Abstract
Anchoring microtubules to the centrosome is critical for cell geometry and polarity, yet the molecular mechanism remains unknown. Here we show that the conserved human Msd1/SSX2IP is required for microtubule anchoring. hMsd1/SSX2IP is delivered to the centrosome in a centriolar satellite-dependent manner and binds the microtubule-nucleator γ-tubulin complex. hMsd1/SSX2IP depletion leads to disorganised interphase microtubules and misoriented mitotic spindles with reduced length and intensity. Furthermore, hMsd1/SSX2IP is essential for ciliogenesis, and during zebrafish embryogenesis, knockdown of its orthologue results in ciliary defects and disturbs left-right asymmetry. We propose that the Msd1 family comprises conserved microtubule-anchoring proteins.
Keywords: centrosome, ciliogenesis, left-right symmetry, microtubule anchoring, spindle orientation
Introduction
The animal centrosome and the fungi spindle pole body (SPB) comprise major microtubule organising centres (MTOCs). The centrosome consists of an orthogonally arranged pair of centrioles and an amorphous electron-dense matrix that surrounds the centrioles, called the pericentriolar material 1. In addition, abundant peripheral foci termed centriolar satellites are present around the centrosome, which are used for dynein-dependent transport of a cohort of centrosomal proteins from the cytoplasm to the centrosome along the microtubule cytoskeleton 2, 3, 4. The yeast SPB, on the other hand, lacks centrioles and comprises laminar configurations on or within the nuclear membrane. The γ-tubulin complex (γ-TuC), which is a universal microtubule nucleator, localises to virtually all forms of the MTOCs 5.
Amongst the multi-layered roles of the centrosome, anchoring of the microtubule minus end is thought to be critical for cell division, cell polarity and the ordered progression of developmental programmes 6, 7. Nonetheless, our knowledge of microtubule anchoring to this organelle remains surprisingly limited. The only molecule to be established as a microtubule-anchoring factor in human cells is Ninein. However, Ninein concentrates at the subdistal appendage of the mother centriole 8, 9, whilst the microtubule-nucleating γ-TuC and several γ-TuC attachment factors are composed of the much broader pericentriolar material 8, 10. Thus Ninein alone does not seem sufficient for physically coupling microtubule nucleation with its anchoring; another anchoring factor(s) may exist.
We previously showed that in fission yeast, a mitotic-specific SPB component, Msd1 (mitotic spindle disanchored 1), is required for anchoring the minus end of spindle microtubules to the SPB 11. In this study, we have identified the human and zebrafish Msd1 orthologues and characterised their roles in microtubule anchoring.
Results and Discussion
Conserved human Msd1/SSX2IP localises to the centrosome and centriolar satellites, and interacts with γ-tubulin and PCM1
Comprehensive homology searches have shown that Msd1 is conserved in many eukaryotes beyond fungi (Fig 1A). All the Msd1 members share three internal coiled-coil domains that exhibit the highest homology (∼30% identity, Fig 1B). Hereafter, we refer to the human orthologue as hMsd1/SSX2IP.
Figure 1.
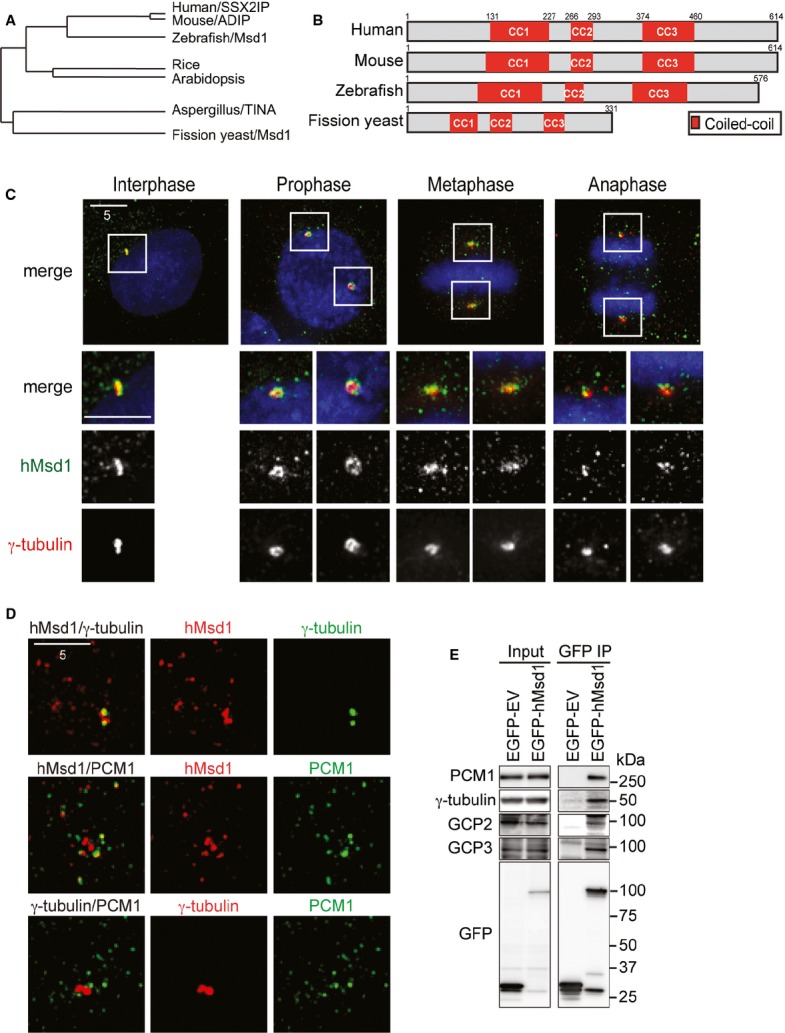
Cellular localisation and protein interactions of conserved hMsd1/SSX2IP.
A, B hMsd1/SSX2IP constitutes a conserved protein family. Phylogenetic dendrogram (A) and a schematic representation of Msd1 members (B) are presented. The most conserved coiled-coil domains are shown in red boxes (CC1, CC2 and CC3).
C, D Cellular localisation of hMsd1 during the cell cycle. HeLa cells were immunostained with indicated antibodies. DNA was stained with DAPI (blue). The bottom three rows in (C) show magnifications of the boxed areas on the top. Scale bars, 5 μm.
E hMsd1 interacts with the γ-tubulin complex and PCM1. Either empty vector or plasmids carrying EGFP-hMsd1 were transfected into HeLa cells. Immunoprecipitation was performed with an anti-GFP antibody. The positions of molecular weight markers (kDa) are shown on the right.
Immunofluorescence microscopy in both HeLa and U2OS cells showed that hMsd1/SSX2IP localised to the centrosome and numerous pericentrosomal foci (Fig 1C and supplementary Fig S1A–C). The appearance of discrete multiple foci around the centrosome is reminiscent of centriolar satellites 3. Indeed, as recently reported 12, these foci (∼70%, n > 500) colocalised with PCM1, the first molecule to be identified as a satellite component 3 (Fig 1D and supplementary Fig S1B and C). Furthermore, the disruption of centriolar satellite structures resulted in the disappearance of hMsd1 foci from the pericentriolar region, but not the centrosome (supplementary Fig S1D–F). hMsd1 coimmunoprecipitated with PCM1 and the γ-tubulin complex (Fig 1E and supplementary Fig S1G and H). These data showed that hMsd1 is a novel component of centriolar satellites and simultaneously localises to the centrosome.
hMsd1/SSX2IP is required for anchoring radial interphase microtubules to the centrosome
To address the functions of hMsd1, we depleted hMsd1 using siRNA oligonucleotides in HeLa and U2OS cells (Fig 2A). In U2OS cells, in which interphase microtubules are mainly composed of radial arrays emanating from the centrosome 7, hMsd1 depletion resulted in marked disorganisation of interphase microtubules with reduced intensities (Fig 2B–D).
Figure 2.
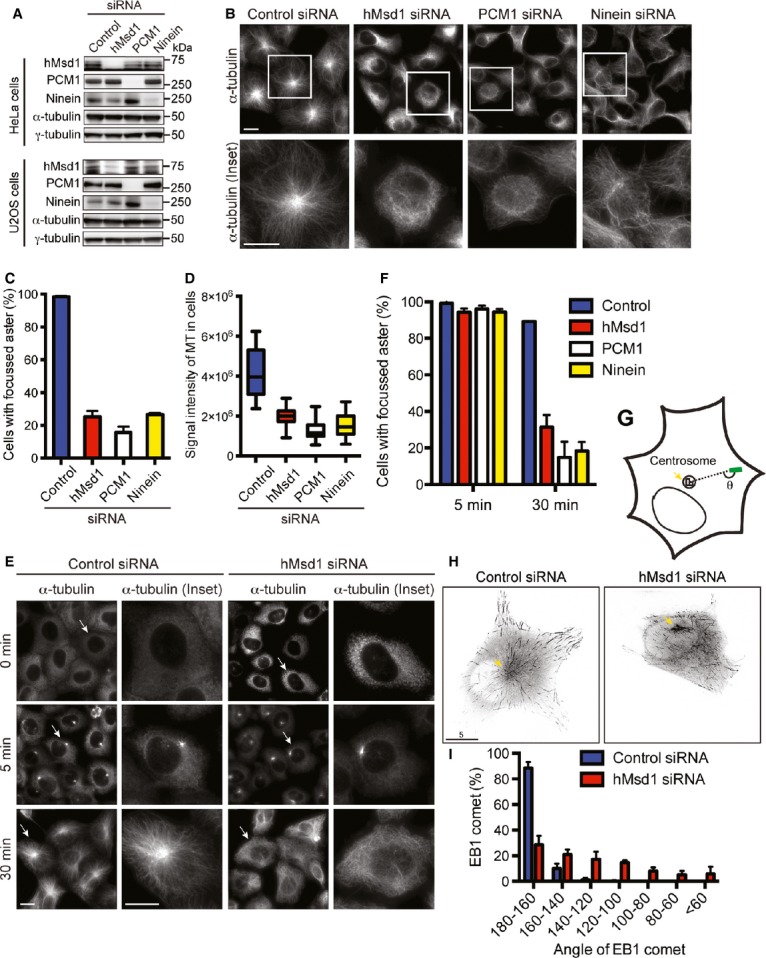
hMsd1/SSX2IP is required for formation of radial interphase microtubule arrays via anchoring microtubule minus ends.
A Evaluation of siRNA-mediated protein depletion.
B–D hMsd1 depletion leads to interphase microtubule disorganisation. U2OS cells were transfected with control or hMsd1 siRNA, and 48 h later were fixed and immunostained with an anti-α-tubulin antibody. Enlarged images are shown on the bottom. The percentage of cells with focussed asters (C) and microtubule intensities (D) were measured.
E, F Microtubule nucleation activities are not defective in hMsd1-depleted cells. Microtubule regrowth assay was performed in control (left) or hMsd1 siRNA cells (right) (E). The percentage of cells with focussed microtubule asters was measured (F).
G–I Measurements of EB1 comet angles. Schematic representation (G), snap shots of EB1-Venus-expressing U2OS cells transfected with control or hMsd1 siRNA (H, arrow points to the centrosome) and quantitative measurements (I) are presented.
Data information: Data represent the mean ± s.d. (C and F, >200 cells, three replicates, n = 3). The box-and-whisker plot indicates the minimum and maximum values, the 25th and 75th percentiles, and the median (D, >200 cells, n = 3). >50 EB1 comets were measured (I, n = 7). Scale bars, 5 μm (B, E, H).
Proper formation of radial microtubule arrays consists of at least two steps: nucleation and anchoring. To precisely determine at which step hMsd1 is involved in radial array formation, we performed a microtubule regrowth assay. hMsd1-depleted cells displayed efficient initial nucleation (Fig 2E and F). In stark contrast, after 30 min of regrowth, whilst control cells formed robust radial microtubule arrays, hMsd1 knockdown cells exhibited disorganised microtubules with no focussed structures. Consistent with normal nucleation activities, microtubule nucleation factors including γ-tubulin, Pericentrin, NEDD1 and CDK5RAP2 5 were retained at the centrosome in hMsd1 knockdown cells (supplementary Fig S2A and B). In line with earlier reports 4, 9, we confirmed that almost identical results were observed in cells depleted for PCM1 or Ninein (Fig 2B–D and F and supplementary Fig S2C). Intriguingly, centrosomal hMsd1 did not colocalise with Ninein and hMsd1 deletion did not lead to reduced levels of Ninein at the centrosome (supplementary Fig S2D and E).
We next measured the angles at which the interphase microtubules lie relative to the centrosome. To this end, we used Venus-tagged EB1, a plus-end tracking protein that forms as a comet-like shape at the growing microtubule tips 13 (θ in Fig 2G). Whilst in most of control cells the EB1 comets displayed θ angles within 20° of this 180° alignment, those in hMsd1-depleted cells showed a much broader range of angles (Fig 2H and I). Taking these results together, we concluded that hMsd1 is not involved in microtubule nucleation, but instead it is a novel microtubule-anchoring factor independent of Ninein recruitment and actively transported via centriolar satellites to the centrosome, where it interacts with the γ-TuC.
Targeting the γ-tubulin binding domain within hMsd1/SSX2IP to the centrosome is enough for its microtubule-anchoring role
We next asked how the hMsd1 protein interacts with the two proteins PCM1 and γ-tubulin, consequently localising to the distinct centrosomal sites (the core centrosome and centriolar satellites). For this purpose, a series of hMsd1 truncation mutants were created by either N- or C-terminal deletions, which were then examined for individual cellular localisation and interactions. Furthermore, in order to assess the functionality of these constructs, all the truncation mutants as well as full-length hMsd1 were created in an siRNA-resistant manner (Fig 3A and supplementary Fig S3A). As expected, siRNA-resistant full-length hMsd1 not only localised to both the centrosome and centriolar satellites, but also interacted with γ-tubulin and PCM1 (Fig 3B and supplementary Fig S3B). It also rescued the microtubule disorganisation phenotype following knockdown of endogenous hMsd1 (Fig 3B and C). On the other hand, the hMsd1 mutant lacking the C-terminal half (hMsd1-N, 1–373 aa) was capable of interacting with PCM1, but not γ-tubulin (Fig 3A and supplementary Fig S3). Consistently, this construct still localised to the centriolar satellites, but not the centrosome (Fig 3B). By contrast, the N-terminally deleted construct (hMsd1-C, 373–614 aa) delocalised from any specific substructures; nonetheless, hMsd1-C was still capable of interacting with γ-tubulin, but not PCM1. Neither the N-terminal or C-terminal half was capable of rescuing microtubule organisation defects (Fig 3B and C).
Figure 3.
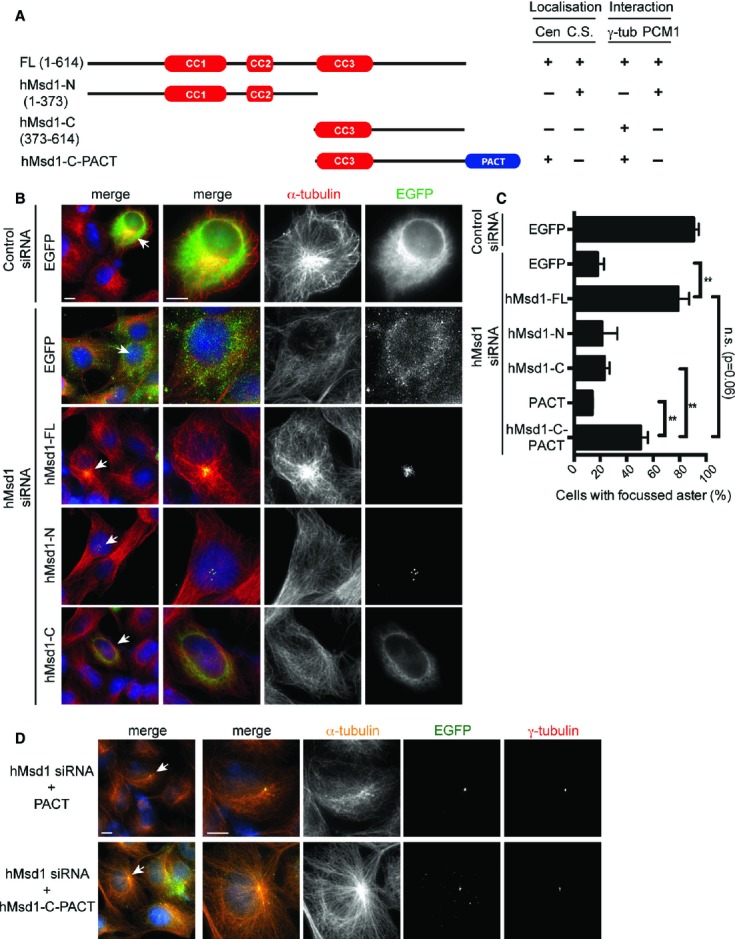
- A schematic presentation of hMsd1 deletion constructs and the summary of their localisation and interaction with γ-tubulin and PCM1. Cen, centrosome; C. S. centriolar satellites.
- Ectopic expression of various hMsd1 truncation mutants. U2OS cells were transfected with control or hMsd1 siRNA, and further transfected with plasmids containing EGFP or individual EGFP-connected, siRNA-resistant hMsd1 constructs. Cells were fixed 24 h after the second transfection and stained with GFP (green) and α-tubulin (red). DNA was stained with DAPI (blue). Enlarged images are shown in the three right-hand side panels corresponding to the cells indicated by arrows on the far left panel. Scale bars, 5 μm.
- Quantification of cells with focused microtubule asters. Data represent the mean ± s.d. (>200 cells, n = 3). Statistical analysis was performed using two-tailed unpaired student's t-tests. **P < 0.001, n.s. not significant.
- Centrosome-targeted C-terminal half of hMsd1 rescues interphase microtubule defects. Transfection experiments similar to (B) were repeated, with the exception that EGFP-tagged PACT or hMsd1-C-PACT were used instead. Scale bars, 5 μm.
We then asked whether the non-functional C-terminal region that displayed mislocalisation would become functional if forced to localise to the centrosome. To address this, we added the centrosomal-targeting motif (the PACT domain) 14 to the hMsd1-C construct (hMsd1-C-PACT). Remarkably, hMsd1-C-PACT rescued microtubule defects in hMsd1-depleted cells as efficiently as full-length hMsd1 (Fig 3C and D). In summary, hMsd1 consists of two structural domains, the N-terminal PCM1-binding and the C-terminal γ-TuC-binding regions. Once at the centrosome, the C-terminal domain is enough to anchor and stabilise the minus end of microtubules.
hMsd1/SSX2IP determines proper spindle orientation parallel to the planar axis
During mitosis, hMsd1 localised to the broad pericentriolar region that encompassed the γ-tubulin signals (supplementary Fig S4A). We then observed spindle organisation in hMsd1-depleted HeLa cells blocked in metaphase. A 3D-reconstruction of spindle microtubules along the z-axis revealed that mitotic spindles were tilted relative to the planar surface (Fig 4A, bottom, side view). Precise measurement of the tilted angles in individual spindles 15 showed that whilst most (88%) of the control cells exhibited angles smaller than 20°, in hMsd1-depleted cells, approximately 60% of spindles displayed angles larger than 20° (θ, Fig 4B and C). Furthermore, we noticed that more than 50% of these mitotic cells showed shorter spindle length (supplementary Fig S4B). The majority of the cells showed a combination of these two spindle defects. In addition, although less abundant, other defects were also observed including misaligned chromosomes, multi-polar or collapsed spindles and centrosome fragmentations (supplementary Fig S4C). Remarkably, similar to interphase microtubule defects (Fig 3D), the C-terminal half of hMsd1 connected to the PACT domain (hMsd1-C-PACT) was capable of supressing spindle orientation defects as efficiently as full-length hMsd1 (Fig 4D and E). These results indicate that hMsd1 function at the centrosome is required not only for anchoring interphase microtubules but also ensuring spindle integrity, in particular for proper spindle orientation and length.
Figure 4.
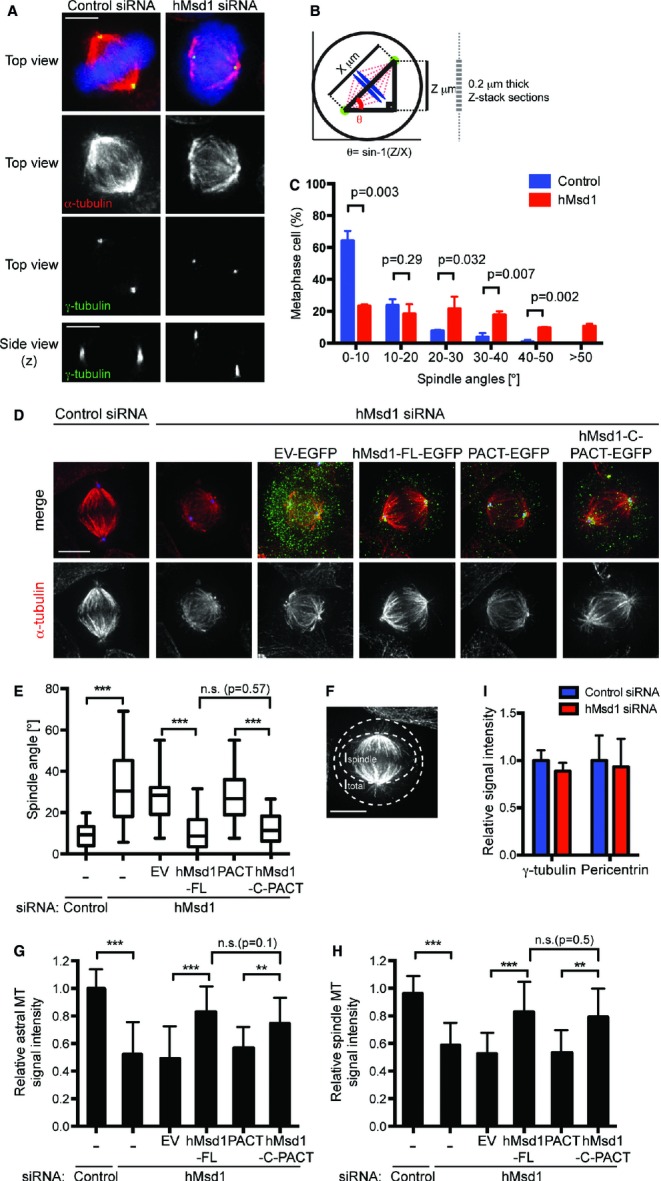
hMsd1/SSX2IP is required for proper spindle orientation by anchoring the astral microtubules to the centrosome.
A Spindle tilt upon hMsd1 depletion. HeLa cells were transfected with control or hMsd1 siRNA, followed by treatment with MG132 (5 μM) for an additional 3 h. Cells were fixed and immunostained with antibodies against α-tubulin (red) and γ-tubulin (green). DNA was stained with DAPI (blue).
B, C Quantification of spindle angles. Detailed procedures are provided in supplementary Data 1.
D, E Centrosome-targeted C-terminal half of hMsd1 rescues spindle tilt and astral microtubule defects. hMsd1 siRNA-treated HeLa cells were transfected with indicated plasmids containing siRNA-resistant hMsd1 constructs. After 24 h, cells were treated with MG132 for 3 h, fixed and immunostained with antibodies against GFP (green), γ-tubulin (blue) and α-tubulin (red). Quantification of spindle angles is shown in (E).
F Schematic overview and quantification of the observed metaphase spindles.
G–I Quantification of signal intensities of astral (G) and spindle microtubules (H). γ-tubulin and Pericentrin signals remain unchanged (I). See supplementary Fig S4F for immunofluorescence images.
Data information: Data represent the mean ± s.d. (C, G-I, >200 cells, n = 3). The box-and-whisker plot indicates the minimum and maximum values, the 25th and 75th percentiles, and the median (E, >200 cells, n = 3). Statistical analysis was performed using two-tailed unpaired student's t-tests. **P < 0.001, ***P < 0.0001, n.s. (not significant) (C, E, G, H). Scale bars, 5 μm (A, D, F).
hMsd1/SSX2IP is required for anchoring mitotic astral microtubules
We next explored why mitotic spindle misorientation was induced by hMsd1 depletion. It is known that for proper spindle orientation, especially an orientation parallel to the substratum, cell adhesion mediated by the interaction between astral microtubules and cortical structures, is critical 15. In fact, spindle orientation defects were also observed in hMsd1-depleted cells grown on a fibronectin-coated coverslip (supplementary Fig S4D). Quantification of signal intensities corresponding to astral microtubules in hMsd1-depleted cells showed a marked reduction of astral microtubules compared with control cells (<50%, Fig 4F and G). In line with this result, intensities as well as the number of EB1 signals were also substantially decreased in hMsd1-depleted cells (supplementary Fig S4E). In addition, intensities of spindle microtubules were also decreased upon hMsd1 depletion (Fig S4H). The centrosome-targeting hMsd1-C-PACT construct was capable of rescuing these spindle defects (Fig. 4G and H).
In sharp contrast to microtubules, the signal intensities of γ-tubulin or Pericentrin were indistinguishable between control and hMsd1-siRNA treated cells (Fig 4I and supplementary Fig S4F). Consistently, upon conducting the microtubule regrowth assay in mitotic cells, we did not see any defects in hMsd1-depleted cells during the initial stage of microtubule nucleation around the centrosome; however after 30 min regrowth, tilted spindle microtubules with compromised astral microtubules were formed (supplementary Fig S4G and H). We therefore envision that the spindle misorientation phenotypes stem from reduced astral microtubules induced by defects in anchoring these microtubules to the centrosome.
hMsd1/SSX2IP is critical for cilia formation
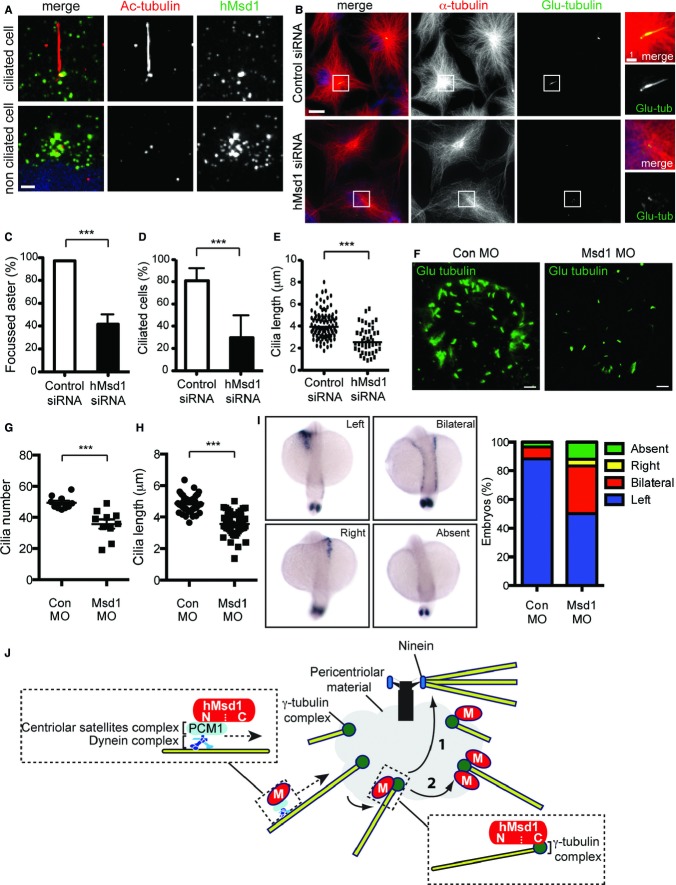
Centriolar satellites play a crucial role in cilia formation and mutations in their components lead to phenotypically related syndromes, which are collectively called ciliopathies 2. To ask whether hMsd1 is required for ciliogenesis, primary cilia were induced by serum starvation in the hTERT-immortalised RPE-1 cells. In ciliated cells hMsd1 localised to the basal body as well as numerous peripheral foci (Fig 5A). In non-ciliated cells, as in HeLa and U2OS cells, this protein localised to both the centrosome and surrounding satellites. Upon hMsd1 depletion, we were able to confirm microtubule disorganisation in hMsd1-depleted RPE-1 cells (Fig 5B and C) and moreover, we found that primary cilia formation was significantly impaired under this condition (Fig 5B and D). In addition, the cilia length was substantially shortened (Fig 5E). Defects in microtubule organisation and ciliogenesis are not attributable to altered centriole number, as no cells displayed more than four centrin-GFP spots (centriole marker, supplementary Fig S5A and B). Centrosome targeting C-terminal hMsd1 (hMsd1-C-PACT) could rescue both microtubule and ciliogenesis defects of hMsd1-depleted cells (supplementary Fig S5C–F). These results indicated that hMsd1 is critical for ciliogenesis, and further suggested that its role in microtubule anchoring to the basal body is required for primary cilia formation.
Figure 5.
Knockdown of Msd1/SSX2IP orthologues impairs cilia formation in human RPE-1 cells and left-right asymmetry during zebrafish embryogenesis.
A hMsd1 localisation in hTERT-RPE-1 cells. Serum-starved (top) or asynchronously growing hTERT-RPE-1 cells (bottom) were fixed and immunostained with indicated antibodies. DNA was stained with DAPI (blue).
B–E hMsd1 depletion leads to microtubule disorganisation and ciliogenesis defects. hTERT-RPE-1 cells transfected with control (top) or hMsd1 siRNA (bottom) were immunostained with indicated antibodies. DNA was stained with DAPI (blue). Enlarged images corresponding to the centrosomal/basal body region (squares, left) are shown in the far right panels. Quantification of the percentage of cells with focussed microtubule asters (C) or cilia (D) and cilia length (E) is shown.
F–H Zebrafish msd1-morphant embryos display cilia formation defects. Kupffer's vesicles were stained with whole-mount immunofluorescence microscopy using antibody against detyrosinated tubulin (Glu-tubulin, green) (F). Quantification of the percentage of ciliated cells (G) and cilia length (H) is shown.
I Defects in left-right asymmetry in msd1-morphant embryos. Four types of spaw expression patterns are presented on the left, and quantification of individual phenotypes is shown on the right. Note that staining in the tailbud was still observed in “Absent”. n = 60 (wild-type); n = 126 (msd1 morpholino).
J Model of microtubule anchoring to the centrosome via hMsd1/SSX2IP. See text for details.
Data information: Data represent the mean ± s.d. (C–E, n = 3, >200 cells; G–I, n = 2, >10 Kupffer's vesicles/embryos). Statistical analysis was performed using two-tailed unpaired student's t-tests. ***P < 0.0001 (C–E, G, H). Scale bars, 10 μm (F), 5 μm (B, left), 1 μm (A,B, right).
Knockdown of the zebrafish Msd1 orthologue causes developmental defects in left-right asymmetry during embryogenesis
Finally, we sought to address the roles of the Msd1 orthologue in the zebrafish embryo. GFP-tagged zebrafish Msd1 (Msd1-GFP) localised to the centrosome in deep cells of the early embryo (supplementary Fig S5G) and to the basal bodies in the Kupffer's vesicles (highly ciliated transient organs, equivalent of mammalian nodes) 16 (supplementary Fig S5H). We next performed msd1 knockdown by injecting morpholino antisense nucleotides. The Kupffer's vesicles contained less cilia (Fig 5F and G) and the length of cilia was also substantially shortened (Fig 5H). We saw no difference in the size of the Kupffer's vesicles upon msd1 knockdown, indicating that zebrafish Msd1 is not required for Kupffer's vesicle formation.
Kupffer's vesicles are essential for the initiation of left-right asymmetry in the zebrafish embryo 16. Markedly, in situ hybridisation against the laterality marker southpaw (spaw) 17 revealed that msd1-morphants exhibited noticeable laterality defects. Whilst approximately 90% of the control embryos showed leftward spaw expression (Left, n = 60, 53/60), msd1-morphants displayed symmetrical expression (Bilateral, n = 126, 42/126), reversed expression (Right, 6/126) or no detectable expression (Absent, 15/126) (Fig 5I). The zebrafish Msd1 orthologue is, therefore, required for the establishment of left-right asymmetry.
hMsd1/SSX2IP is a novel microtubule-anchoring factor
Recently, Barenz et al proposed that hMsd1/SSX2IP is a centrosome maturation factor 12. We did observe similar centrosomal defects such as centrosome fragmentation upon hMsd1/SSX2IP depletion, but its phenotypic appearance is time-dependent; approximately 40% after prolonged siRNA treatment (96 h) compared to approximately 20% under conditions in this study (48 h, supplementary Fig S6). We envisage that compromised centrosome integrity is induced as a secondary phenotype that stems from preceding microtubule-anchoring defects. Alternatively, albeit not mutually exclusive, hMsd1/SSX2IP might be directly involved in centrosome maturation to some extent.
We hypothesise that hMsd1 plays a vital role in microtubule anchoring via either of the following two mechanisms (Fig 5J). In the first scenario (an indirect mediator model), microtubules nucleating from the γ-TuC are delivered to the subdistal appendage of the mother centriole via hMsd1, where the microtubule minus end is captured and tethered by Ninein. In the second scenario (a direct anchor model), hMsd1 tethers microtubules to the pericentriolar material by directly interacting with the γ-TuC. In this case, hMsd1 may spatially and physically link the microtubule minus end to the nucleation machinery. Proper orientation of mitotic spindles is vital for spatial control of cell division and differentiation programmes, in which spindle misorientation promotes tumour formation 18. hMsd1/SSX2IP reportedly accelerates the invasion and metastasis of hepatocellular carcinoma 19, 20. The misregulation of hMsd1 levels and/or activities is expected to lead to ciliopathies and/or cancers, which further investigation will enlighten in the near future.
Materials and Methods
Cell cultures, synchronisation, and reagents
Human cervical cancer HeLa cells and osteosarcoma U2OS cells were cultured in high-glucose DMEM (Invitrogen) supplemented with 10% fetal bovine serum (FBS). Immortalised human pigment epithelial cells hTERT-RPE1 were cultured in DMEM/F12 (Invitrogen) supplemented with 10% FBS and 1% non-essential amino acids. All cells were cultured in a humidified 5% CO2 incubator at 37°C.
RNA interference
Double-stranded siRNA oligonucleotides were synthesised with the sequences 5′-GACAGACAGUUACAAUGUA-3′ (hMsd1 siRNA; Dharmacon), 5′-GGCUUUAACUAAUUAUGGA-3′ (PCM1 siRNA; Dharmacon), or 5′-CGGUACAAUGAGUGUAGAA-3′ (Ninein siRNA; Dharmacon). Control depletion was carried out using siGENOME non-targeting siRNA (Dharmacon).
Immunofluorescence microscopy
Standard procedures for immunofluorescence microscopy were followed (see the supplementary Data 1 for details).
Antibodies
See the supplementary Data 1.
Other experimental details are provided in the supplementary Data 1.
Acknowledgments
We thank Michel Bornens, Andrew Fry, Fanni Gergely, Toshiyuki Habu, Alexey Khodjakov, Tomohiro Matsumoto, Takahiro Matsusaka, Andreas Merdes, Sarah McClelland, Sean Munro, Miho Ohsugi and Richard Vallee for their generous gift of reagents used in this study and useful advice. We are grateful to Val Wood and Penelope Coggill for first letting us know of the existence of the Msd1 family, and to Hisashi Tatebe, Kazuhiro Shiozaki, Aengus Stewart, and Probir Chakravarty for helping perform the phylogenetic analysis. We thank Kathleen Scheffler for her contributions to this work during the initial stage. We are grateful to Risa Mori and Peter Parker for critical reading of the manuscript. A.H. and C.I. were supported by fellowships from the Daiichi-Sankyo Foundation of Life Science and the Uehara Memorial Foundation, respectively. The zebrafish work was supported by the UCL Cell Biology Unit funded by Medical Research Council and Cancer Research UK programme grant (M.T.). T.T. was supported by Cancer Research UK.
Author contributions
The experiments were designed by AH. and TT, AH performed the majority of experiments and data analysis, and AH and CI performed zebrafish experiments under the supervision of MT, AH and TT wrote the paper with suggestions from other authors.
Conflict of interest
The authors declare that they have no conflict of interest.
Supplementary information for this article is available online: http://embor.embopress.org
References
- Nigg EA, Stearns T. The centrosome cycle: centriole biogenesis, duplication and inherent asymmetries. Nat Cell Biol. 2011;13:1154–1160. doi: 10.1038/ncb2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes CA, Prosser SL, Romio L, Hirst RA, O'Callaghan C, Woolf AS, Fry AM. Centriolar satellites are assembly points for proteins implicated in human ciliopathies, including oral-facial-digital syndrome 1. J Cell Sci. 2011;124:600–612. doi: 10.1242/jcs.077156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Sasaki H, Yuba-Kubo A, Tsukita S, Shiina N. Centriolar satellites: molecular characterization, ATP-dependent movement toward centrioles and possible involvement in ciliogenesis. J Cell Biol. 1999;147:969–980. doi: 10.1083/jcb.147.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159:255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat Rev Mol Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–R624. doi: 10.1016/s0960-9822(03)00530-x. [DOI] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113:3013–3023. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nat Cell Biol. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Toya M, Sato M, Haselmann U, Asakawa K, Brunner D, Antony C, Toda T. γ-Tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- Barenz F, Inoue D, Yokoyama H, Tegha-Dunghu J, Freiss S, et al. The centriolar satellite protein SSX2IP promotes centrosome maturation. J Cell Biol. 2013;202:81–95. doi: 10.1083/jcb.201302122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer JS, Salmon ED, Mitchison TJ. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol Biol Cell. 2004;15:1776–1784. doi: 10.1091/mbc.E03-11-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima F, Nishida E. Integrin-mediated adhesion orients the spindle parallel to the substratum in an EB1- and myosin X-dependent manner. EMBO J. 2007;26:1487–1498. doi: 10.1038/sj.emboj.7601599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Neumuller RA, Knoblich JA. Dividing cellular asymmetry: asymmetric cell division and its implications for stem cells and cancer. Genes Dev. 2009;23:2675–2699. doi: 10.1101/gad.1850809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Lin Y, Zhang Y, Zhu Z, Huo K. SSX2IP promotes metastasis and chemotherapeutic resistance of hepatocellular carcinoma. J Transl Med. 2013;11:52. doi: 10.1186/1479-5876-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinn BA, Bullinger L, Thomas NS, Mills KI, Greiner J. SSX2IP expression in acute myeloid leukaemia: an association with mitotic spindle failure in t(8;21), and cell cycle in t(15;17) patients. Br J Haematol. 2008;140:250–251. doi: 10.1111/j.1365-2141.2007.06892.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.