exhibited the highest levels. Kidney was the only extrahepatic organ that expressed detectable levels of CYP2E1 and this expression was male-specific. Conversely, CYP3A25 was only detected in adult nonpregnant female kidney lysates and CYP2D26 was predominantly expressed in female kidney lysates, including expression in kidney isolated from pregnant mice. Interestingly, the kidney exhibited the greatest sex-specific trends in P450 protein levels. The number of P450 enzymes identified in lung tissue was second only to liver expression: 15 of the 27 proteins screened were detected in lung tissue (Fig. 2). CYPs 4A12A, 2F2, and 2B9 were among the most highly expressed P450 proteins in lung protein lysates. In contrast to the kidney, no sex-, age-, or pregnancy-specific trends were identified in lung tissue. Although CYPs 2C67 and 4V2 were expressed in lung they were not detectable in other extrahepatic tissues. Besides liver, only brain could be isolated from fetuses in sufficient quantities. CYP2B9 was the sole P450 enzyme that was detectable in all of the Balb/c brain lysates analyzed (Fig. 2). Similar to what was observed for liver microsomes, CYP2B9 was found in higher abundance in the male and female mice that were 3–4 weeks of age than in older mice. In addition to CYP2B9, fetal brain lysate expressed CYPs 3A25 and 4A12A, whereas CYP2C29 was detected in the brain lysates of 8- to10-month-old female Balb/c mice. Proteins detected in the greatest number of intestinal lysates analyzed in these studies were CYPs 2C29, 4A12A, and 3A25 (Fig. 2). CYPs 3A13, 2J5, 2C37, 4B1, and 4A10 were also detected. CYP4B1 was found at higher levels in pregnant mice than in age-matched nonpregnant female intestinal lysates. CYPs 2C70 and 2J5 were the most frequently detected P450 enzymes in heart lysates (Fig. 2). CYPs 4A12A, 2F2, 3A25, and 4B1 were also detected. No sex-, age-, or pregnancy-specific trends were identified through the analysis of the heart lysates.
Discussion
Here we describe the development of a targeted proteomics assay, a powerful tool that can be broadly applied to probe the impact of small molecules and pathophysiological states on the expression of murine P450 enzymes. To demonstrate the application of this assay, we present an analysis of P450 protein expression patterns in male and female mice over a range of ages in several organs. Previous reports have relied overwhelmingly on mRNA analyses; therefore, our studies provide novel insight into murine P450 distribution at the protein level. Genomic analyses have identified 102 murine P450 genes; however, it is not currently known how many P450 proteins are expressed in mice. In the current study, we found 27 murine P450 enzymes were readily detectable and distinguishable using mass spectrometry. CYPs 4B1, 3A25, 4A12A, and 2C29 were the most consistently expressed among the mouse organs analyzed, although a number of age-, sex- and pregnancy-dependent trends were noted in the expression of these and other P450s.
Among the consistently expressed P450s, CYP4B1 expression was elevated in adult male kidney compared with age-matched females, and this is consistent with previous reports of dimorphic expression of CYP4B1 mRNA (Renaud et al., 2011). Further, CYP3A25 mRNA has been shown to be expressed most prominently in liver and intestines and this finding is in line with our proteomics results (Renaud et al., 2011). Similar to the results presented here, CYP2C29 mRNA has been shown to be mainly expressed in liver and lung but also in many other organs (Choudhary et al., 2003). CYP2C29 detoxifies α- and β-unsaturated aldehydes, found endogenously in food and in the environment, which can otherwise react with genomic material (Amunom et al., 2011). Our data identified CYP2B9 as the most abundant murine P450 in brain, and sexually dimorphic expression was not observed. In addition, CYP2B9 protein was identified here in both fetal liver and brain.
Striking differences in P450 expression were noted between organs. The kidney exhibited several instances of differential P450 expression in males versus females, including CYPs 2D26, 2B9, and 2E1. This trend was most striking for CYP2E1, which appeared to be expressed only in the kidneys of male mice. Interestingly, this pattern of expression was not observed in liver. Levels of CYP2E1 in pregnant liver were markedly lower than in age-matched female Balb/c mice. P450 protein expression in lung parallels previously observed expression patterns for human enzymes. In particular, Bieche et al., (2007) determined that human CYP2F1 was predominantly expressed in lung compared with other extrahepatic tissues. In our study, we found that murine CYP2F2 was one of the more abundant P450 enzymes in lung. Drug-metabolizing enzymes in lung are particularly important for environmental toxins and drugs delivered by inhalation. Since the intestine can contribute to first-pass metabolism following oral administration of a drug, understanding the expression of P450 enzymes in this organ is of interest. In the present study, enzymes belonging to the CYP3A and CYP2C subfamilies were identified in intestine, and this is consistent with previous human studies (Ding and Kaminsky, 2003). The CYP4A, CYP4B, and CYP2J subfamilies were also identified in mouse intestine, and interestingly, these enzymes have not been previously shown to be expressed in either human or mouse intestine (Zhang et al., 2003). CYP2J5 was readily detectable in all mouse liver microsomes except for those isolated from pregnant mice. In addition, this enzyme was undetectable in the heart of 8–10-month-old female mice as well as pregnant mice. Localization of human CYP2J2 in heart is well characterized (Bieche et al., 2007), as is its role in xenobiotic metabolism and the pathogenesis of heart disease (Xu et al., 2013); however, our results raise the intriguing possibility that CYP2J protein may be differentially expressed during pregnancy. The assay developed here can facilitate investigations into the impact of pregnancy on the regulation of CYP2J protein. Human subfamily CYP2C is known to contribute to ischemic heart disease, and although CYP2C mRNA is not particularly abundant in murine heart (Renaud et al., 2011), the proteomic results presented in our work suggest expression of a murine CYP2C, namely CYP2C70, at all ages in heart. Knowledge of fetal P450 expression can lend insight to our understanding of the capacity of neonates to metabolize and clear xenobiotics. Whereas CYP2B9 was detectable in the brains of all mice, CYPs 3A25 and 4A12A were detected only in fetal brain, indicating that neural expression of these enzymes decreases after birth.
The limitations of this study include the use of whole-organ homogenates (for brain, kidney, lung, heart, and intestine), the small sample size (use of only four animals), the use of pooled samples, and the examination of only a small sampling of ages, including one gestation stage. The small number of animals used in addition to the pooling of samples does not allow for the analysis of interindividual differences in the expression of P450 enzymes in mice; however, the use of pooled samples in the present study enabled the broad proof-of-concept application of the assay described here for use in screening murine P450 expression in multiple tissues and age-groups. Subsequent studies using a larger sample number are required to gain a more detailed understanding of quantitative differences in expression among these groups. In addition, because organs were homogenized, the expression levels of P450 enzymes that we estimated do not reflect cell-type–specific levels of P450 expression within the tissues analyzed; however, the method that we developed could be applied to protein lysates isolated from specific cell types of interest. Finally, to gain a comprehensive understanding of P450 expression during pregnancy, fetal development, and development after birth, mice would need to be examined at gestational stages and ages beyond those in this study.
Mouse P450 activity toward the anti–HIV drug EFV revealed significant differences between mice of varying age, sex, and pregnancy status. Microsomes prepared from pregnant mouse livers formed more 8-OHEFV than those isolated from age-matched female mice. Although humans and mice may metabolize drugs differently, it is interesting to note that pregnant women taking EFV demonstrate increased metabolism of EFV in comparison with after birth (Cressey et al., 2012). In addition, because formation of 8-OHEFV and 5-OHEFV using liver microsomes isolated from pregnant mice were significantly increased and decreased, respectively, more than one murine P450 enzyme may contribute to the metabolism of EFV. Further, we detected a metabolite that we propose to be 5-OHEFV. This identification was based on the fragmentation pattern of this analyte and the difference in retention time compared with authentic standards for EFV metabolites oxygenated at the 7- or 8-position. This product has not been noted in previous reports of EFV metabolism; however, none of these studies has examined metabolism of EFV in mice.
In humans, multiple enzymes contribute to the metabolism of EFV, but human CYP2B6 is known to contribute the most to the formation of the primary metabolite, 8-OHEFV. Although we are not able to conclude which murine P450 enzyme(s) catalyzed the metabolism of EFV in our mouse liver microsome experiments, murine CYP2B9 and CYP2B10 are homologous to human CYP2B6. It is interesting to note that while our proteomics studies demonstrated that CYP2B9 was expressed in fetal liver microsomes, EFV was not metabolized by these microsomes. Further, our proteomic profiles did not reveal age-, sex- or pregnancy-related patterns of CYP2B10 expression that alone would be commensurate with the differential metabolism of EFV across these groups. Taken together, these data indicate that enzymes beyond CYP2B9 and CYP2B10 may contribute to EFV metabolism in mice. In addition, through these data, we were able to demonstrate that the microsomes analyzed in the proteomics studies exhibited metabolic activity and also showed age-, sex-, and pregnancy-dependent differences in activity.
In summary, we have developed a targeted proteomics assay that can be used to simultaneously measure the expression of 27 murine P450s belonging to subfamilies that play key roles in drug metabolism. Through the application of this assay, we have performed a comprehensive analysis of the expression of these P450s across age, sex, and pregnancy statuses. This assay broadly enables investigation of the impact of xenobiotics, pathophysiological stimuli, and genetic modification on P450 protein expression in mice.
Supplementary Material
Acknowledgments
This work is dedicated to the memory of our mentor and pioneering mass spectrometrist, Dr. Robert J. Cotter.
Abbreviations
- 5-OHEFV
5-hydroxyEFV
- 8-OHEFV
8-hydroxyEFV
- EFV
efavirenz
- FWHM
full width at half-maximum
- LC-MS
liquid chromatography–mass spectrometry
- MS/MS
tandem mass spectrometry
- NCBI
National Center for Biotechnology Information
- P450
cytochrome P450
- SRM
selected reaction monitoring
Authorship Contributions
Participated in research design: Hersman, Bumpus.
Conducted experiments: Hersman.
Performed data analysis: Hersman, Bumpus.
Wrote or contributed to the writing of the manuscript: Hersman, Bumpus.
Footnotes
This work was supported by the National Institutes of Health [Grants F31-AG041609 (to E.M.H.) and R01-GM103853 (to N.N.B.)].
 This article has supplemental material available at jpet.aspetjournals.org.
This article has supplemental material available at jpet.aspetjournals.org.
References
- Amunom I, Dieter LJ, Tamasi V, Cai J, Conklin DJ, Srivastava S, Martin MV, Guengerich FP, Prough RA. (2011) Cytochromes P450 catalyze the reduction of α,β-unsaturated aldehydes. Chem Res Toxicol 24:1223–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery LB, VanAusdall JL, Hendrix CW, Bumpus NN. (2013) Compartmentalization and antiviral effect of efavirenz metabolites in blood plasma, seminal plasma, and cerebrospinal fluid. Drug Metab Dispos 41:422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bièche I, Narjoz C, Asselah T, Vacher S, Marcellin P, Lidereau R, Beaune P, de Waziers I. (2007) Reverse transcriptase-PCR quantification of mRNA levels from cytochrome (CYP)1, CYP2 and CYP3 families in 22 different human tissues. Pharmacogenet Genomics 17:731–742 [DOI] [PubMed] [Google Scholar]
- Choudhary D, Jansson I, Schenkman JB, Sarfarazi M, Stoilov I. (2003) Comparative expression profiling of 40 mouse cytochrome P450 genes in embryonic and adult tissues. Arch Biochem Biophys 414:91–100 [DOI] [PubMed] [Google Scholar]
- Cressey TR, Stek A, Capparelli E, Bowonwatanuwong C, Prommas S, Sirivatanapa P, Yuthavisuthi P, Neungton C, Huo Y, Smith E, et al. IMPAACT P1026s Team (2012) Efavirenz pharmacokinetics during the third trimester of pregnancy and postpartum. J Acquir Immune Defic Syndr 59:245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Kaminsky LS. (2003) Human extrahepatic cytochromes P450: function in xenobiotic metabolism and tissue-selective chemical toxicity in the respiratory and gastrointestinal tracts. Annu Rev Pharmacol Toxicol 43:149–173 [DOI] [PubMed] [Google Scholar]
- Guengerich FP. (2008) Cytochrome P450 and chemical toxicology. Chem Res Toxicol 21:70–83 [DOI] [PubMed] [Google Scholar]
- Guengerich FP, Cheng Q. (2011) Orphans in the human cytochrome P450 superfamily: approaches to discovering functions and relevance in pharmacology. Pharmacol Rev 63:684–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange V, Picotti P, Domon B, Aebersold R. (2008) Selected reaction monitoring for quantitative proteomics: a tutorial. Mol Syst Biol 4:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ. (2010) Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26:966–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. (2004) Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics 14:1–18 [DOI] [PubMed] [Google Scholar]
- Ogburn ET, Jones DR, Masters AR, Xu C, Guo Y and Desta Z (2010) Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 2A6 as the principal catalyst of efavirenz 7-hydroxylation. Drug Metab Dispos 38:1218–1229. [DOI] [PMC free article] [PubMed]
- Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. (2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138:795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucy JL, Lasker JM. (1991) Isolation of P450 enzymes from human liver. Methods Enzymol 206:577–587 [DOI] [PubMed] [Google Scholar]
- Renaud HJ, Cui JY, Khan M, Klaassen CD. (2011) Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci 124:261–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BA, Gorski JC, Jones DR, Hall SD, Flockhart DA, Desta Z. (2003) The cytochrome P450 2B6 (CYP2B6) is the main catalyst of efavirenz primary and secondary metabolism: implication for HIV/AIDS therapy and utility of efavirenz as a substrate marker of CYP2B6 catalytic activity. J Pharmacol Exp Ther 306:287–300 [DOI] [PubMed] [Google Scholar]
- Wiśniewski JR, Zougman A, Nagaraj N, Mann M. (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362 [DOI] [PubMed] [Google Scholar]
- Xu M, Ju W, Hao H, Wang G, Li P. (2013) Cytochrome P450 2J2: distribution, function, regulation, genetic polymorphisms and clinical significance. Drug Metab Rev 45:311–352 [DOI] [PubMed] [Google Scholar]
- Zhang QY, Dunbar D, Kaminsky LS. (2003) Characterization of mouse small intestinal cytochrome P450 expression. Drug Metab Dispos 31:1346–1351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.