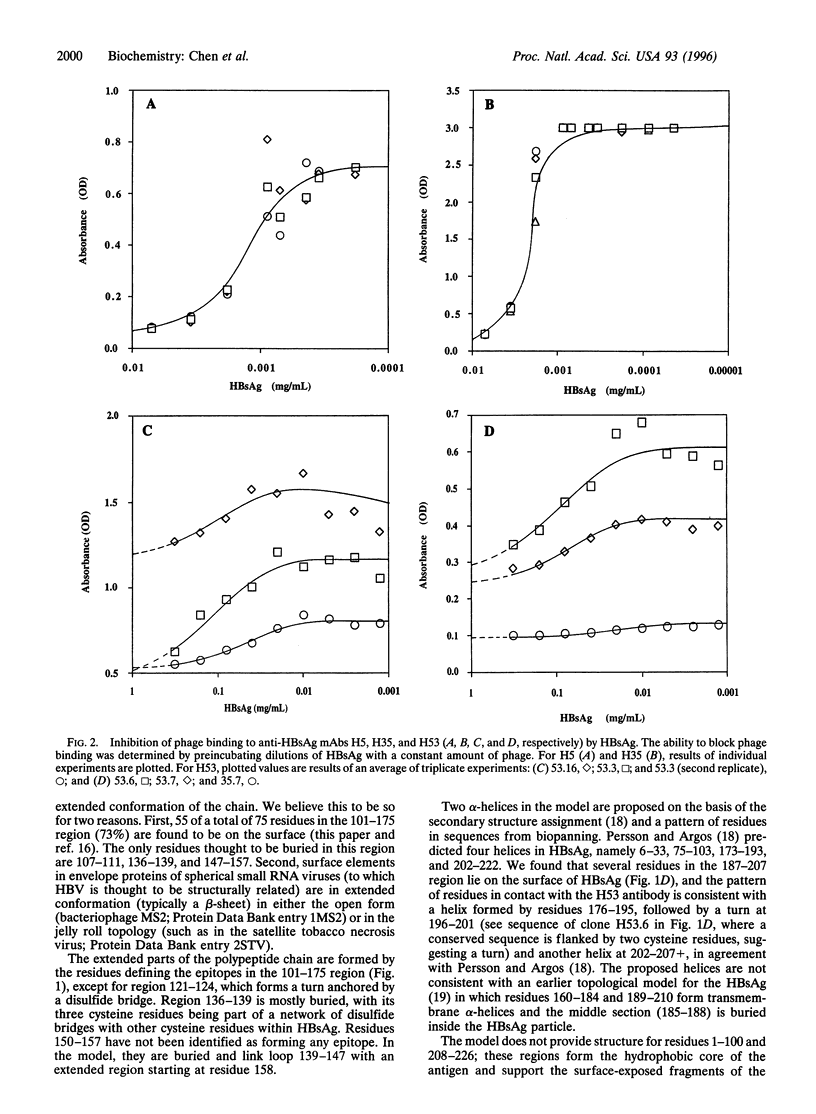
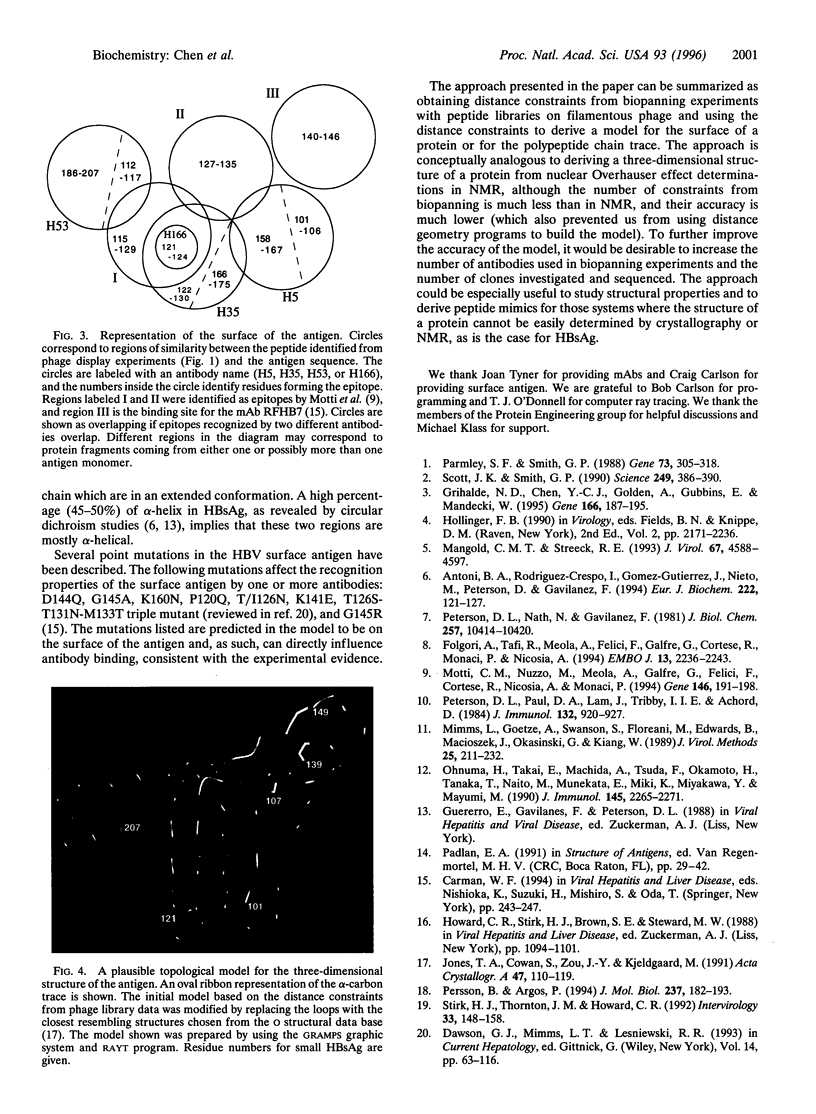
Abstract
The structure of the small hepatitis B virus surface antigen (HBsAg) was investigated by epitope mapping of four anti-HBsAg monoclonal antibodies (mAbs). Amino acid sequences of epitopes were derived from affinity-enrichment experiments (biopanning) using a filamentous phage peptide library. The library consists of 10(9) different clones bearing a 30-residue peptide fused to gene III. Sequence homologies between peptides obtained from panning the library against the antibodies and the native HBsAg sequence allowed for precise description of the binding regions. Three of four mAbs were found to bind to distinct discontinuous epitopes between amino acid residues 101 and 207 of HBsAg. The fourth mAb was demonstrated to bind to residues 121-124. The sequence data are supported by ELISA assays demonstrating the binding of the HBsAg-specific peptides on filamentous phage to mAbs. The sequence data were used to map the surface of HBsAg and to derive a topological model for the alpha-carbon trace of the 101-207 region of HBsAg. The approach should be useful for other proteins for which the crystal structure is not available but a representative set of mAbs can be obtained.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Antoni B. A., Rodríguez-Crespo I., Gómez-Gutiérrez J., Nieto M., Peterson D., Gavilanes F. Site-directed mutagenesis of cysteine residues of hepatitis B surface antigen. Analysis of two single mutants and the double mutant. Eur J Biochem. 1994 May 15;222(1):121–127. doi: 10.1111/j.1432-1033.1994.tb18849.x. [DOI] [PubMed] [Google Scholar]
- Folgori A., Tafi R., Meola A., Felici F., Galfré G., Cortese R., Monaci P., Nicosia A. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994 May 1;13(9):2236–2243. doi: 10.1002/j.1460-2075.1994.tb06501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grihalde N. D., Chen Y. C., Golden A., Gubbins E., Mandecki W. Epitope mapping of anti-HIV and anti-HCV monoclonal antibodies and characterization of epitope mimics using a filamentous phage peptide library. Gene. 1995 Dec 12;166(2):187–195. doi: 10.1016/0378-1119(95)00658-3. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Zou J. Y., Cowan S. W., Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991 Mar 1;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- Mangold C. M., Streeck R. E. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol. 1993 Aug;67(8):4588–4597. doi: 10.1128/jvi.67.8.4588-4597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimms L., Goetze A., Swanson S., Floreani M., Edwards B., Macioszek J., Okasinski G., Kiang W. Second generation assays for the detection of antibody to HBsAg using recombinant DNA-derived HBsAg. J Virol Methods. 1989 Aug;25(2):211–231. doi: 10.1016/0166-0934(89)90034-7. [DOI] [PubMed] [Google Scholar]
- Motti C., Nuzzo M., Meola A., Galfré G., Felici F., Cortese R., Nicosia A., Monaci P. Recognition by human sera and immunogenicity of HBsAg mimotopes selected from an M13 phage display library. Gene. 1994 Sep 2;146(2):191–198. doi: 10.1016/0378-1119(94)90292-5. [DOI] [PubMed] [Google Scholar]
- Ohnuma H., Takai E., Machida A., Tsuda F., Okamoto H., Tanaka T., Naito M., Munekata E., Miki K., Miyakawa Y. Synthetic oligopeptides bearing a common or subtypic determinant of hepatitis B surface antigen. J Immunol. 1990 Oct 1;145(7):2265–2271. [PubMed] [Google Scholar]
- Parmley S. F., Smith G. P. Antibody-selectable filamentous fd phage vectors: affinity purification of target genes. Gene. 1988 Dec 20;73(2):305–318. doi: 10.1016/0378-1119(88)90495-7. [DOI] [PubMed] [Google Scholar]
- Persson B., Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994 Mar 25;237(2):182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Peterson D. L., Nath N., Gavilanes F. Structure of hepatitis B surface antigen. Correlation of subtype with amino acid sequence and location of the carbohydrate moiety. J Biol Chem. 1982 Sep 10;257(17):10414–10420. [PubMed] [Google Scholar]
- Peterson D. L., Paul D. A., Lam J., Tribby I. I., Achord D. T. Antigenic structure of hepatitis B surface antigen: identification of the "d" subtype determinant by chemical modification and use of monoclonal antibodies. J Immunol. 1984 Feb;132(2):920–927. [PubMed] [Google Scholar]
- Scott J. K., Smith G. P. Searching for peptide ligands with an epitope library. Science. 1990 Jul 27;249(4967):386–390. doi: 10.1126/science.1696028. [DOI] [PubMed] [Google Scholar]
- Stirk H. J., Thornton J. M., Howard C. R. A topological model for hepatitis B surface antigen. Intervirology. 1992;33(3):148–158. doi: 10.1159/000150244. [DOI] [PubMed] [Google Scholar]




