Abstract
The circulatory system is the first hierarchically ordered network to form during the development of vertebrates as it is an indispensable means of adequate oxygen and nutrient delivery to developing organs. During the initial phase of vascular development, endothelial lineage-committed cells differentiate, migrate, and coalesce to form the central large axial vessels and their branches. The subsequent phase of vessel expansion (i.e., angiogenesis) involves a cascade of events including endothelial cell migration, proliferation, formation of an immature capillary structure, recruitment of mural cells and deposition of a basement membrane to yield a functional vasculature. These series of events are tightly regulated by the coordinated expression of several angiogenic, morphogenic and guidance factors. The extracellular matrix (ECM) is synthesized and secreted by embryonic cells at the earliest stages of development and forms a pericellular network of bioactive stimulatory and inhibitory angiogenesis regulatory factors. Here we describe the role of a subset of inducible immediate-early gene-encoded, ECM-associated integrin- and heparin-binding proteins referred to as CCN1 (or Cyr61) and CCN2 (or CTGF) and their function in the development of the vascular system. Gene-targeting experiments in mice have identified CCN1 and CCN2 as critical rate-limiting determinants of endothelial cell differentiation and quiescence, mural cell recruitment and basement membrane formation during embryonic vascular development. Emphasis will be placed on the regulation and function of these molecules and their contextual mode of action during vascular development. Further understanding of the mechanisms of CCN1- and CCN2-mediated blood vessel expansion and remodeling would enhance the prospects that these molecules provide for the development of new treatments for vascular diseases.
Keywords: extracellular matrix, CCN1, CCN2, blood vessels, development, angiogenesis
INTRODUCTION
“CCN” is an acronym that refers to the initials of the first three members of a subset of extracellular matrix (ECM) proteins namely cysteine-rich 61 (Cyr61 or CCN1), connective tissue growth factor (CTGF or CCN2), and nephroblastoma overexpressed (Nov or CCN3) [1]. There are three other members of this family labeled Wnt-inducible secreted proteins (WISP-1 or CCN4; WISP-2 or CCN5; and WISP-3 or CCN6) based on their induction by Wnt ligands. The six members of this family of proteins share structural features but they all are functionally distinct. By far, the first 2 members, CCN1 and CCN2, exhibit more critical functions in vascular tissue development and diseases, and they will be the main focus of this review [2].
CCN1 and CCN2 are non-structural bioactive ECM molecules which bridge the functional divide between structural macromolecules and growth factors, cytokines, proteases, and other related proteins [3]. As such, these molecules have been classified as members of the matricellular protein family which also includes diverse ECM proteins such as thrombospondins, tenascins, osteopentin and osteonectin. Overall, matricellular proteins do not subserve a physical role in the extracellular environment like the collagens, proteoglycans and glycoproteins which essentially provide the mechanical scaffolding within which cells and tissues are built [4]. Instead, they influence the cell fate and function and modulate signals emanating from the extracellular environment. Different functions and roles in vascular development and/or pathology have been attributed to each matricellular protein. Experimental perturbations in in vitro and in vivo systems modeling cardiovascular diseases and cancer have shown that the altered matricellular gene expression affected organ susceptibility/vulnerability to pathogenic factors, which underscored their roles in myocardium and vascular system remodeling, angiogenesis, developmental synaptogenesis, connective tissue organization and dynamics of wound repair [5, 6].
However, CCN1 and CCN2 exhibit several distinctive characteristics. First, while other matricellular proteins are ubiquitously and constitutively expressed, CCN1 and CCN2 proteins are encoded by inducible immediate-early genes whose expression is associated with developmental or pathological events only [3]. Second, most matricellular proteins function as direct modulators of specific ECM fibril systems regulating the organization or higher order assembly of basement membranes and collagen fibrils [7, 8, 9]. Instead, CCN proteins seem to act as adaptors or scaffolds that can bring cytokines and growth factors into close proximity to the cell surface by binding integrins, heparane sulfate proteoglycans (HSPGs) and receptor tyrosine kinases [10, 11, 12]. However, the molecular status of CCN1 and CCN2 in situ is still unknown. Third, the CCN1 and CCN2 genes are essential for viability as either CCN1- or CCN2-deficient mice exhibited severe deficiencies in new blood vessel formation and/or skeletogenesis causing embryonic lethality [13]. Conversely, deficiency in mice of matricellular genes other than CCN1 and CCN2 resulted in superficially mild phenotypes [4]. The lack of lethal phenotypes in those cases is probably a consequence of gene family expansion that led to a balance of functional redundancy and specialization. As extracellular proteins that interact with and regulate the expression of other ECM proteins, proteases and cytokines, CCN1 and CCN2 are necessary for the creation of a balanced ECM environment, the disruption of which can affect fundamental aspects of cell differentiation and tissue growth and development.
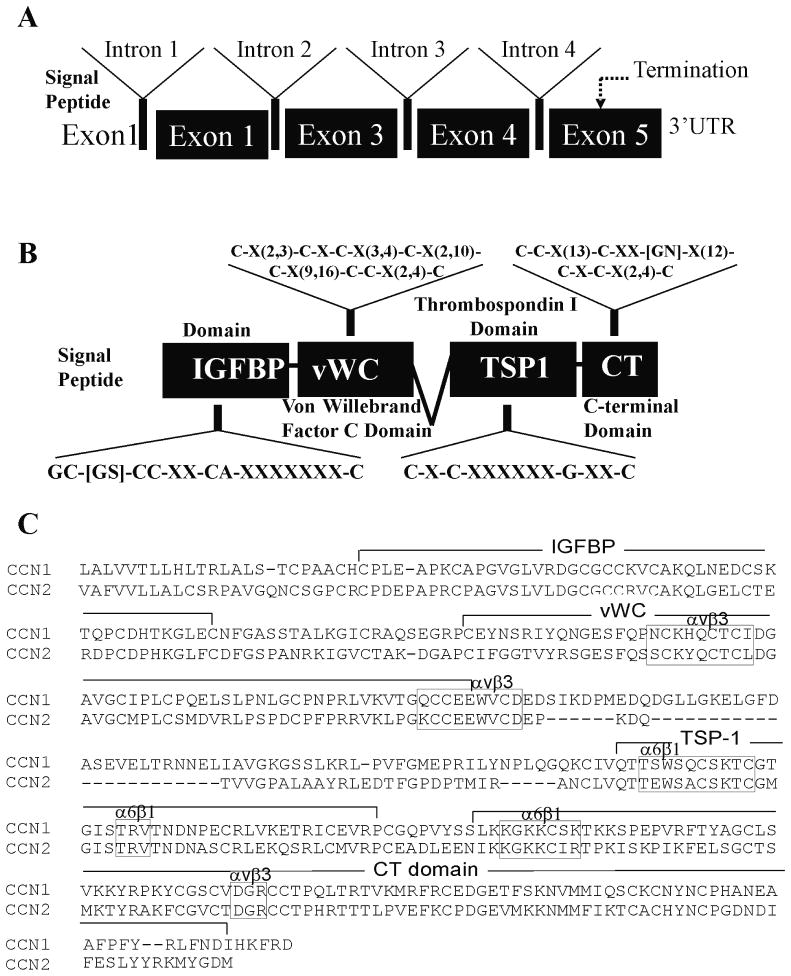
Structurally, CCN1 and CCN2 are organized as multimodular molecules composed of four distinct individual cysteine-rich motifs [14, 15], each of which is encoded by a separate exon (Fig. 1). These multimodlular proteins consist of an N-terminal secretory peptide followed by (1) an insulin-like growth factor-binding protein (IGFBP) homology domain, (2) a von Willebrand factor type C domain (vWC), (3) a thrombospondin type 1 repeat homology domain (TSP1), and (4) a C-terminal domain (CT) containing oligomerization and heparin binding motifs. The primary sequence contains 38 cysteines accounting for 17 disulphide bonds spread throughout the 4 domains. CCN proteins may have been formed during evolution by exon shuffling or other recombination mechanisms from ancestral domains. Each domain consists of a consensus sequence that has evolved independently from the ancestral motif following its transfer gradually changing its structure and acquiring new functions.
Fig. 1.
Gene and modular domain structure of CCN1 and CCN2. A and B: diagrams of CCN1 and CCN2 gene structure and modular domains of the encoded proteins. The consensus motifs found in each modular domain are indicated. C: aligned amino acid sequences of the CCN1 and CCN2 proteins. The conserved consensus sequences are framed within rectangles.
CCN1 and CCN2 gene expression is regulated at the transcriptional, post-transcriptional, translational and post-translational levels in response to mitogenic stimuli such as vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), platelet-derived growth factor (PDGF), transforming growth factor (TGF)-β, sphingosine 1-phosphate, endothelin-1, angiotensin II and lipopolysaccharides as well as physical and environmental inputs such as shear stress, mechanical stretch and oxygen deprivation [16, 3, 17]. The transcriptional requirements of the CCN1 and CCN2 genes do not involve new protein synthesis but only transcriptional activators with a PDZ-binding motif such as Yap1 and serum response factor-CarG box tertiary complexes [18]. This is consistent with the immediate early-gene expression pattern of these genes [18, 19]. Conversely, CCN1 expression has been found to be downregulated during tissue involution, in avascular tissues and under conditions associated with vasoobliteration which is consistent with a potential role of this protein in vessel formation, stabilization and integrity [20, 21]. The transcription factor, FOXO3a, is a negative transcriptional regulator of the CCN1 gene and the suppression of CCN1 is among several mechanisms by which FOXO3a inhibits vascular smooth muscle cell proliferation [22].
Like CCN1, the CCN2 gene is transcriptionally activated in vascular cells in response to mitogenic and environmental stimuli [17, 23]. Under hypoxic conditions, the expression of the CCN2 gene is induced both transcriptionally through hypoxia-inducible factor (HIF)-1α activation of the CCN2 promoter and post-transcriptionally through enhanced mRNA stability through an AUA-rich 3′UTR sequence [24, 25]. CCN2 is also a predicted and validated target of the miR-17-92 microRNA cluster which regulates tumor neovascularization as well [26]. However, the half life of the CCN2 protein is relatively low (half of that of CCN1) because its affinity for heparin is much lower than that of CCN1, significantly reducing its interactivity with the ECM proteins [27].
Functionally, the CCN1 protein regulates many cellular activities such as cell adhesion, migration, proliferation, differentiation, survival, senescence and apoptosis while the primary biological roles of CCN2 appear to be in the formation of connective tissues during embryonic development and their maintenance and homeostasis in the adult, and in coordinating tissue repair following injury [28, 29, 30]. As such, both proteins have emerged as critical regulators of developmental vessel formation as demonstrated by the lethal angiogenic and vascular deficiencies upon their targeted deletions in mice [31]. We will use the implications of CCN1 and CCN2 knockout mice as a springboard to analyze the role of these molecules in vascular development.
Formation and developmental control of blood vessels
Formation of the earliest primitive vascular tubes is achieved by in situ differentiation of mesenchymal cell-derived haemangiogenic stem cells that further differentiate into angioblasts, the progenitors of endothelial cells (ECs) and to haemangioblasts, the progenitors of haematopoietic cells [32, 33]. Upon their differentiation, endothelial precursors coalesce into cords and then form a lumen. This vasculogenic process is the major mechanism of initial formation of blood island vessels, dorsal aorta, endocardium, and vitelline vessels in the embryo [34]. On the other hand, the predominant means of vessel expansion in all organ systems is angiogenesis, which is based on endothelial sprouting or intussusceptive microvascular growth [35, 36]. The formation of the vasculature by vasculogenesis or angiogenesis depends on the proportions of the angioblast progenitors in each developing organ. However, both processes are not mutually exclusive inasmuch as angioblasts can be incorporated into expanding pre-existing blood vessels [37, 38]. During vascular repair, circulating endothelial precursors populate a growing vascular network from within blood vessels [37].
The first vascular structures (i.e., endothelial cells) appear in the extraembryonic yolk sac at the gastrulation stage as blood islands/angiogenic clusters lined by ECs and perfused by early erythrocytes [39]. An immature vascular network then forms by fusion of blood islands and a subsequent vascular remodeling leads to the formation of the complex yolk sac vasculature. The intraembryonic vasculature proper forms further as angioblasts differentiate into ECs followed by an active angiogenic sprouting of blood vessels [40, 41]. As the embryonic blood vessels become established and various tissues differentiate, ECs continue to undergo numerous changes, generating functionally distinct, tissue specific vascular beds including the complex vascular structures of the dorsal aorta and vitelline vessels, and primary plexuses of lungs, spleen, and heart regions. The dorsal aorta develops at the same time as the early cardiac structure and produces branches that supply blood to the rest of the embryo. For venous circulation, three paired veins form to drain into the sinus venosus which later becomes incorporated into the right atrium [42]. These include (1) the vitelline veins which return poorly oxygenated blood from the yolk sac, (2) the umbilical vein which carries well oxygenated blood from the primordial placenta and (3) the common cardinal veins which return poorly oxygenated blood from the embryo’s body.
The newly formed embryonic vessels further specialize into arteries, veins, and capillaries, which have distinct characteristics based on the presence and amount of smooth muscle cells and specific ECM. Capillaries, the business end of the cardiorespiratory system, are the microvessels responsible for most of the peripheral exchange. These structures consist of a single layer of ECs surrounded by a basement membrane within which mural cells (e.g., pericytes) are embedded. The appropriate guidance of vessels and development of tissue-specific differences is critical for proper tissue function. Specific sprout guidance factors are spacially organized within certain tissues (e.g., nervous system including retina) and yield highly stereotypical blood vessel networks. In other tissues (e.g., mouse yolk sac), the absence of guidance cues leads to a freely branched vessel network [43]. The loss of these regulatory factors often leads to vascular lethal defects during fetal development or an abnormal neovascularization and a variety of pathological conditions in the adult [44, 45]. Many excellent reviews cover vascular development more globally and the mechanisms involved and the readers are referred to them for more detailed information [46, 47, 48].
There is some information on the cellular and molecular processes that control where and when blood vessels form. The primary stimulus for angiogenesis is often a mismatch between the growing metabolic demand of the developing tissue and substrate delivery [49, 50]. A complex network of cytokines of the VEGF, FGF, PDGF, TGF-β, insulin-like growth factor (IGF) and angiopoietin systems has been identified as the ultimate responsive factors that palliate the metabolic and oxygen demands in tissues [51, 52, 53]. In avian models for instance, FGF is involved in the induction of angioblasts while VEGF supports the assembly and patterning of vessels [54, 55]. The relevant VEGF receptors, the tyrosine kinases VEGFR2 and VEGFR3, are activated on EC membrane surface upon ligand binding, which triggers downstream signaling including activation of the mitogen-activated kinase (MAPK) pathway, phosphoinositide kinase-3 (PI3K), Akt, phospholipase C and small GTPases such as Rac1 [56, 57]. How these signals or their selective activation translates into different biological responses as diverse as EC differentiation or proliferation, or angiogenesis is not fully understood. The Notch pathway, which is well known for its roles in cell fate determination and differentiation processes, is upregulated by VEGF and is responsible for EC specification in sprouting vessels [58, 59]. Sprouts are headed by migrating endothelial tip cells, which dynamically extend long filopodial protrusions in a polarized way, migrate into the ECM, and sense their environment for attractive and repulsive signals for guidance. In the embryonic spinal cord and the retina, the heparane sulfate anchored isoform of VEGF-A (VEGF164 in mice, VEGF165 in humans) promotes the polarization of tip cells and the directional extension of filopodia [60, 61]. The Delta-like 4 (Dll4)/Notch pathway establishes an adequate ratio between stalk and tip cell populations by restricting tip cell formation through “lateral inhibition” in response to a VEGF gradient. Thus, specificity may be provided by the spaciotemporal expression patterns of VEGF and its receptors.
Following this angiogenic remodeling, the endothelium secretes PDGF, which induces the recruitment and differentiation of vascular smooth muscle cells [62, 63]. Subsequently, the vascular smooth muscle cells secrete angiopoietins, which ensure proper interaction between endothelial and mural cells [64, 65]. Finally, the vascular smooth muscle cells deposit matrix proteins, such as elastin, that inhibit vascular smooth muscle cell proliferation thereby stabilizing the mature vessel [66]. Modulation of the activity of these and other molecular intermediates of vessel formation may represent one of the underlying molecular mechanism(s) leading to the CCN protein-dependent regulation of vascular development.
Expression and molecular control of vascular development by CCN1
CCN1 is most highly and dynamically expressed in the second and third trimesters of pregnancy, especially in non-proliferating interstitial extravillous trophoblastic giant cells, vascular ECs, and mesenchymal and stromal cells of the placental villi [67]. CCN1 was found to be continuously expressed during the formation of and in the fully developed placenta in 12.5-day post coitum mouse embryos particularly in angiogenic cell types such as the giant trophoblasts and the overlying compact deciduas [68]. These angiogenic structures produce the essential factors (e.g., VEGF, basic (b) FGF and PDGF) required for maternal vessel growth towards the developing embryo [69]. Meanwhile, a prominent cardiac expression of CCN1 was seen as early as E8.5 in mice expressing lacZ under the control of the endogenous CCN1 promoter [70]. CCN1 expression continued at E10.5 especially in the trunk arteriosus, which later divides to form the aorta and pulmonary trunks [71]. CCN1 expression was also found in all major arteries branching from the heart of the developing fetal circulatory system. These include the aortic and pulmonary trunks which are branches of the aortic arches that are of neural crest origin and in the dorsal aorta and umbilical artery which are of mesodermal origin. Mesodermal cells ultimately differentiate into smooth muscle. Of interest is that structures originating from the neural crest also give rise to cartilaginous tissue; therefore, all regions of mouse embryo involving cartilage formation expressed CCN1 as well [72]. CCN1 was shown to function as a soluble chemotactic signal in human mesenchymal stem cells and induce a site-directed cell recruitment in vivo [20, 73]. Whether CCN1 affects the linear commitment of mesenchymal stem cells and/or regulates cell and/or further cell differentiation is yet to be determined.
Our group has examined the vascular localization of CCN1 during development of the retinal vasculature, as the morphology of the primitive plexus at the edge of the developing retinal vasculature is highly similar to some of the first blood vessels in early vertebrate embryos [13]. In the mouse, the retina is avascular at birth and vessels grow concentrically from the center toward the periphery following an existing network of astrocytes [74, 75, 76]. Once a primary plexus is established, sprouts dive vertically deep into the retina and form intermediary and deeper vascular layers. Using transgenic mice expressing the green fluorescent protein (GFP) gene under the control of the CCN1 promoter, we found that CCN1 was dynamically expressed postnatally in the mouse eye and that its expression was transient and largely confined to the advancing vascular front of the developing vasculature. CCN1 appeared to be dynamically expressed in both endothelial tip and stalk cells suggesting its potential role in the phenotypic plasticity of ECs. Interestingly, CCN1 expression is also associated with hyaloid vessels as well, especially in macrophages and shifts to the retinal vasculature emerging from the optic disc [13]. Hyaloid vessels form a complex of intraocular vessels that penetrate the retina at the optic disc and branch anteriorly through the vitreous to the lens. Hyaloid vessels then regress progressively by apoptosis as the retinal vasculature develops by angiogenesis in a synchronized manner suggesting that overlapping CCN1 signals may control both processes simultaneously.
The generation of CCN1 mutant mice provided a unique animal model for addressing the role of CCN1 in vascular development. A large majority of CCN1 null embryos died of hemorrhage and/or placental defects between E11.5 and E14.5 while a fraction of embryos died earlier as a result of defects in chorioallantoic fusion [77]. Both cardiac and vascular defects were the most prominent alterations observed. They seemed to originate at the chorioallantoic junction, where the allantois failed to fuse with the placenta, resulting in severe undervascularization in the placental labyrinth or from defects of large vessel bifurcation resulting in a poor vascular coverage of the chorionic plate. Large fetal vessels such as the aorta appeared dilated as in large aneurysm. Vascular cells such as ECs and smooth muscle cells were not confined within their layers as ECs were mislocalized in the media layer of the aorta. Subsequently, CCN1-deficient embryos suffer from compromised integrity and hemorrhage of arterial vessels in which the most expression of CCN1 occurs during development.
Meanwhile, severe atrioventricular septal defects (AVSD) have also been observed in CCN1-deficient embryos [71]. AVSDs are characterized by the complete or partial absence of partitioning of the atrioventricular valve. These are common genetic disorders that cause congenital heart disease in humans.
Regulation of vascular cell function and behavior by CCN1
Once secreted, the CCN1 protein regulates many aspects of vascular cell function including adhesion, migration, proliferation, differentiation and survival [10, 78]. Many of these activities derive from CCN1 interaction with integrin receptors localized in the vWC domain which contains binding sites bearing structural homologies to those involved in the interaction with αvβ3, αvβ5 and α2β3 integrins [78, 79, 80, 81]. CCN1 signaling requires HSPG as a co-receptor and involves either interaction with a single integrin type or the concerted binding to several integrins [82]. The interaction between αvβ3 integrins and the vWC domain was shown to be largely responsible for CCN1-mediated endothelial cell adhesion, migration and survival while CCN1 binding to integrin α6β1 mediates capillary-like tubule formation of non-activated ECs [83, 84]. Interestingly, CCN1-deficient mutants phenocopied chorioallantoic and vascular defects previously reported in α4 and αv intergin-deficient mice respectively, which underscores the importance of CCN1-integrin signaling [85]. Defective signaling through CCN1-integrin interaction may, at least in part, account for the chorioallantoic and vascular defects associated with CCN1 deletion in mice.
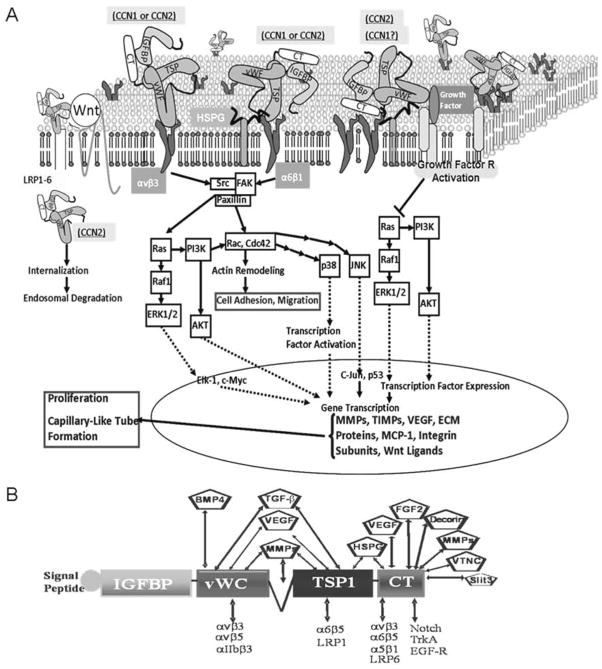
Fig. 2 provides a simplified pathway map of CCN1- (and CCN2)-dependent intracellular signaling and their putative interacting partners. CCN1-integrin interaction culminates into the activation of small GTPases such as RhoA GTPases, Rac and Cdc42, which leads to cytoskeletal actin reorganization, formation of lamelilpodia/filipodia and cell migration [86, 87]. CCN1 also activates the Ras pathway axis through signaling molecules in focal complexes, MAPK (ERK1/2), and/or Akt/PKB signaling [88, 28], and enhances the expression and/or stability of key regulatory proteins that promote cell cycle progression, adhesion and capillary-like tube formation [89]. A broad spectrum of genes that are responsive to EC stimulation by CCN1 includes but is not limited to matrix metalloproteinases (MMPs), tissue inhibitor of metalloproteinases (TIMPs), integrins and VEGF [90, 91, 92]. In particular, VEGF-C, which is predominantly expressed by embryonic endothelium and macrophages, was downregulated in CCN1-null mouse embryo [77]. The spatial distribution and temporal qualities of the dynamics of CCN1 targets vary with the cell context, enabling the cell to program an appropriate biological response. The adhesive and chemotactic activities of CCN1 on endothelial and smooth-muscle cells may underlie the loss of vessel integrity upon CCN1 gene deletion in mice.
Fig. 2.
A: Simplified pathway map of CCN1 and CCN2-dependent outside-in signaling pathways regulating endothelial cell function and behavior. B: Interaction of CCN1 and CCN2 proteins via their constituent domains with extracellular ligands and transmembrane proteins.
Not only does CCN1 initiate its own intracellular signaling, it also is able to modulate the activity of other unrelated molecules (e.g., growth factors, cytokines, ECM proteins) by potentiating their crosstalk with cell membrane and growth factor receptors (Fig. 2B). For instance, in Xenopus embryos, CCN1 inhibits bone morphogenic protein (BMP) signaling through interference with BMP interaction with its receptor [93, 94]. CCN1 may constitute one of the extracellular fine-tuning modulators of BMP signaling during vascular development essentially by shaping gradients of BMP activity in areas of uniform ligand expression.
CCN1 also directly interacts via its CT domain with fibronectin and vitronectin and facilitates the activation of αv integrins by these proteins [95, 96, 93]. CCN1 physically binds to the somatomedin B domain of vitronectin which is also the functional region that mediates interactions with plasminogen activator inhibitor-1, uPAR, and integrins αvβ3/αvβ5. Such interaction may affect the angiogenic tonus conveyed by those molecules. Along these lines, binding of CCN1 to ECM proteins displaces bFGF from the matrix and increases both its availability to interact with its receptor on ECs and its angiogenic properties [79]. Similarly, VEGF has been shown to bind to fibronectin via its fibronectin-like domain and promotes EC growth and migration [97]. As the vWC domain of CCN1 bears similarities with fibronectin-like domain, and may have evolutionarily preceded it (i.e., the latter has evolved as a truncation of the vWC domain), raises the possibility that functional similarities may, at least in part, exist between these two types of modules. Therefore, the angiogenic activity of CCN1 may depend on the interaction with and/or modulation of VEGF activity. Our group has shown that both the vWC and TSP1 domains of CCN1 containing stretches of amino acid residues involved in integrin and growth factors binding form a synergy region located on the same face of the molecule [98]. This suggests that the simultaneous interaction of growth factors and integrins with this region may be sterically hindered and thus, mutually exclusive.
Expression and molecular control of vascular development by CCN2
Since its initial discovery, CCN2 has been considered an important determinant factor of extracellular matrix synthesis and remodeling controlling the formation of connective tissue-rich compartments [99, 100, 29]. However, direct tissue-specific overexpression of the CCN2 transgene in fibrosis-prone organs (e.g., heart, liver, kidney) failed to induce any fibrotic reaction in the target tissues [101, 102, 103]. Interestingly, lung-specific CCN2 transgene overexpression showed evidence of an impaired formation of the alveolar vascular network characterized by fibrosis in and around blood vessels, alveolar septa and bronchioles [104]. This clearly suggests that CCN2 affects blood vessel formation but only at specific stages of their development.
During mouse embryogenesis, CCN2 is widely but dynamically expressed especially within embryonic tissues that give rise to the cardiovascular, musculoskeletal and nervous systems [105]. Precisely, CCN2 was detected as early as E7.5 in the parietal endoderm and crypt cells. It further continued in the notochord, cephalic mesenchymal cells, first bronchial pouch and in the roof and floor palate of the neural tube. At E.8, CCN2 expression was prominent in the heart myocardium and in the common cardinal vein, bronchial arch arteries, and the dorsal aorta. Later during fetal development, CCN2 levels increased in all large blood vessels including the aorta, pulmonary artery, and particularly in the ductus arteriosus and pulmonary trunks. CCN2 localized at the abluminal surface of the endothelium and in mural cells in blood vessels and capillaries. Although CCN2 expression decreased postnatally, it remained detectable in all large blood vessels. The persistence of basal CCN2 gene expression during adulthood suggests that CCN2 may be involved in vessel stability and/or maintenance.
CCN2 localized pericellularly in arteries, veins and capillaries. ECs and pericytes were the major sources of CCN2. Interestingly, CCN2 is also highly expressed in skeletal tissues during endochondral ossification primarily in the cartilage, within maturing chondrocytes and in the developing ribs and long bones [106]. CCN2-null mice die shortly after birth from respiratory failure due to disruption of basic lung development (i.e., lungs were hypoplastic with reduced cell proliferation and increased apoptosis) and failed thoracic expansion [107]. The mutant phenotype essentially revealed a role for CCN2 in growth plate angiogenesis [108]. Hypertrophic zones of CCN2 mutant growth plates were expanded, and endochondral ossification was impaired. A dramatic decrease of the capillary network in the growth plate was evident in CCN2 mutants which accounted for defective replacement of cartilage by bone during endochondral ossification. No obvious vascular defects during the initial formation of the large vasculature from E9.5 to E13.5 were observed probably due to compensatory pathways mediated by the related CCN1 protein [109]. However, after E14.5, the large vessels in CCN2-null embryos were enlarged with localized edema. There was no deficiency in mural cell recruitment to these vessels but the cells in the media layer appeared heterogeneous in size and dysmorphic. Conversely, poor coverage of microvessels by pericytes was evident in CCN2 mutant mice. Surprisingly, arterial-venous identity appeared to be maintained in CCN2 mutants suggesting that CCN2 has no effect on arteriovenous specification markers.
Regulation of vascular cell function and behavior by CCN2
CCN2 was characterized as a growth factor inducible, immediate early gene-encoded matricellular protein sharing the multimodular structure of CCN1 and a 52% identity with its primary sequence [15]. The CCN2 protein elicits a strong angiogenic response comparable to that of either CCN1 or bFGF when tested in the chorioallantoic membrane assay of chick embryos or when injected subcutaneously in mice [110, 27, 111]. Exogenously added CCN2 to cultured ECs effectively activated mitogenic signaling cascades (e.g., ERK1/2, p38 MAPK) and promoted cell growth, clearly indicating that CCN2 has a strong intrinsic angiogenic activity [112].
Like CCN1, the function of CCN2 is contextual and depends on the availability of membrane receptors and/or interacting partners in the cellular microenvironment (Fig. 2). Through direct binding to integrin αvβ3, CCN2 promotes endothelial cell adhesion, migration, proliferation, and tubule formation, thus, recapitulating typical angiogenic events [80, 113, 114]. CCN2 also promotes survival of ECs deprived of growth factors, a condition that otherwise induces apoptosis [83]. CCN2-integrin interaction activates FAK and other intracellular signaling molecules such as MAPK, PI 3-kinase and Rho GTPases [115, 116, 81, 117]. The latter signaling pathway culminates into changes in cytoskeletal actin organization, filopodia and lamellipodia formation and ECM protein deposition (e.g., fibronectin, fibrillar collagens, proteoglycans). CCN2 binds to a variety of other integrins expressed by different cells, e.g., αIIbβ3 on activated platelets [118], α6β1 on skin fibroblasts [116] and αMβ2 in blood moocytes [114]. In fibroblasts, CCN2 binds to integrins α5β1 and α4β1, and syndecan 4 [116]. Such interaction is critical for cell spreading and adhesion. CCN2 also interacts with the low-density lipoprotein receptor proteins (LRP) although such interaction was shown to only lead to CCN2 internalization and degradation in the endosomes [119].
CCN2 signaling was shown to modulate growth factor and ECM protein gene expression as well [120, 121, 122]. CCN2 induced PDGF-B expression in ECs, and potentiated PDGF-B-mediated Akt signaling in mural (e.g., vascular smooth muscle/pericytes) cells [123, 124]. PDGF signaling and the establishment of the endothelial basement membrane are required for pericyte recruitment and retention [125, 126]. These processes appeared to be defective in CCN2-deficient mice [109]. In addition, CCN2 induced the production of endothelial basement membrane components in vitro, and was required for their expression in vivo [106, 127]. Blood vessel leakage and altered basement membrane integrity were attributed to CCN2 deletion in vivo while basement membrane thickening was linked to excessive production of CCN2 in glomerular capillaries of transgenic animal models [108, 128]. Thus, CCN2 is required for the normal production of the vascular basement membrane.
In addition to the aforementioned interactions, CCN2 may exert its effects through direct physical interaction via one of its domains with bioactive molecules and/or one or more cell surface tyrosine kinase receptors. In particular, Inoki et al. have shown that CCN2 directly interacts with VEGF164 both in vitro and in vivo via its modular domain, TSP1, which is highly conserved between CCN1 and CCN2 [129]. Such interaction hinders VEGF164 binding to its receptor (VEGFR2) and reduced VEGF-mediated angiogenesis. Proteolysis of the complex CCN2-VEGF by MMPs released the bound VEGF in an active form and enhanced its angiogenic potential [113, 130]. Thus, CCN2 may act by fine-tuning the bioavailability of VEGF, releasing it for angiogenic action only in contexts where MMPs are being secreted and activated. Meanwhile, a recent study has shown that CCN2 physically interacts with the epidermal growth factor receptor (EGFR) and tropomyosin receptor kinase A (TrkA) via its carboxy terminal domain leading to MAPK activation and proinflammatory factor gene upregulation [131]. CCN2 also has broad affinity to a series of other angiogenic regulators with common CT motifs including Slit3, von Willebrand factor, PDGF-B and FGF [132, 133]. Mutation of the CT domain of CCN2 abolished its interactivity with bioactive molecules with common CT motifs and reduced their proangiogenic properties in vitro. However, whether these in vitro findings have physiological significance is still unknown and warrant further investigations.
CONCLUSION
Overall, CCN1 and CCN2 share many similarities in their activities and patterns of expression, and exhibit overlapping and non overlapping functions. With so many potential modes of action and interacting partners it is conceivable that CCN1 and CCN2 influence different vascular developmental events. Given their context- and cell type-dependent biological activities that vary based on the presence of specific receptors and interacting partners in the cell microenvironment, the function of these proteins may vary under physiological and disease conditions.
Despite significant advances in recent years, many questions concerning the mechanisms whereby CCN1 and CCN2 regulate vascular development remain to be addressed. Vascular growth and morphogenesis are initiated, at least in part, by VEGF-derived signals and ECM-integrin-cytoskeletal signaling axis which causes simultaneous sprouting (i.e., cord formation) and lumen formation events that eventually lead to interconnecting networks of EC-lined tubes [134]. These and other processes appeared to be defective in CCN1- and/or CCN2-deficient mouse embryo despite the presence of VEGF. In both normal and pathological angiogenesis, sprouts are headed by migrating endothelial tip cells, which dynamically extend long filopodial protrusions in a polarized way, migrate into the ECM, and sense their environment for attractive and repulsive signals for guidance [135]. Therefore, are CCN1 and CCN2-derived signals important for the control of endothelial tip and stalk cell specification and endothelial tip cell protrusive activity? As Notch and Wnt signaling pathways control multiple aspects of endothelial cell biology, are CCN1 and CCN2 important in engaging these signaling pathways at various stages of vascular development? Are they indispensable in rearrangement and migration of ECs and mural cells to ensure a coordinated outgrowth of vascular sprouts while maintaining integrity of nascent vessels? How do CCN1 and CCN2 help tissues cope with or succumb to hypoxic stress? A deeper understanding of these fundamental principles will undoubtedly aid in the development of new treatments for blood vessel-related pathologies.
Acknowledgments
I thank Lulu Yan and Jinog Choi for their technical contribution and helpful comments. This work was supported by a grant from the National Eye Institute of the National Institutes of Health (EY022091-01).
Footnotes
CONFLICT OF INTEREST STATEMENT
The author has no conflict of interest to disclose.
References
- 1.Brigstock DR, Goldschmeding R, Katsube KI, Lam SC, Lau LF, Lyons K, Naus C, Perbal B, Riser B, Takigawa M, Yeger H. Mol Pathol. 2003;56:127–128. doi: 10.1136/mp.56.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leask A, Abraham DJ. J Cell Sci. 2006;119:4803–4810. doi: 10.1242/jcs.03270. [DOI] [PubMed] [Google Scholar]
- 3.Chaqour B, Goppelt-Struebe M. FEBS J. 2006;273:3639–3649. doi: 10.1111/j.1742-4658.2006.05360.x. [DOI] [PubMed] [Google Scholar]
- 4.Bornstein P, Sage EH. Curr Opin Cell Biol. 2002;14:608–616. doi: 10.1016/s0955-0674(02)00361-7. [DOI] [PubMed] [Google Scholar]
- 5.Adams JC, Lawler J. Int J Biochem Cell Biol. 2004;36:961–968. doi: 10.1016/j.biocel.2004.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kyriakides TR, Bornstein P. Thromb Haemost. 2003;90:986–992. doi: 10.1160/TH03-06-0399. [DOI] [PubMed] [Google Scholar]
- 7.Bornstein P, Kyriakides TR, Yang Z, Armstrong LC, Birk DE. J Investig Dermatol Symp Proc. 2000;5:61–66. doi: 10.1046/j.1087-0024.2000.00005.x. [DOI] [PubMed] [Google Scholar]
- 8.Bradshaw AD. J Cell Commun Signal. 2009;3:239–246. doi: 10.1007/s12079-009-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Z, Kyriakides TR, Bornstein P. Mol Biol Cell. 2000;11:3353–3364. doi: 10.1091/mbc.11.10.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau LF. J Cell Commun Signal. 2012;6:121–123. doi: 10.1007/s12079-012-0169-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Du XY. J Cell Biochem. 2007;100:1337–1345. doi: 10.1002/jcb.21194. [DOI] [PubMed] [Google Scholar]
- 12.Wolf N, Yang W, Dunk CE, Gashaw I, Lye SJ, Ring T, Schmidt M, Winterhager E, Gellhaus A. Endocrinology. 2010;151:2835–2845. doi: 10.1210/en.2009-1195. [DOI] [PubMed] [Google Scholar]
- 13.Yan L, Chaqour B. J Cell Commun Signal. 2013;7:253–263. doi: 10.1007/s12079-013-0206-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel J. Curr Opin Cell Biol. 1991;3:779–785. doi: 10.1016/0955-0674(91)90050-9. [DOI] [PubMed] [Google Scholar]
- 15.Holbourn KP, Perbal B, Ravi AK. J Cell Commun Signal. 2009;3:25–41. doi: 10.1007/s12079-009-0048-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brigstock DR. J Endocrinol. 2003;178:169–175. doi: 10.1677/joe.0.1780169. [DOI] [PubMed] [Google Scholar]
- 17.Chowdhury I, Chaqour B. Eur J Biochem. 2004;271:4436–4450. doi: 10.1111/j.1432-1033.2004.04382.x. [DOI] [PubMed] [Google Scholar]
- 18.Hanna M, Liu H, Amir J, Sun Y, Morris SW, Siddiqui MA, Lau LF, Chaqour B. J Biol Chem. 2009;284:23125–23136. doi: 10.1074/jbc.M109.019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pobbati AV, Hong W. Cancer Biol Ther. 2013;14:390–398. doi: 10.4161/cbt.23788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hasan A, Pokeza N, Shaw L, Lee HS, Lazzaro D, Chintala H, Rosenbaum D, Grant MB, Chaqour B. J Biol Chem. 2011;286:9542–9554. doi: 10.1074/jbc.M110.198689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y, Kim HP, Ifedigbo E, Lau LF, Choi AM. Am J Respir Cell Mol Biol. 2005;33:297–302. doi: 10.1165/rcmb.2005-0144OC. [DOI] [PubMed] [Google Scholar]
- 22.Lee HY, Chung JW, Youn SW, Kim JY, Park KW, Koo BK, Oh BH, Park YB, Chaqour B, Walsh K, Kim HS. Circ Res. 2007;100:372–380. doi: 10.1161/01.RES.0000257945.97958.77. [DOI] [PubMed] [Google Scholar]
- 23.Huang BL, Dornbach LM, Lyons KM. J Cell Commun Signal. 2007;1:17–32. doi: 10.1007/s12079-007-0003-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins DF, Biju MP, Akai Y, Wutz A, Johnson RS, Haase VH. Am J Physiol Renal Physiol. 2004;287:F1223–F1232. doi: 10.1152/ajprenal.00245.2004. [DOI] [PubMed] [Google Scholar]
- 25.Kondo S, Kubota S, Mukudai Y, Moritani N, Nishida T, Matsushita H, Matsumoto S, Sugahara T, Takigawa M. Oncogene. 2006;25:1099–1110. doi: 10.1038/sj.onc.1209129. [DOI] [PubMed] [Google Scholar]
- 26.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, Furth EE, Lee WM, Enders GH, Mendell JT, Thomas-Tikhonenko A. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kireeva ML, Latinkic BV, Kolesnikova TV, Chen CC, Yang GP, Abler AS, Lau LF. Exp Cell Res. 1997;233:63–77. doi: 10.1006/excr.1997.3548. [DOI] [PubMed] [Google Scholar]
- 28.Chen CC, Lau LF. Int J Biochem Cell Biol. 2009;41:771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leask A, Parapuram SK, Shi-Wen X, Abraham DJ. J Cell Commun Signal. 2009;3:89–94. doi: 10.1007/s12079-009-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou D, Herrick DJ, Rosenbloom J, Chaqour B. J Appl Physiol. 2005;98:2344–2354. doi: 10.1152/japplphysiol.01093.2004. [DOI] [PubMed] [Google Scholar]
- 31.Mosher DF, Adams JC. Matrix Biol. 2012;31:155–161. doi: 10.1016/j.matbio.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Hanahan D. Science. 1997;277:48–50. doi: 10.1126/science.277.5322.48. [DOI] [PubMed] [Google Scholar]
- 33.Ribatti D, Nico B, Crivellato E, Artico M. Anat Rec B New Anat. 2006;289:3–8. doi: 10.1002/ar.b.20087. [DOI] [PubMed] [Google Scholar]
- 34.Risau W, Flamme I. Annu Rev Cell Dev Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- 35.Davis GE, Senger DR. Circ Res. 2005;97:1093–1107. doi: 10.1161/01.RES.0000191547.64391.e3. [DOI] [PubMed] [Google Scholar]
- 36.Wacker A, Gerhardt H. Curr Opin Cell Biol. 2011;23:676–685. doi: 10.1016/j.ceb.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 37.Asahara T. Ernst Schering Res Found Workshop. 2005;54:111–129. doi: 10.1007/3-540-37644-5_8. [DOI] [PubMed] [Google Scholar]
- 38.Auerbach R, Huang H, Lu L. Stem Cells. 1996;14:269–280. doi: 10.1002/stem.140269. [DOI] [PubMed] [Google Scholar]
- 39.Zambidis ET, Oberlin E, Tavian M, Peault B. Trends Cardiovasc Med. 2006;16:95–101. doi: 10.1016/j.tcm.2006.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Downs KM, Gifford S, Blahnik M, Gardner RL. Development. 1998;125:4507–4520. doi: 10.1242/dev.125.22.4507. [DOI] [PubMed] [Google Scholar]
- 41.Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, Breitman ML, Schuh AC. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 42.Sato Y. Dev Growth Differ. 2013;55:113–129. doi: 10.1111/dgd.12010. [DOI] [PubMed] [Google Scholar]
- 43.Hogan KA, Ambler CA, Chapman DL, Bautch VL. Development. 2004;131:1503–1513. doi: 10.1242/dev.01039. [DOI] [PubMed] [Google Scholar]
- 44.Bautch VL. Cold Spring Harb Perspect Med. 2012;2:a006452. doi: 10.1101/cshperspect.a006452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chappell JC, Bautch VL. Curr Top Dev Biol. 2010;90:43–72. doi: 10.1016/S0070-2153(10)90002-1. [DOI] [PubMed] [Google Scholar]
- 46.Arora R, Papaioannou VE. Blood. 2012;120:2562–2572. doi: 10.1182/blood-2012-03-390070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coultas L, Chawengsaksophak K, Rossant J. Nature. 2005;438:937–945. doi: 10.1038/nature04479. [DOI] [PubMed] [Google Scholar]
- 48.Hogan KA, Bautch VL. Curr Top Dev Biol. 2004;62:55–85. doi: 10.1016/S0070-2153(04)62003-5. [DOI] [PubMed] [Google Scholar]
- 49.Adair TH, Gay WJ, Montani JP. Am J Physiol. 1990;259:R393–R404. doi: 10.1152/ajpregu.1990.259.3.R393. [DOI] [PubMed] [Google Scholar]
- 50.Hudlicka O, Brown MD. J Vasc Res. 1996;33:266–287. doi: 10.1159/000159155. [DOI] [PubMed] [Google Scholar]
- 51.Battegay EJ. J Mol Med (Berl) 1995;73:333–346. doi: 10.1007/BF00192885. [DOI] [PubMed] [Google Scholar]
- 52.Dobrescu G. Rev Med Chir Soc Med Nat Iasi. 1997;101:31–39. [PubMed] [Google Scholar]
- 53.Smith SK. Hum Reprod Update. 1998;4:509–519. doi: 10.1093/humupd/4.5.509. [DOI] [PubMed] [Google Scholar]
- 54.Poole TJ, Finkelstein EB, Cox CM. Dev Dyn. 2001;220:1–17. doi: 10.1002/1097-0177(2000)9999:9999<::AID-DVDY1087>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 55.Tamura K, Amano T, Satoh T, Saito D, Yonei-Tamura S, Yajima H. Mech Dev. 2003;120:199–209. doi: 10.1016/s0925-4773(02)00411-2. [DOI] [PubMed] [Google Scholar]
- 56.Howe GA, Addison CL. Vasc Cell. 2012;4:1. doi: 10.1186/2045-824X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olsson AK, Dimberg A, Kreuger J, Claesson-Welsh L. Nat Rev Mol Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 58.Phng LK, Potente M, Leslie JD, Babbage J, Nyqvist D, Lobov I, Ondr JK, Rao S, Lang RA, Thurston G, Gerhardt H. Dev Cell. 2009;16:70–82. doi: 10.1016/j.devcel.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Roca C, Adams RH. Genes Dev. 2007;21:2511–2524. doi: 10.1101/gad.1589207. [DOI] [PubMed] [Google Scholar]
- 60.Benedito R, Roca C, Sorensen I, Adams S, Gossler A, Fruttiger M, Adams RH. Cell. 2009;137:1124–1135. doi: 10.1016/j.cell.2009.03.025. [DOI] [PubMed] [Google Scholar]
- 61.Hellstrom M, Phng LK, Hofmann JJ, Wallgard E, Coultas L, Lindblom P, Alva J, Nilsson AK, Karlsson L, Gaiano N, Yoon K, Rossant J, Iruela-Arispe ML, Kalen M, Gerhardt H, Betsholtz C. Nature. 2007;445:776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 62.Stratman AN, Schwindt AE, Malotte KM, Davis GE. Blood. 2010;116:4720–4730. doi: 10.1182/blood-2010-05-286872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Winkler EA, Bell RD, Zlokovic BV. Mol Neurodegener. 2010;5:32. doi: 10.1186/1750-1326-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Armulik A, Abramsson A, Betsholtz C. Circ Res. 2005;97:512–523. doi: 10.1161/01.RES.0000182903.16652.d7. [DOI] [PubMed] [Google Scholar]
- 65.Distler JH, Hirth A, Kurowska-Stolarska M, Gay RE, Gay S, Distler O. Q J Nucl Med. 2003;47:149–161. [PubMed] [Google Scholar]
- 66.Simonavicius N, Ashenden M, van WA, Lax S, Huso DL, Buckley CD, Huijbers IJ, Yarwood H, Isacke CM. Blood. 2012;120:1516–1527. doi: 10.1182/blood-2011-01-332338. [DOI] [PubMed] [Google Scholar]
- 67.Gellhaus A, Schmidt M, Dunk C, Lye SJ, Kimmig R, Winterhager E. Mol Hum Reprod. 2006;12:389–399. doi: 10.1093/molehr/gal044. [DOI] [PubMed] [Google Scholar]
- 68.O’Brien TP, Lau LF. Cell Growth Differ. 1992;3:645–654. [PubMed] [Google Scholar]
- 69.Schwenke M, Knofler M, Velicky P, Weimar CH, Kruse M, Samalecos A, Wolf A, Macklon NS, Bamberger AM, Gellersen B. PLoS One. 2013;8:e54336. doi: 10.1371/journal.pone.0054336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kireeva ML, Latinkic BV, Kolesnikova TV, Chen CC, Yang GP, Abler AS, Lau LF. Exp Cell Res. 1997;233:63–77. doi: 10.1006/excr.1997.3548. [DOI] [PubMed] [Google Scholar]
- 71.Mo FE, Lau LF. Circ Res. 2006;99:961–969. doi: 10.1161/01.RES.0000248426.35019.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wong M, Kireeva ML, Kolesnikova TV, Lau LF. Dev Biol. 1997;192:492–508. doi: 10.1006/dbio.1997.8766. [DOI] [PubMed] [Google Scholar]
- 73.Schutze N, Schenk R, Fiedler J, Mattes T, Jakob F, Brenner RE. BMC Cell Biol. 2007;8:45. doi: 10.1186/1471-2121-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dorrell MI, Aguilar E, Friedlander M. Invest Ophthalmol Vis Sci. 2002;43:3500–3510. [PubMed] [Google Scholar]
- 75.Fruttiger M. Angiogenesis. 2007;10:77–88. doi: 10.1007/s10456-007-9065-1. [DOI] [PubMed] [Google Scholar]
- 76.Chaqour B. J Ophthalmic Vis Res. 2013;8:77–82. [PMC free article] [PubMed] [Google Scholar]
- 77.Mo FE, Muntean AG, Chen CC, Stolz DB, Watkins SC, Lau LF. Mol Cell Biol. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu H, Yang R, Tinner B, Choudhry A, Schutze N, Chaqour B. Endocrinology. 2008;149:1666–1677. doi: 10.1210/en.2007-1415. [DOI] [PubMed] [Google Scholar]
- 79.Kolesnikova TV, Lau LF. Oncogene. 1998;16:747–754. doi: 10.1038/sj.onc.1201572. [DOI] [PubMed] [Google Scholar]
- 80.Chen N, Leu SJ, Todorovic V, Lam SC, Lau LF. J Biol Chem. 2004;279:44166–44176. doi: 10.1074/jbc.M406813200. [DOI] [PubMed] [Google Scholar]
- 81.Grzeszkiewicz TM, Lindner V, Chen N, Lam SC, Lau LF. Endocrinology. 2002;143:1441–1450. doi: 10.1210/endo.143.4.8731. [DOI] [PubMed] [Google Scholar]
- 82.Leu SJ, Chen N, Chen CC, Todorovic V, Bai T, Juric V, Liu Y, Yan G, Lam SC, Lau LF. J Biol Chem. 2004;279:44177–44187. doi: 10.1074/jbc.M407850200. [DOI] [PubMed] [Google Scholar]
- 83.Leu SJ, Lam SC, Lau LF. J Biol Chem. 2002;277:46248–46255. doi: 10.1074/jbc.M209288200. [DOI] [PubMed] [Google Scholar]
- 84.Leu SJ, Liu Y, Chen N, Chen CC, Lam SC, Lau LF. J Biol Chem. 2003;278:33801–33808. doi: 10.1074/jbc.M305862200. [DOI] [PubMed] [Google Scholar]
- 85.Bader BL, Rayburn H, Crowley D, Hynes RO. Cell. 1998;95:507–519. doi: 10.1016/s0092-8674(00)81618-9. [DOI] [PubMed] [Google Scholar]
- 86.Kim KH, Chen CC, Monzon RI, Lau LF. Mol Cell Biol. 2013;33:2078–2090. doi: 10.1128/MCB.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lobel M, Bauer S, Meisel C, Eisenreich A, Kudernatsch R, Tank J, Rauch U, Kuhl U, Schultheiss HP, Volk HD, Poller W, Scheibenbogen C. Cell Mol Life Sci. 2012;69:3101–3113. doi: 10.1007/s00018-012-0981-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen CC, Chen N, Lau LF. J Biol Chem. 2001;276:10443–10452. doi: 10.1074/jbc.M008087200. [DOI] [PubMed] [Google Scholar]
- 89.Murphy LO, MacKeigan JP, Blenis J. Mol Cell Biol. 2004;24:144–153. doi: 10.1128/MCB.24.1.144-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen CC, Mo FE, Lau LF. J Biol Chem. 2001;276:47329–47337. doi: 10.1074/jbc.M107666200. [DOI] [PubMed] [Google Scholar]
- 91.Qin Z, Fisher GJ, Quan T. J Biol Chem. 2013;288:12386–12394. doi: 10.1074/jbc.M112.424358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yang R, Amir J, Liu H, Chaqour B. Physiol Genomics. 2008;36:1–14. doi: 10.1152/physiolgenomics.90291.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Latinkic BV, Mercurio S, Bennett B, Hirst EM, Xu Q, Lau LF, Mohun TJ, Smith JC. Development. 2003;130:2429–2441. doi: 10.1242/dev.00449. [DOI] [PubMed] [Google Scholar]
- 94.Mercurio S, Latinkic B, Itasaki N, Krumlauf R, Smith JC. Development. 2004;131:2137–2147. doi: 10.1242/dev.01045. [DOI] [PubMed] [Google Scholar]
- 95.Francischetti IM, Kotsyfakis M, Andersen JF, Lukszo J. PLoS One. 2010;5:e9356. doi: 10.1371/journal.pone.0009356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jim Leu SJ, Sung JS, Huang ML, Chen MY, Tsai TW. Biochem Biophys Res Commun. 2013;434:885–891. doi: 10.1016/j.bbrc.2013.04.045. [DOI] [PubMed] [Google Scholar]
- 97.Wijelath ES, Rahman S, Namekata M, Murray J, Nishimura T, Mostafavi-Pour Z, Patel Y, Suda Y, Humphries MJ, Sobel M. Circ Res. 2006;99:853–860. doi: 10.1161/01.RES.0000246849.17887.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Choi J, Lin A, Shrier E, Lau LF, Grant MB, Chaqour B. J Biol Chem. 2013;288:23075–23089. doi: 10.1074/jbc.M113.475418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Blom IE, Goldschmeding R, Leask A. Matrix Biol. 2002;21:473–482. doi: 10.1016/s0945-053x(02)00055-0. [DOI] [PubMed] [Google Scholar]
- 100.Grotendorst GR, Duncan MR. FASEB J. 2005;19:729–738. doi: 10.1096/fj.04-3217com. [DOI] [PubMed] [Google Scholar]
- 101.Panek AN, Posch MG, Alenina N, Ghadge SK, Erdmann B, Popova E, Perrot A, Geier C, Dietz R, Morano I, Bader M, Ozcelik C. PLoS One. 2009;4:e6743. doi: 10.1371/journal.pone.0006743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tong ZY, Brigstock DR. J Endocrinol. 2006;188:R1–R8. doi: 10.1677/joe.1.06719. [DOI] [PubMed] [Google Scholar]
- 103.Yokoi H, Mukoyama M, Mori K, Kasahara M, Suganami T, Sawai K, Yoshioka T, Saito Y, Ogawa Y, Kuwabara T, Sugawara A, Nakao K. Kidney Int. 2008;73:446–455. doi: 10.1038/sj.ki.5002722. [DOI] [PubMed] [Google Scholar]
- 104.Wu S, Platteau A, Chen S, McNamara G, Whitsett J, Bancalari E. Am J Respir Cell Mol Biol. 2010;42:552–563. doi: 10.1165/rcmb.2009-0068OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chuva de Sousa Lopes SM, Feijen A, Korving J, Korchynskyi O, Larsson J, Karlsson S, ten DP, Lyons KM, Goldschmeding R, Doevendans P, Mummery CL. Dev Dyn. 2004;231:542–550. doi: 10.1002/dvdy.20162. [DOI] [PubMed] [Google Scholar]
- 106.Hall-Glenn F, Lyons KM. Cell Mol Life Sci. 2011;68:3209–3217. doi: 10.1007/s00018-011-0782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Baguma-Nibasheka M, Kablar B. Dev Dyn. 2008;237:485–493. doi: 10.1002/dvdy.21433. [DOI] [PubMed] [Google Scholar]
- 108.Ivkovic S, Yoon BS, Popoff SN, Safadi FF, Libuda DE, Stephenson RC, Daluiski A, Lyons KM. Development. 2003;130:2779–2791. doi: 10.1242/dev.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hall-Glenn F, De Young RA, Huang BL, van HB, Hofmann JJ, Chen TT, Choi A, Ong JR, Benya PD, Mikkola H, Iruela-Arispe ML, Lyons KM. PLoS One. 2012;7:e30562. doi: 10.1371/journal.pone.0030562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Babic AM, Chen CC, Lau LF. Mol Cell Biol. 1999;19:2958–2966. doi: 10.1128/mcb.19.4.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shimo T, Nakanishi T, Nishida T, Asano M, Kanyama M, Kuboki T, Tamatani T, Tezuka K, Takemura M, Matsumura T, Takigawa M. J Biochem. 1999;126:137–145. doi: 10.1093/oxfordjournals.jbchem.a022414. [DOI] [PubMed] [Google Scholar]
- 112.Kubota S, Kawaki H, Kondo S, Yosimichi G, Minato M, Nishida T, Hanagata H, Miyauchi A, Takigawa M. Biochimie. 2006;88:1973–1981. doi: 10.1016/j.biochi.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 113.Dean RA, Butler GS, Hamma-Kourbali Y, Delbe J, Brigstock DR, Courty J, Overall CM. Mol Cell Biol. 2007;27:8454–8465. doi: 10.1128/MCB.00821-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Schober JM, Chen N, Grzeszkiewicz TM, Jovanovic I, Emeson EE, Ugarova TP, Ye RD, Lau LF, Lam SC. Blood. 2002;99:4457–4465. doi: 10.1182/blood.v99.12.4457. [DOI] [PubMed] [Google Scholar]
- 115.Nagai N, Klimava A, Lee WH, Izumi-Nagai K, Handa JT. Invest Ophthalmol Vis Sci. 2009;50:1903–1910. doi: 10.1167/iovs.08-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen Y, Abraham DJ, Shi-Wen X, Pearson JD, Black CM, Lyons KM, Leask A. Mol Biol Cell. 2004;15:5635–5646. doi: 10.1091/mbc.E04-06-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kiwanuka E, Andersson L, Caterson EJ, Junker JP, Gerdin B, Eriksson E. Exp Cell Res. 2013;319:2938–2946. doi: 10.1016/j.yexcr.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 118.Jedsadayanmata A, Chen CC, Kireeva ML, Lau LF, Lam SC. J Biol Chem. 1999;274:24321–24327. doi: 10.1074/jbc.274.34.24321. [DOI] [PubMed] [Google Scholar]
- 119.Gao R, Brigstock DR. Hepatol Res. 2003;27:214–220. doi: 10.1016/s1386-6346(03)00241-9. [DOI] [PubMed] [Google Scholar]
- 120.James LR, Le C, Doherty H, Kim HS, Maeda N. PLoS One. 2013;8:e70441. doi: 10.1371/journal.pone.0070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsoutsman T, Wang X, Garchow K, Riser B, Twigg S, Semsarian C. J Mol Cell Cardiol. 2013;62:164–178. doi: 10.1016/j.yjmcc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 122.Wang JC, Sonnylal S, Arnett FC, De CB, Zhou X. Int J Immunopathol Pharmacol. 2011;24:595–601. doi: 10.1177/039463201102400305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 124.Praidou A, Androudi S, Brazitikos P, Karakiulakis G, Papakonstantinou E, Dimitrakos S. Curr Diabetes Rev. 2010;6:304–312. doi: 10.2174/157339910793360815. [DOI] [PubMed] [Google Scholar]
- 125.Hellstrom M, Gerhardt H, Kalen M, Li X, Eriksson U, Wolburg H, Betsholtz C. J Cell Biol. 2001;153:543–553. doi: 10.1083/jcb.153.3.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hoffmann J, Feng Y, Vom HF, Hillenbrand A, Lin J, Erber R, Vajkoczy P, Gourzoulidou E, Waldmann H, Giannis A, Wolburg H, Shani M, Jaeger V, Weich HA, Preissner KT, Hoffmann S, Deutsch U, Hammes HP. FASEB J. 2005;19:2035–2036. doi: 10.1096/fj.04-2109fje. [DOI] [PubMed] [Google Scholar]
- 127.Kantarci A, Nseir Z, Kim YS, Sume SS, Trackman PC. J Dent Res. 2011;90:887–893. doi: 10.1177/0022034511404703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuiper EJ, van ZR, Roestenberg P, Lyons KM, Goldschmeding R, Klaassen I, Van Noorden CJ, Schlingemann RO. J Histochem Cytochem. 2008;56:785–792. doi: 10.1369/jhc.2008.950980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Inoki I, Shiomi T, Hashimoto G, Enomoto H, Nakamura H, Makino K, Ikeda E, Takata S, Kobayashi K, Okada Y. FASEB J. 2002;16:219–221. doi: 10.1096/fj.01-0332fje. [DOI] [PubMed] [Google Scholar]
- 130.Hashimoto G, Inoki I, Fujii Y, Aoki T, Ikeda E, Okada Y. J Biol Chem. 2002;277:36288–36295. doi: 10.1074/jbc.M201674200. [DOI] [PubMed] [Google Scholar]
- 131.Rayego-Mateos S, Rodrigues-Diez R, Morgado-Pascual JL, Rodrigues Diez RR, Mas S, Lavoz C, Alique M, Pato J, Keri G, Ortiz A, Egido J, Ruiz-Ortega M. J Mol Cell Biol. 2013;5:323–335. doi: 10.1093/jmcb/mjt030. [DOI] [PubMed] [Google Scholar]
- 132.Aoyama E, Kubota S, Takigawa M. FEBS Lett. 2012;586:4270–4275. doi: 10.1016/j.febslet.2012.10.038. [DOI] [PubMed] [Google Scholar]
- 133.Pi L, Shenoy AK, Liu J, Kim S, Nelson N, Xia H, Hauswirth WW, Petersen BE, Schultz GS, Scott EW. FASEB J. 2012;26:3365–3379. doi: 10.1096/fj.11-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gariano RF. Prog Retin Eye Res. 2003;22:295–306. doi: 10.1016/s1350-9462(02)00062-9. [DOI] [PubMed] [Google Scholar]
- 135.Gerhardt H. Organogenesis. 2008;4:241–246. doi: 10.4161/org.4.4.7414. [DOI] [PMC free article] [PubMed] [Google Scholar]