Abstract
Oncogenic kinase activity and the resulting aberrant growth and survival signaling is a common driving force of cancer. Accordingly, many successful molecularly targeted anti-cancer therapeutics are directed at inhibiting kinase activity. To assess kinase activity in minute patient samples, we have developed an immunocapture-based in vitro kinase assay on an integrated polydimethylsiloxane (PDMS) microfluidics platform that can reproducibly measure kinase activity from as few as 3,000 cells. For this platform, we adopted the standard radiometric 32P-ATP labeled phosphate transfer assay. Implementation on a microfluidic device required us to develop methods for repeated trapping and mixing of solid-phase affinity micro beads. We also developed a solid state beta-particle camera imbedded directly below the microfluidic device for real-time quantitative detection of the signal from this and other microfluidic radio bioassays. We demonstrate that the resulting integrated device can measure ABL kinase activity from BCR-ABL positive leukemia patient samples. The low sample input requirement of the device creates new potential for direct kinase activity experimentation and diagnostics on patient blood, bone marrow, and needle biopsy samples.
Keywords: Cell Signaling, Protein serine-threonine kinases, Protein tyrosine kinases, Molecular diagnosis and prognosis, Methodology for proteomics, Novel assay technology
INTRODUCTION
As principal signal transduction components, kinases regulate as many as 50% of intracellular proteins. Aberrant kinase function is involved in the etiology of many diseases and most forms of cancer (1-3). The 518 known human kinases represent one of the largest classes of drug targets pursued by pharmaceutical companies (1, 3), spurred in part by successful targeted inhibition of kinases by antibodies (4) and small molecule kinase inhibitors (5). Kinase-related signaling measurements directly from patient samples often involve detecting the consequences of activity, for example the resulting downstream phosphorylation patterns (6-8). Direct measurement of kinase activity from patient samples can greatly complement these phosphoprofiling techniques (9). There are currently numerous kinase assay technologies with various requirements and ranges of application (10), including a fluorescence-based microfluidic format for screening of kinase inhibition using purified kinase (11). Radiometric kinase assays represent the earliest technology and are generally considered a high standard for determining basic enzyme properties, in part because radiolabeling of reaction substrates does not alter their intrinsic biochemical and physical properties (10).
Microfluidics offer a prime operation platform for implementation of chemical reactions (12-15) and biological assays (16-18) in a miniaturized fashion due to their inherent advantages of sample and reagent economy, operation fidelity, high throughput, automated operation and precise control over the microenvironment. The advent of poly(dimethylsiloxane) (PDMS)-based microfluidic devices has enabled the systems-level assembly of individual microfluidic functional modules. These integrated devices provide digitally controlled operation of complicated chemical (19, 20) and biological processes (21, 22) in stand-alone platforms. PDMS devices are fabricated by soft-lithography-based technology (23) in which two layers of fluid and control channels (24) can be designed to create integrated valves and other functional components (15, 19, 21, 22).
Readout directly from microfluidic devices has been accomplished using microelectronic devices coupled to chemical and physical changes in the fluidic space (25), using an integrated nuclear magnetic resonance (NMR) sensor (26), and via detection of fluorescent light using avalanche photodiode (APD) detectors (27, 28) or CCD optical camera imaging coupled to microscopy (29) or a scanner (30). However, a wide range of traditional biological and enzymatic assays take advantage of the high detection sensitivity of radioisotopes and use radioactivity as a means to label without altering the chemical properties of the substrates and enzymes. To readout radioactivity distributions directly from a microfluidic chip we developed a solid-state beta particle camera using a position sensitive avalanche photodiode (PSAPD) charged-particle detector. The beta camera allows real-time quantitative monitoring of the radio assay performance with high sensitivity and low background.
To aid the study of diseases driven by aberrant signaling and the development of kinase inhibition therapies, our goal was to create a miniaturized kinase assay that allows for activity measurements from small patient samples. We therefore developed a microfluidic assay platform that is directly coupled to a beta camera for sensitive and quantitative readout of kinase activity via measurement of 32P incorporation. We developed our microfluidic in vitro kinase radio assay (μ-ivkra) using BCR-ABL oncogenic kinase-positive leukemia samples, since this molecularly defined disease has offered many insights into kinase-driven tumorigenesis (31), is one of the first successful molecular targets of small molecule kinase inhibitors, yet still remains a clinical challenge for a subset of cases (32). Our stand-alone benchtop device can measure kinase activity from as few as 3,000 cells and opens new possibilities for experimentation directly on minute patient samples from blood draws, bone marrow aspirates and needle biopsies.
MATERIALS AND METHODS
Microfluidic platform fabrication and control
The integrated microfluidic chip was fabricated using an established two-layer soft lithography process and was controlled using pneumatic manifolds digitally instructed through a computer interface (33). Technical details are provided in the Supplementary Data.
Beta camera
The ‘beta camera’ quantitative radioactivity imaging sensor is comprised of a position sensitive avalanche photodiode (PSAPD) silicon semiconductor device with an active area of 14 × 14 mm2 (Model P1305-P; RMD Radiation Monitoring Devices, Watertown, MA), custom electronic readout circuitry, and a computer based data acquisition card driven by LabVIEW image acquisition software (National Instruments). Charged particles that interact within the depletion region of the PSAPD convert a portion of their kinetic energy into electron-hole pairs. The PSAPD is operated at high voltage reverse bias (+1750 V) which accelerates the electron-hole pairs and amplifies the signal by 1,000 fold through an avalanche effect. The PSAPD readout uses a five channel output with four position channels and one sum channel. The relative amplitudes of the four position channels are used to determine the location of signal along 2 dimensions. The imaging system also includes a CCD optical camera and reference points that allow for spatial co-registration of the beta camera radioactivity images with a photographic image of the microfluidic chip. Further details of the beta camera are in the Supplemental Data and will be published elsewhere (N.T.V., A.F.C.).
Cell culture
Pro-B, lymphoid, Ba/F3 cells transformed with BCR-ABL (p210 isoform) have been described previously and were provided by Charles Sawyers (UCLA) (34). These cells display similar levels of BCR-ABL expression and signaling as patient leukemia primary samples that are positive for the Philadelphia chromosome (Ph+), the chromosomal translocation that results in expression of the BCR-ABL fusion protein. The K562 (Ph+) and U937 (Ph−) human leukemic cell lines were provided by John Colicelli (UCLA). BCR-ABL expression (or lack of expression) was verified by the presence (or absence) of a 210 kDa anti-c-ABL reactive protein (p210 BCR-ABL isoform; antibody clones K-12, Santa Cruz Biotechnology and OP20, EMD Chemicals, Gibbstown, NJ) and elevated (or baseline) pan-specific anti-phosphotyrosine levels (clone 4G10, Millipore, Temecula, CA). Cells were maintained in RPMI 1640 (Cellgro, Mediatech Inc, Manassas, VA) with 10% fetal bovine serum (Omega Scientific, Tarzana, CA). Cells were lysed in mRIPA buffer (10 mM Beta-glycerophosphate, 50 mM Tris pH 7.4, 1% NP-40, 0.25% Na deoxycolate, 1 mM EDTA, 150 mM NaCl, 1 mM Vanadate, with freshly added 1 mM PMSF, 20 μg/ml leupeptin, 20 μg/ml aprotinin).
Human leukemia patient samples and mouse xenograft system
Primary cells from the peripheral blood or bone marrow of pre-B-cell acute lymphoblastic leukemia (pre-B-ALL) patients were injected into sublethally irradiated (250 cGy) immune-deficient NOD/SCID mice and serially passaged no more than 3 times (35, 36). The human leukemia cells create a leukemia-like disease burden in the mice and become the dominant subpopulation in the bone marrow, peripheral blood, and spleen. Disease burden was monitored by measuring the percent of human leukemic cells in the peripheral blood or spleen using hCD45 flow cytometry. Spleen samples were collected by immediate lysis of scalpel-dissected spleen cells in mRIPA buffer using the frosted ends of glass slides to promote cell dissociation, on ice. The human cells in the Ph+ samples were uniformly Ph+ based on cytogenetics. The Ph-sample was a pre-B-ALL with normal karyotype.
Off-chip and on-chip in vitro kinase radio assays
In vitro kinase assays were performed using established protocols for bead-based immunoaffinity capture of a specific kinase followed by kinase reactions in the presence of a kinase-specific peptide substrate and 32P labeled ATP (10, 34). The microfluidic on-chip assay was developed using reagents that were compatible and efficient in the context of microfluidic channels and valves. For the microfluidic assay, 0.1% n-dodecyl-beta-D-maltoside (DDM) was added to all buffers to prevent bead clumping (indicated by /DDM in the buffer name). 0.1% DDM had no detectable effect on BCR-ABL kinase activity (data not shown). For BCR-ABL kinase immunocapture, the off-chip assay used agarose protein A/G beads (40-160 μm; Pierce, Rockford, IL) and the on-chip assay used smaller and more sturdy Protein G polystyrene beads (6.7 μm; Spherotech Inc., Lake Forest, CA). The large average size and fragility of the agarose beads precluded their use on-chip, and the small size of the polystyrene beads made them impractical for the pipeting-based off-chip assay. Both bead types were coated with anti-c-ABL antibody (OP20, EMD Chemicals). Off-chip assays were performed using 400 μl (20 μg/μl, or 4×107 cell equivalents) of cell lysate, and on-chip assays used the amounts indicated in the figure legends. Abltide-biotin conjugate peptide was used as substrate (Millipore), and following the reaction the radio-labeled and unlabeled peptide was captured in off-chip assays using SAM2 Biotin Capture Membrane squares (Promega, Madison, WI) or in the on-chip case using streptavidin-coated polystyrene beads (6.7 μm; Spherotech Inc.). Further details for the off-chip and on-chip assays are in the Supplemental Data and Supplementary Figure 1.
RESULTS
Overall configuration of the microfluidic kinase assay device
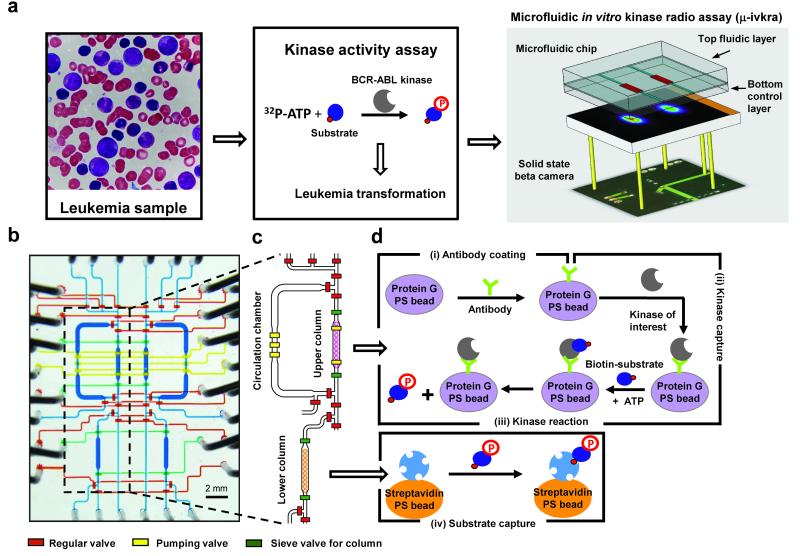
To develop a microfluidic in vitro kinase radio assay (μ-ivkra) to directly measure kinase activity in minute patient samples, we designed and fabricated a polydimethylsiloxane (PDMS)-based integrated microfluidic chip (24) that performs an immunocapture-based kinase assay (Fig. 1). To enable radioactivity-based readout of the assay, we integrated a position sensitive avalanche photodiode detector (PSAPD) (37) designed to function as a camera for imaging charged beta particles. The beta camera is imbedded directly below the microfluidic chip, allows real-time monitoring of the radioactivity distribution during the assay, and quantifies the final amount of radioactivity incorporated into the substrate (Fig. 1a). Control and readout of the microfluidic device and beta camera are performed via custom electronics and a personal computer. The resulting device performs an automated multi-step kinase reaction assay with coupled readout in a single unit, complete from sample loading to final quantitative data.
Figure 1. Schematic of the microfluidic immunocapture-based in vitro kinase radio assay (μ-ivkra).
(a) Leukemia smear from a BCR-ABL-driven chronic myelogenous leukemia (CML) patient (left). In the kinase activity assay the radio-labeled phosphate from 32P-ATP is transferred to a peptide substrate and the kinase activity is quantified based on isotope incorporation (center). Configuration of the μ-ivkra device (right). A microfluidics platform is directly coupled to a highly sensitive charged particle camera (beta camera, PSAPD), enabling on-chip measurements of radioisotope-based kinase assays. (b) Micrograph of a μ-ivkra chip loaded with colored dyes. The microfluidic chip includes two symmetric, isolated reaction units under synchronous digital control. The two layer microfluidic chip has a top fluidic and a bottom control layer. The liquid coloring indicates the delegated functional responsibilities of various components: blue for fluidic channels; and in the bottom control layer, red for regular isolation valves, green for sieve valves to trap beads, and yellow for peristaltic pump valves driving fluidic circulation. The scale bar represents 2 mm. Additional photographs of the chip platform are in the supplement (Supplementary Fig. 2). (c) Drawing of the core part of a single unit. (d) Schematic of the core steps in the microfluidic in vitro kinase radio assay: (step i) antibody capture using protein G-conjugated polystyrene (PS) beads (6.7 μm diameter), (step ii) kinase capture using antibody-coated beads, (step iii) kinase reaction, and (step iv) biotin-labeled peptide substrate capture using 6.7 μm diameter streptavidin-coated polystyrene beads. Steps i-iii are performed in the upper column and circulation chamber where beads can be trapped in the column, released into the circulation chamber (370 nL volume each), mixed to homogeneity, and re-trapped multiple times. The sieve value-mediated bead trapping approach facilitates solution exchange and bead washing (Supplementary Video 1). Capture of substrate (step iv) and washing of unincorporated radioisotope (32P-ATP) occurs in the lower column prior to quantitative radiometric detection imaging using the beta camera.
Microfluidic device design and operation
The standard in vitro kinase radio assay involves immunocapture of the kinase of interest from cell lysate to a solid-phase support, typically using antibody-coated beads. After washing away other kinases and proteins, 32P-ATP and a peptide substrate with specificity to the kinase of interest are incubated with the captured kinase to allow kinase-catalyzed transfer of radio-labeled phosphate from the ATP to the substrate. The peptide substrate (both 32P-labeled and unlabeled) is captured using an affinity resin (e.g. biotin-conjugated peptides captured with a streptavidin solid support membrane) and unincorporated radioisotope (as 32P-ATP) is washed away. To assess kinase activity, the amount of radio-labeled phosphate incorporated into the substrate is quantitatively measured, typically using liquid scintillation.
Our chip design contains two isolated and symmetric fluidic modules for individual kinase reactions run in parallel. The fluidic layer channels of both modules are simultaneously controlled by the pressure actuations of their shared bottom control layer – specifically, each bottom layer control channel regulates a valve in one fluidic module, crosses the midline of the device, and also regulates the corresponding valve in the second module (Fig. 1b, Supplementary Fig. 2). Each module has both i) an upper circulation chamber and bead-trapping column for the immunocapture and kinase reaction steps and ii) a lower bead-trapping column for the final substrate capture step (Fig. 1c-d). The upper circulation chamber is designed for manipulating the immunocapture beads through multiple trap and release steps (Fig. 2a, steps i-iii).
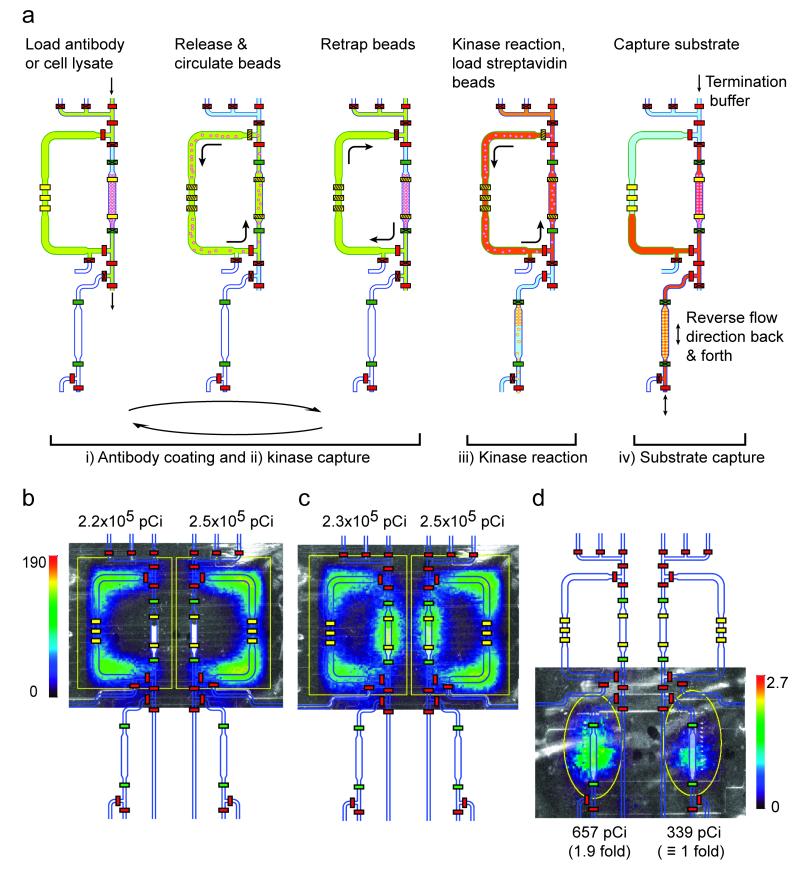
Figure 2. Schematic diagrams of key steps of the operation of the on-chip kinase radio assay.
(a) Roman numerals correspond to the steps in Figure 1d. (steps i-ii) Loading anti-kinase antibody solution or cell lysate into the circulation chamber (370 nL volume). Releasing protein-G beads (6.7 μm diameter, polystyrene) into the circulation chamber for antibody coating (30 min at room temperature (RT)) (step i) or kinase capture (30 min, RT) (step ii); the solution and beads are circulated around the upper chamber driven by two serial sets of peristaltic pump valves. The hatch lines on the yellow pump valves indicate an active pumping mechanism. The pumping direction is reversed every 80 sec to prevent bead clumping. Re-trapping beads in the capture column of the upper circulation chamber between steps (5 min) to allow for bead washing and solution exchange in the circulation chamber. (step iii) Kinase reaction with solution and bead circulation in the upper chamber (15 min, RT). The operational steps here are the same as in steps i-ii, except that while waiting for the kinase reaction to finish, the streptavidin polystyrene (PS) beads are independently loaded into the lower column (80 nL volume) from below. (step iv) Capturing and washing the labeled (and unlabeled) peptide substrate in the lower streptavidin bead substrate-capture column using a flow reversal protocol, prior to washing away any unincorporated radioisotope (32P-ATP) and using the beta camera (PSAPD) for imaging and quantitation. Different channel colors represent different loaded solutions. (b-d) Mid-experiment monitoring of device and assay performance and final signal quantitation using the beta camera. Overlaid images of the CCD camera optical image, the false color beta camera radioactivity image, and the chip AutoCAD drawing during various assay steps: (b-c) loaded radioactive (32P-ATP) kinase reaction buffer prior to (b) and after (c) circulation demonstrating equal loading, and (d) the final captured radiolabeled peptide substrate in the lower column showing the anticipated 2-fold difference in kinase activity between 6,000 and 3,000 Ba/F3 + BCR-ABL cells. The color bars represent the beta camera image scales in counts per second per mm2. Total detected activity (pCi) for each ROI (region of interest, yellow outline) is indicated. See Supplemental Methods for how the ROI was chosen.
To miniaturize the kinase radio assay to a microfluidic format, we first developed methods for the reproducible manipulation of solid-support micro beads. In our assay, protein G-conjugated beads are loaded into the chip, coated with anti-ABL antibody, used to capture BCR-ABL kinase from cellular lysates, and then the captured kinase is incubated with 32P-ATP and peptide substrate for the kinase reaction. These steps require three sieve valve-mediated cycles of bead trapping, release and resuspension for the required solution exchanges and a final re-trapping of the immunocapture beads (Supplementary Video 1). In the two-layer integrated PDMS microfluidic platform, we found that polystyrene beads were rigid enough for use in conjunction with the microfluidic valves, and that 6.7 μm diameter beads could be readily trapped behind sieve-style valves (19) and subsequently released and resuspended homogenously. When closed, the sieve values block most of the channel cross-section and thus block bead passage, yet liquid can flow through the remaining opening (further described in Supplementary Methods). For the bead trapping and re-mixing steps, we optimized our methods to prevent bead clumping and subsequent clogging of the channels. To avert clumping our final device uses two sets of imbedded peristaltic pump valves per chamber to drive mixing, a pumping protocol with a 2.5 Hz pumping frequency, reversal of the pumping direction every 80 sec (Supplementary Video 1), and a mild surfactant (0.1% n-dodecyl-beta-D-maltoside (DDM)) in the solutions.
At the end of the assay, the kinase reaction mixture is passed through a lower substrate-capture column. This column is pre-loaded with 6.7 μm streptavidin-coated polystyrene beads to capture the 32P-labeled (and unlabeled) biotin-conjugated peptide substrate while washing away unincorporated radioisotope (32P-ATP) (Fig. 2a, step iv). This single use column is kept homogeneous using a flow reversal protocol that promotes efficient capture of the peptide substrate. The flow protocol involves passing the peptide substrate-containing liquid through the bead column in both directions alternatively three times, followed by washing. For this step the pumping is driven by the non-column peristaltic pump in the upper circulation chamber using a pumping frequency of 5 Hz, and the adjoining chip channels are used as reservoirs during the flow reversals. Upon peptide substrate capture in the lower column, the microfluidic steps of the assay are complete and the 32P incorporated into the peptides is quantitatively measured using the beta camera.
Assay readout via the beta camera
The beta camera serves as the radioactivity readout device for the microfluidic kinase assay. The detector in the beta camera is a 14 × 14 mm2 position sensitive avalanche photodiode (PSAPD) with a very low inherent background count rate (1.5 counts/hour/mm2). Benefits of the PSAPD detector include its monolithic and thus rugged design, and position decoding via a five channel analog readout. The PSAPD was originally designed for detection of scintillation light produced in nuclear imaging applications (37). We modified the PSAPD with a Mylar passivation layer to block visible light. When the PSAPD is placed in close contact with a radioactive source, it has a high avalanche-mediated sensitivity for detecting emitted charged particles. Using known levels of 32P, we established a calibration curve and determined the absolute sensitivity of the integrated microfluidic beta camera to be 29% (see Supplementary Fig. 3). The beta camera itself has high intrinsic sensitivity to 32P particles that traverse through the PSAPD detection region. The geometric configuration of the integrated microfluidic beta camera device reduces the overall sensitivity due to i) half of the beta particles being emitted away from the planar detector and ii) attenuation of beta particles by the control layer of PDMS material (100 μm) and the glass slide (150 μm) between the microfluidic channel and the beta camera.
The beta camera is position sensitive and thus a single detector is used for multiple simultaneous readouts from adjacent columns (Fig. 2b-d). In addition to the final readout, the beta camera can be used for qualitative and quantitative operational checkpoints. For example, during the assay we monitor in real time the radioactivity distribution and intensity to test for uniform chip operation during fluid and bead manipulation. These checkpoints verify equal loading, uniform mixing, proper valve operation and chip channel integrity (Fig. 2b-c and Supplementary Table 1). These checkpoints were useful during the development phase of the microfluidic device and protocol (see the kinase autophosphorylation example below).
On-chip in vitro BCR-ABL kinase radio assay
Using the μ-ivkra integrated microfluidic and beta camera platform described above and in Figures 1-2, we have successfully developed an on-chip in vitro BCR-ABL kinase radio assay. The assay adheres to all of the steps of traditional immunocapture-based in vitro kinase radio assays (Fig. 1d). The most notable differences are in terms of scale, including reduced lysate and reagent input, smaller solid-phase bead components, and shorter incubation times (Fig. 2a).
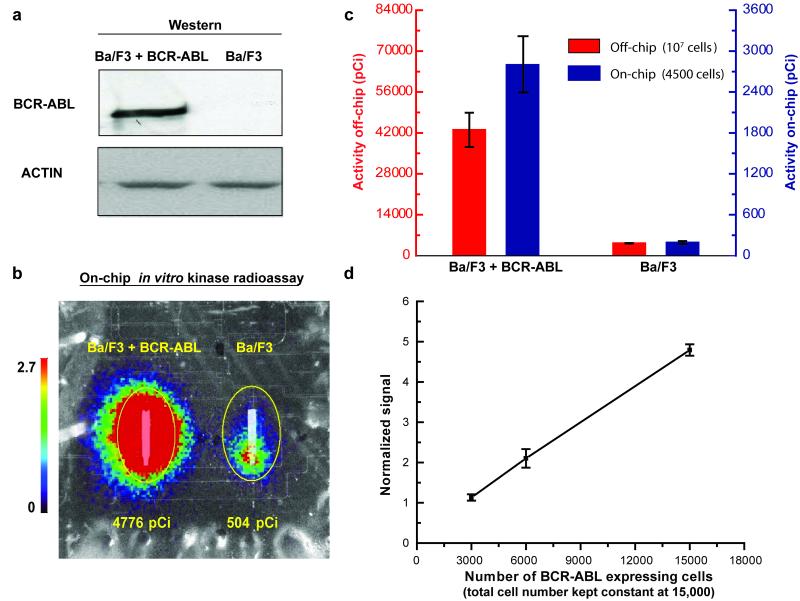
For validation of the microfluidic assay, we performed both a traditional off-chip BCR-ABL kinase radio assay and our on-chip kinase assay using the same cell lysates (Fig. 3). These assays were done using Ba/F3 cells, a pro-B murine cell line, transfected with either a BCR-ABL expression plasmid or an empty vector control (Fig. 3a). The off-chip kinase assay was performed using millions of cells while the on-chip assay required only 4,500 cells. We observed similar order of magnitude ‘BCR-ABL versus control’ fold-change values between the off-chip (10.2-fold, ±1.2 standard deviation, n=3) and on-chip (14.6±0.7-fold, n=3) assays (Fig. 3b-c), confirming that the on-chip assay was performing correctly. These absolute quantitation assays were performed using the same lysate split into aliquots, and the low standard deviation in fold change and raw radioactivity signal (Fig. 3c) reflects the reproducibility of the assay performed on different days using different chips. The difference in exact fold change between on- and off-chip results is likely primarily due to differences in the unsubtracted background signal between the two assays and its effect on the accurate measurement of the low ABL activity in the BCR-ABL-free control cells, see further discussion below related to the linearity range of the platform.
Figure 3. Development of an on-chip in vitro BCR-ABL kinase radio assay.
(a) Anti-ABL immunoblot of the Ba/F3 cell line with and without expression of constitutively active BCR-ABL kinase (loading control, anti-Actin immunoblotting). (b) The on-chip BCR-ABL kinase reaction was performed with 12,000 Ba/F3 + BCR-ABL cells (left column) and parental Ba/F3 cells (right column). Final results are represented by an overlay of optical and beta camera false-color images (20 min acquisition) of the lower substrate capture columns. The color bar scale (counts per second per mm2) and total detected activity (pCi) for each ROI (region of interest, yellow circle) are indicated. (c) Comparison between traditional test tube-based (off-chip) and on-chip in vitro BCR-ABL kinase radio assays. Off-chip assays were performed with 107 cells and on-chip assays with 4,500 cells. Results reflect absolute quantitation, and the error bars represent the standard deviation for three assays performed on different days using aliquots of the same lysate with different chips. (d) Demonstration of linearity using BCR-ABL expressing Ba/F3 cells. In each experiment, samples were diluted with parental non-BCR-ABL-expressing cells as necessary to keep the total cell number constant at 15,000. The graph incorporates data from a total of 16 sample aliquots (8 pair-wise chip experiments performed on different chips using mixed aliquots of the same lysates). The results from each experiment were normalized such that the mean detection signal was set equal to the mean number of cells in the experiment divided by 3000. Non-specific background signal was not subtracted. Thus, the results represent relative quantitation designed to identify the linear region of the platform. The R-squared value from a linear regression fit is 0.997. Error bars represent the standard deviation.
We next tested the linearity of the on-chip in vitro BCR-ABL kinase radio assay. In these experiments we loaded different amounts of lysate from BCR-ABL expressing Ba/F3 cells into each of the microfluidic chip reaction units (with parental Ba/F3 cell lysate used to dilute the amount of BCR-ABL kinase while keeping the total cell number constant). These relative quantitation experiments demonstrated linearity over a 5-fold range of input (Fig. 3d, Supplementary Fig. 4). The lower limit was due mainly to non-specific background signal from radioisotope bound to the streptavidin beads (the beta camera itself has very low inherent background). Future device designs will use smaller volumes of capture reagents to reduce the background, and will incorporate additional reaction units to allow direct determination and subtraction of background to extend this linear range.
We are also able to detect autophosphorylation of BCR-ABL bound to the antibody-coated beads in the upper kinase reaction module at the end of the kinase reaction (Supplementary Fig. 5). During the development of the assay this readout was used to verify that the reaction had proceeded as anticipated with stronger signal coming from the sample with higher BCR-ABL levels.
The two 30 min binding steps of our on-chip assay are shorter than those used in conventional immunocapture-based kinase radio assays, which are often reported as from 1-4 hours to overnight (10, 38). In total the on-chip assay can be completed in 4 hours. Only 1 hour elapses from lysate addition to the end of the kinase reaction, thus reducing the potential for decay of kinase activity. This and other advantages of the microfluidic platform, such as efficient, immediate and consistent mixing, improve the net on-chip kinase activity efficiency by approximately an order of magnitude compared to off-chip assays performed with the same lysates (Supplementary Table 2).
Application of the microfluidic kinase assay on different leukemic systems
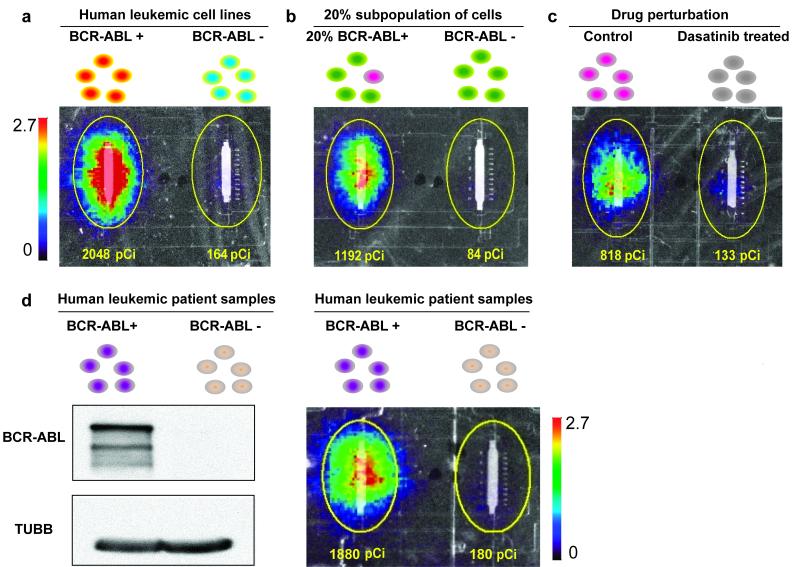
We next tested our microfluidic assay on additional BCR-ABL-expressing leukemia systems. We were able to detect substantial BCR-ABL kinase activity differences between patient-derived leukemic cell lines with endogenous BCR-ABL expression (K562) compared to leukemic cell lines without BCR-ABL expression (U937) (Fig. 4a).
Figure 4. On-chip in vitro BCR-ABL kinase radio assays in leukemic systems.
(a) Detection of BCR-ABL activity from BCR-ABL expressing human cell lines. The on-chip in vitro BCR-ABL kinase radio assay was performed on lysates from the human leukemic cell line K562 (with leukemia patient derived BCR-ABL expression) and U937 (with no BCR-ABL expression). (b) Qualitative detection of BCR-ABL activity from a subpopulation of BCR-ABL expressing cells as a model for heterogeneous patient samples. BCR-ABL activity was detected from a 1 in 5 subpopulation of BCR-ABL expressing cells (4,500 Ba/F3 + BCR-ABL : 18,000 Ba/F3 cells versus 22,500 Ba/F3 cells). (c) Detection of sensitivity to BCR-ABL-targeted drug inhibition. Ba/F3 + BCR-ABL (p210) cells were treated with 125nM dasatinib or DMSO solvent alone (control) for 2 h and then BCR-ABL kinase activity was measured in the microfluidic platform (4,500 cell input). (d) Detection of BCR-ABL activity from Ph+ leukemic patient samples in a mouse xenograft system. Mouse spleens with more than 95% hCD45 Ph+ cells or more than 90% hCD45 Ph− cells were harvested, lysed and analyzed by anti-c-ABL immunoprecipitation and immunoblotting and anti-TUBB immunoblotting (left) and by the on-chip in vitro BCR-ABL kinase radio assay (4,500 cell input) (right). All results are represented by overlays of optical images of the lower substrate capture columns and beta camera false color images (20 min acquisition). The color bar scale (counts per second per mm2) and total detected activity (pCi) for each ROI (region of interest, yellow circle) are indicated. The size of the ROI is uniform across all experiments.
Since leukemic patient samples are often ‘contaminated’ with normal cells that do not have elevated kinase activity, we tested whether we can measure BCR-ABL kinase activity from a subpopulation of BCR-ABL-expressing cells. We made a 1:4 mixture of BCR-ABL- to non-BCR-ABL-expressing cells using the Ba/F3 system. The signal from this 20% BCR-ABL-expressing subpopulation was clearly detectable and distinct compared to the background signal from a control non-BCR-ABL-expressing population (Fig. 4b).
One potential application of the on-chip radioactive in vitro kinase assay is to measure the inhibition of kinases by molecularly targeted therapeutics directly on patient and animal model samples. To demonstrate this use, we treated BCR-ABL-expressing Ba/F3 cells with 125 nM dasatinib, a well-known BCR-ABL inhibitor (39), and then measured BCR-ABL kinase activity from 4,500 cells. Upon treatment, the BCR-ABL activity was inhibited substantially, with the detected assay signal reduced to background levels (Fig. 4c).
Application of the microfluidic kinase assay on patient samples
Finally, we tested our platform for detecting BCR-ABL kinase activity from clinical samples. We used pre-B-cell acute lymphoblastic leukemia (pre-B-ALL) primary patient cells that were both positive and negative for the Philadelphia chromosome (Ph) and thus also for BCR-ABL protein. We injected these patient cells into NOD/SCID mice in which they proliferate to create a leukemia-like disease burden. Ultimately the human leukemic cells make up the vast majority of the peripheral blood, bone marrow and spleen cells (35, 36). Fresh spleens from these leukemia-burdened xenograft mice were processed and loaded onto the microfluidic platform to measure BCR-ABL kinase activity. Signal comparable to our cell line results was obtained from the Ph+ patient samples, while the negative control Ph− samples had only background levels of signal (Fig. 4d).
DISCUSSION
By coupling a microfluidic platform to a solid-state beta camera, we have developed an integrated platform for immunocapture-based in vitro kinase radio assays on minute cancer samples. The device executes an automated, multi-step solid-phase binding and enzymatic reaction and provides imaging-based final quantitative readout of the assay. In the realm of microfluidics, we have developed new techniques for efficient handling of solid-phase micro-beads that allow for the multiple bind, wash, and solvent exchange cycles required by many affinity-based bioassays. In particular we found that bead trapping, release, and homogenization using sieve valves and peristaltic pumps is an efficient and reproducible approach. Nonetheless, the full development of a clinical device may require further engineering of bead manipulation techniques, or the introduction of redundancy measures, to ensure the robust performance required in a clinical setting.
The integration of the solid-state beta camera directly underneath the microfluidic platform allows imaging the radioisotope distribution both as a final readout and for real-time monitoring of the operational steps of the assay (Fig. 2b-d, Supplementary Table 1). This ability to troubleshoot in real time proved useful in our assay-development and protocol testing phase (Supplementary Fig. 6). The PSAPD-based imaging camera has high sensitivity and very low inherent background (1.5 counts/hour/mm2), making feasible the acquisition of very weak signals over time periods longer than the 20 minutes used here. Many bioassays are radioisotope-based and therefore this sensitive imaging device facilitates the miniaturization of these assays to a microfluidic platform.
There are significant benefits from reducing the radiometric kinase assay from the macro-scale to the micro-scale. First, we reduce the amount of cell input 2-3 orders of magnitude compared to conventional and 96-well format assays (38, 40). Similar reduction in the amount of other reagents and radioactivity are another benefit, especially in the context of radiation safety. By porting the assay onto a microfluidic platform, we improved reproducibility of the assay by introducing digital operation and a well-controlled microenvironment. The microfluidic environment also generally increases net binding and reaction efficiencies (19, 41, 42), as is the case for our assay (Supplementary Table 2). The overall improvements in efficiency and sensitivity expand possibilities for kinase activity measurements in experimentation on and diagnostics of minute patient samples. Future chip designs will increase the number of reaction units per chip to increase the number of samples or kinases that can be assayed simultaneously, and will couple the device to upstream microfluidic modules for efficient sample delivery or automated cell treatment and lysis (43-45), towards full experimental lab-on-a-chip aspirations (46, 47).
In addition to the assay reported here, there are two other recent advances in high sensitivity kinase assays that complement one another. One approach involves a phosphorylation-sensitive fluorescent chemosensor readout coupled to an electro concentration microfluidic device (17). The second approach uses quantitative mass spectrometry as a readout for phosphorylation, allowing for several tens of kinases to be monitored simultaneously using distinct and kinase-specific peptide substrates (48, 49) As currently developed, both of these assays rely on the peptide sequence for determining kinase specificity and do not benefit from the additional specificity afforded by kinase immunocapture. Both of the microfluidic format assays have the potential to be developed into inexpensive benchtop stand alone units – for the microfluidic radio assay this is in part made possible by the compact beta camera device. Disadvantages of the different assays include the need to develop functional chemosensors specific to each kinase for the fluorescent assay, the need of a sophisticated equipment and associated data analysis for the mass spectrometry assay, and the use of radioactivity (although substantially minimized by the microfluidic scale) for our assay. Radio-, fluorescent chemosensor-, and mass spectrometry-based kinase assays will likely continue to be used towards specific applications.
Taken together, the reduced sample input, decreased assay time, and digitally controlled reproducibility of our microfluidic kinase radio assay facilitates direct experimentation on clinical samples that are either precious or perishable. Here we demonstrate this potential using a patient sample and mouse xenograft system. Future experiments will develop reproducible sample collection and measurement conditions for primary patient samples. Other applications include profiling of patient and animal model samples for their kinase inhibitor drug sensitivity, or measurement of kinase activity from stem cells, cancer stem cells, rare immune cells and other subpopulations, for example following flow cytometry- or microfluidic-based sorting.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Jonathan Said (UCLA) for the leukemia smear micrograph and Brian Skaggs for guidance on the BCR-ABL kinase assay. This work was supported by a Jonsson Cancer Center Foundation (JCCF)/UCLA Transdisciplinary Team Grant to T.G.G, H.-R.T. and A.F.C. N.V.T. is a postdoctoral fellow supported through the National Institute of Biomedical Imaging and Bioengineering (5T32EB002101). Y.- M.K. is supported by the Jean Perkins Foundation and Stop Cancer. T.G.G. is an Alfred P. Sloan Research Fellow.
REFERENCES
- 1.Cohen P. The development and therapeutic potential of protein kinase inhibitors. Curr Opin Chem Biol. 1999;3:459–65. doi: 10.1016/S1367-5931(99)80067-2. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. Signaling--2000 and beyond. Cell. 2000;100:113–27. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 3.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 4.Pegram MD, Konecny G, Slamon DJ. The molecular and cellular biology of HER2/neu gene amplification/overexpression and the clinical development of herceptin (trastuzumab) therapy for breast cancer. Cancer Treat Res. 2000;103:57–75. doi: 10.1007/978-1-4757-3147-7_4. [DOI] [PubMed] [Google Scholar]
- 5.Druker BJ, Sawyers CL, Kantarjian H, et al. Activity of a Specific Inhibitor of the BCR-ABL Tyrosine Kinase in the Blast Crisis of Chronic Myeloid Leukemia and Acute Lymphoblastic Leukemia with the Philadelphia Chromosome. N Engl J Med. 2001;344:1038–42. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 6.Irish JM, Kotecha N, Nolan GP. Mapping normal and cancer cell signalling networks: towards single-cell proteomics. Nat Rev Cancer. 2006;6:146–55. doi: 10.1038/nrc1804. [DOI] [PubMed] [Google Scholar]
- 7.Gorre ME, Mohammed M, Ellwood K, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293:876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- 8.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 9.Janes KA, Albeck JG, Gaudet S, Sorger PK, Lauffenburger DA, Yaffe MB. A systems model of signaling identifies a molecular basis set for cytokine-induced apoptosis. Science. 2005;310:1646–53. doi: 10.1126/science.1116598. [DOI] [PubMed] [Google Scholar]
- 10.Jia Y, Quinn CM, Kwak S, Talanian RV. Current in vitro kinase assay technologies: the quest for a universal format. Curr Drug Discov Technol. 2008;5:59–69. doi: 10.2174/157016308783769414. [DOI] [PubMed] [Google Scholar]
- 11.Dunne J, Reardon H, Trinh V, Li E, Farinas J. Comparison of On-Chip and Off-Chip Microfluidic Kinase Assay Formats. ASSAY and Drug Development Technologies. 2004;2:121–9. doi: 10.1089/154065804323056468. [DOI] [PubMed] [Google Scholar]
- 12.Watts P, Haswell SJ. The application of micro reactors for organic synthesis. Chemical Society Reviews. 2005;34:235–46. doi: 10.1039/b313866f. [DOI] [PubMed] [Google Scholar]
- 13.deMello AJ. Control and detection of chemical reactions in microfluidic systems. Nature. 2006;442:394–402. doi: 10.1038/nature05062. [DOI] [PubMed] [Google Scholar]
- 14.Lin W-Y, Wang Y, Wang S, Tseng H-R. Integrated microfluidic reactors. Nano Today. 2009;4:470–81. doi: 10.1016/j.nantod.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Sui G, Mocharla VP, et al. Integrated microfluidics for parallel screening of an in situ click chemistry library. Angew Chem Int Ed Engl. 2006;45:5276–81. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]
- 16.Fan AC, Deb-Basu D, Orban MW, et al. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15:566–71. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JH, Cosgrove BD, Lauffenburger DA, Han J. Microfluidic Concentration-Enhanced Cellular Kinase Activity Assay. Journal of the American Chemical Society. 2009;131:10340–1. doi: 10.1021/ja902594f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun J, Masterman-Smith M, Graham N, et al. A microfluidic platform for systems pathology: multiparameter single-cell signaling measurements of clinical brain tumor specimens. Cancer Research. 2010 doi: 10.1158/0008-5472.CAN-10-0076. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee CC, Sui G, Elizarov A, et al. Multistep synthesis of a radiolabeled imaging probe using integrated microfluidics. Science. 2005;310:1793–6. doi: 10.1126/science.1118919. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Lin WY, Liu K, et al. An integrated microfluidic device for large-scale in situ click chemistry screening. Lab Chip. 2009;9:2281–5. doi: 10.1039/b907430a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen CL, Skordalakes E, Berger JM, Quake SR. A robust and scalable microfluidic metering method that allows protein crystal growth by free interface diffusion. Proc Natl Acad Sci U S A. 2002;99:16531–6. doi: 10.1073/pnas.262485199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ottesen EA, Hong JW, Quake SR, Leadbetter JR. Microfluidic digital PCR enables multigene analysis of individual environmental bacteria. Science. 2006;314:1464–7. doi: 10.1126/science.1131370. [DOI] [PubMed] [Google Scholar]
- 23.Xia YN, Whitesides GM. Soft lithography. Annual Review of Materials Science. 1998;28:153–84. [Google Scholar]
- 24.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Monolithic microfabricated valves and pumps by multilayer soft lithography. Science. 2000;288:113–6. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 25.Vykoukal DM, Stone GP, Gascoyne PR, Alt EU, Vykoukal J. Quantitative detection of bioassays with a low-cost image-sensor array for integrated microsystems. Angew Chem Int Ed Engl. 2009;48:7649–54. doi: 10.1002/anie.200901814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee H, Sun E, Ham D, Weissleder R. Chip-NMR biosensor for detection and molecular analysis of cells. Nat Med. 2008;14:869–74. doi: 10.1038/nm.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruger J, Singh K, O’Neill A, Jackson C, Morrison A, O’Brien P. Development of a microfluidic device for fluorescence activated cell sorting. Journal of Micromechanics and Microengineering. 2002;12:486–94. [Google Scholar]
- 28.Chabinyc ML, Chiu DT, McDonald JC, et al. An Integrated Fluorescence Detection System in Poly(dimethylsiloxane) for Microfluidic Applications. Analytical Chemistry. 2001;73:4491–8. doi: 10.1021/ac010423z. [DOI] [PubMed] [Google Scholar]
- 29.Chung K, Crane MM, Lu H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat Methods. 2008;5:637–43. doi: 10.1038/nmeth.1227. [DOI] [PubMed] [Google Scholar]
- 30.Ozawa T, Kinoshita K, Kadowaki S, et al. MAC-CCD system: a novel lymphocyte microwell-array chip system equipped with CCD scanner to generate human monoclonal antibodies against influenza virus. Lab Chip. 2009;9:158–63. doi: 10.1039/b810438g. [DOI] [PubMed] [Google Scholar]
- 31.Wong S, Witte ON. The BCR-ABL Story: Bench to Bedside and Back. Annual Review of Immunology. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. [DOI] [PubMed] [Google Scholar]
- 32.Ramirez P, DiPersio JF. Therapy options in imatinib failures. Oncologist. 2008;13:424–34. doi: 10.1634/theoncologist.2007-0170. [DOI] [PubMed] [Google Scholar]
- 33.Duffy DC, McDonald JC, Schueller OJA, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane) Analytical Chemistry. 1998;70:4974–84. doi: 10.1021/ac980656z. [DOI] [PubMed] [Google Scholar]
- 34.Skaggs BJ, Gorre ME, Ryvkin A, et al. Phosphorylation of the ATP-binding loop directs oncogenicity of drug-resistant BCR-ABL mutants. Proc Natl Acad Sci U S A. 2006;103:19466–71. doi: 10.1073/pnas.0609239103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trageser D, Iacobucci I, Nahar R, et al. Pre-B cell receptor-mediated cell cycle arrest in Philadelphia chromosome-positive acute lymphoblastic leukemia requires IKAROS function. J Exp Med. 2009;206:1739–53. doi: 10.1084/jem.20090004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lock RB, Liem N, Farnsworth ML, et al. The nonobese diabetic/severe combined immunodeficient (NOD/SCID) mouse model of childhood acute lymphoblastic leukemia reveals intrinsic differences in biologic characteristics at diagnosis and relapse. Blood. 2002;99:4100–8. doi: 10.1182/blood.v99.11.4100. [DOI] [PubMed] [Google Scholar]
- 37.Shah KS, Farrell R, Grazioso R, Harmon ES, Karplus E. Position-sensitive avalanche photodiodes for gamma-ray imaging. Ieee Transactions on Nuclear Science. 2002;49:1687–92. [Google Scholar]
- 38.Janes KA, Albeck JG, Peng LX, Sorger PK, Lauffenburger DA, Yaffe MB. A High-throughput Quantitative Multiplex Kinase Assay for Monitoring Information Flow in Signaling Networks: Application to Sepsis-Apoptosis. Mol Cell Proteomics. 2003;2:463–73. doi: 10.1074/mcp.M300045-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 40.Wu D, Mand MR, Veach DR, Parker LL, Clarkson B, Kron SJ. A solid-phase Bcr-Abl kinase assay in 96-well hydrogel plates. Anal Biochem. 2008;375:18–26. doi: 10.1016/j.ab.2007.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coti KK, Wang Y, Lin WY, et al. A dynamic micromixer for arbitrary control of disguised chemical selectivity. Chem Commun (Camb) 2008:3426–8. doi: 10.1039/b804530e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zimmermann M, Delamarche E, Wolf M, Hunziker P. Modeling and optimization of high-sensitivity, low-volume microfluidic-based surface immunoassays. Biomed Microdevices. 2005;7:99–110. doi: 10.1007/s10544-005-1587-y. [DOI] [PubMed] [Google Scholar]
- 43.Huang S-B, Lee G-B. Pneumatically driven micro-dispenser for sub-micro-liter pipetting. Journal of Micromechanics and Microengineering. 2009;19:035027. [Google Scholar]
- 44.Du WB, Fang Q, He QH, Fang ZL. High-throughput nanoliter sample introduction microfluidic chip-based flow injection analysis system with gravity-driven flows. Anal Chem. 2005;77:1330–7. doi: 10.1021/ac048675y. [DOI] [PubMed] [Google Scholar]
- 45.Liu K, Xia C, Shen CK-F, van Dam RM. Automated delivery of small fluid volumes through tubing to microfluidic chips. Proceedings of MicroTAS. 2008:62–4. [Google Scholar]
- 46.Hong JW, Quake SR. Integrated nanoliter systems. Nat Biotechnol. 2003;21:1179–83. doi: 10.1038/nbt871. [DOI] [PubMed] [Google Scholar]
- 47.El-Ali J, Sorger PK, Jensen KF. Cells on chips. Nature. 2006;442:403–11. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 48.Kubota K, Anjum R, Yu Y, et al. Sensitive multiplexed analysis of kinase activities and activity-based kinase identification. Nat Biotechnol. 2009;27:933–40. doi: 10.1038/nbt.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu Y, Anjum R, Kubota K, Rush J, Villen J, Gygi SP. A site-specific, multiplexed kinase activity assay using stable-isotope dilution and high-resolution mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:11606–11. doi: 10.1073/pnas.0905165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.